Abstract
In patients with celiac disease, inflammatory T cell responses to HLA-DQ2-bound gluten peptides are thought to cause disease. Two types of HLA-DQ2 molecules exist, termed HLA-DQ2.5 and HLA-DQ2.2. Whereas HLA-DQ2.5 predisposes to celiac disease, HLA-DQ2.2 does not. We now provide evidence that the disease-associated HLA-DQ2.5 molecule presents a large repertoire of gluten peptides, whereas the non-disease-associated HLA-DQ2.2 molecule can present only a subset of these. Moreover, gluten presentation by HLA-DQ2 homozygous antigen-presenting cells was superior to presentation by HLA-DQ2/non-DQ2 heterozygous antigen-presenting cells in terms of T cell proliferation and cytokine secretion. Gluten presentation by HLA-DQ2.5/2.2 heterozygous antigen-presenting cells induced intermediate T cell stimulation. These results correlated with peptide binding to the antigen-presenting cells. Finally, we demonstrate that HLA-DQ trans dimers formed in HLA-DQ2.5/2.2 heterozygous individuals have properties identical with HLA-DQ2.5 dimers. Our findings explain the strongly increased risk of disease development for HLA-DQ2.5 homozygous and HLA-DQ2.2/2.5 heterozygous individuals, and they are indicative of a quantitative model for disease development, where HLA-DQ expression and the available number of T cell-stimulatory gluten peptides are critical limiting factors. This model may have important implications for disease prevention.
Celiac disease (CD) is the most common food-induced enteropathy in humans. Patients display a permanent intolerance toward the gluten proteins in wheat. About 95% of CD patients are HLA-DQ2+ (DQA1*0501, DQB1*0201, termed HLA-DQ2.5 hereafter). HLA-DQ molecules bind and present peptides to antigen-specific T cells. It is now commonly accepted that HLA-DQ2.5 can bind and present gluten peptides and that these HLA-DQ-peptide complexes induce inflammatory T cell responses, causing disease. The HLA-DQ2.5 molecule preferentially binds peptides with negatively charged amino acids at anchor positions (1, 2). Whereas gluten peptides contain few negative charges, these charges can be introduced by the enzyme tissue transglutaminase (tTG) that selectively deamidates glutamine residues in gluten peptides (3-5).
A second HLA-DQ2 molecule exists (DQA1*0201, DQB1*0202, termed HLA-DQ2.2 hereafter) with peptide-binding properties that are virtually identical with the properties of HLA-DQ2.5 (6). In fact, HLA DQ2.2 has been shown to bind and present a gluten peptide to T cells (7). Yet, HLA-DQ2.2 does not predispose for CD unless it is expressed together with HLA-DQ2.5 (8). Individuals homozygous for the disease-associated HLA-DQ2.5 genotype or HLA-DQ2.2/2.5 heterozygous have the highest risk for developing CD. In contrast, HLA-DQ2.5/non-DQ2.2 heterozygous individuals have an only slightly increased risk (9-11). To determine the mechanism underlying these observations we have now compared the gluten-specific T cell response in the context of the HLA-DQ2.5 and HLA-DQ2.2 molecules.
Materials and Methods
Synthetic Peptides. Peptides were synthesized by standard fluorenylmethoxycarbonyl (Fmoc) chemistry on a SyroII peptide synthesizer (MultiSynTech, Witten, Germany). The integrity of the peptides was checked by RP-HPLC and mass spectrometry. The gluten epitopes were synthesized with glutamic acid residues at positions that are deamidated by tTG.
T Cell Proliferation Assays and Cytokine Expression. Proliferation assays were performed in triplicate in 150 μl of Iscove's modified Dulbecco's medium (GIBCO) supplemented with 10% human serum in 96-well flat-bottom plates (Falcon) by using 104 gluten-specific T cells stimulated with a panel of 105 irradiated HLA-DQ2+ allogeneic peripheral blood mononuclear cells [3,000 rads (30 Gy)] in the presence or absence of tTG-treated gluten or gluten peptides (0.2-20 μg/ml). After 48 h at 37°C, cultures were pulsed with 0.5 μCi (1 Ci = 37 GBq) of [3H]thymidine and harvested 18 h thereafter. T cell proliferation experiments with the HLA-DQ2-transduced B cells were performed with 104 gluten-specific T cells stimulated, with 5·103 B cells that were pretreated with mitomycin C (100 μg/ml, M 0503, Sigma) for 1 h, and subsequently preincubated in the presence or absence of tTG-treated gluten peptides (0.2-20 μg/ml) for 24 h. Further experimental details are as described above.
Cytokines (IFN-γ, tumor necrosis factor α, IL-10, IL-5, IL-4, and IL-2) were measured with the human Th1/Th2 Cytokine Bead Array technique (BD Biosciences) in the culture supernatants of the T cell proliferation assays 48 h after stimulation.
Peptide-Binding and Competition Assay. For the peptide-binding assay Epstein-Barr virus (EBV)-transformed B cell lines were used: AVL and Ducaf (HLA-DR3DQ2), Douglas and Brussel (HLA-DR3/7DQ2), FK (HLA-DR3/4DQ2/8), MHG (HLADR3/4DQ2/3), Pitout and EKR (HLA DR7DQ2), CJO (HLADR1DQ1). Flat-bottom plates (Nunc) were coated with the HLA-DQ-specific mAb SPV-L3 (12), 2 μg per well in 100 μl of 50 mM sodium carbonate buffer, pH 9.6, for 2 h at 37°C. Subsequently, the wells were incubated with PBS/gelatin 0.2% (wt/vol) overnight at 4°C and washed before use.
Lysates were prepared of PBS-washed cells (2·106 per ml) in 20 mM Tris·HCl (pH 7.5)/5 mM MgCl2/1% Nonidet P-40 and protease inhibitors (Complete, EDTA-free tablets, Roche Diagnostics). Cell debris was removed by centrifugation at 10,000 × g for 15 min, and 100 μl of the lysates was applied to the coated wells, followed by incubation overnight at 4°C. Next, the plates were washed, and biotin-labeled indicator peptides, MHCIα(46-63) and glia-α9(57-68), were applied in a concentration range (0-5 μM) in binding buffer [5% DMSO/0.05% Nonidet P-40/0.05% Tween 20/16.8 mM citric acid/36 mM Na2HPO4, pH 5.5, and protease inhibitors (Complete, EDTA-free tablets, Roche Molecular Biochemicals)] and incubated for 48 h at 37°C. Plates were washed and incubated with 100 μl of streptavidin-europium diluted 1,000× in assay buffer (both Wallac, Turku, Finland) for 45 min at room temperature. After washing, enhancement solution was applied (Wallac) for 15 min, and the plates were measured in a time-resolved fluorometer (1234, Wallac). Fluorescence-activated cell sorter (FACS) analysis was performed to determine the level of DQ expression on the individual B cell lines.
Competition experiments were performed in the same experimental setup by coincubation of the MHCIα(46-63) indicator peptide (0.6 μM), which binds equally to HLA-DQ2.5 and HLA-DQ2.2 (6), and a concentration range of the competitor peptide (0- to 500-fold excess). All gluten peptides were tested with glutamic acid at the positions that are deamidated by tTG. IC50 values were determined and indicate the concentration of the competitor peptide required for half-maximal inhibition of the binding of the indicator peptide.
Construction of Cell Lines Expressing HLA-DQ2 Cis and Trans Dimers. The HLA-DQ2 α- and β-chains were cloned into retroviral expression vectors as described (13, 14). The HLA-DQ2 (α1*0501, β1*0201, and β1*0202) chains were amplified from cDNA of EBV-transformed lymphoblastoid cell lines HAR and Pitout, respectively. For the HLA α0501, the primers 5′-CGGGATCCACCATGATCCTAAACAAAGCTC-3′ and 5′-TGTGAATCCCATCCTGGCTCGAGTGA-3′ were used. The HLA β0201 was amplified with the primers 5′-CCGGAATTCACCATGTCTTGGAAAAAGGCTTTG-3′ and 5′-CTCCTCGAGTCTCAGGAGTCAGTGCAGG-3′, and the HLA β0202 chain was amplified with the primers 5′-CCGGAATTCACCATGTCTTGGAA(A/G)AAG(G/T)CTTTG-3′ and 5′-CTCCTCGAGTCTCAGGAGTCAGTGCAG(G/A)AGC-3′. Subsequent cloning was performed with the BamHI and XhoI fragments of the DQ2 α-chain and the EcoRI-XhoI fragments of the β-chains. The constructs were transfected into ϕ-NX-A (15) as described (16). Subsequent retroviral transduction of the HLA-DQ2 α- and β-chains into HLA class II-negative cells (EBV-transformed lymphoblastoid cell line SJO) was performed as described (16). HLA-DQ2 expression was measured with SPV-L3 (HLA-DQ) (12) and XIII358.4 (HLA-DQ2) (17) antibodies.
Results
Peptide-Binding Characteristics of HLA-DQ2.5 Favor Gluten Presentation. The evidence is conclusive that a large number of T cell-stimulatory gluten peptides exist (5, 18-24), and, for several of these peptides, the minimal epitopes required for T cell stimulation have been determined (refs. 19 and 22; W.V. and F.K., unpublished data). Alignment of these sequences indicated that five of eight gluten epitopes contain a proline residue at relative position P3 (Table 1). Although the peptide-binding properties of HLA-DQ2.5 and HLA-DQ2.2 are almost identical, a striking difference is that a proline at P3 has an adverse effect on peptide binding to HLA-DQ2.2 but not HLA-DQ2.5 (6). We have therefore determined the binding properties of the eight gluten epitopes to both HLA-DQ2 molecules in peptide competition assays (Table 1). All gluten epitopes bound to HLA-DQ2.5 with high to intermediate affinity. In contrast, only the three gluten epitopes that lack a proline at P3 (glia-γ1, glt-17, and glia-γ30) bound to HLA-DQ2.2 with high affinity, whereas the others bound with low (glia-α2 and glia-γ2) to undetectable (glia-α9, glia-α20, and glu-5) affinity.
Table 1. Peptide-binding registers, IC50 values, and T cell recognition of gluten epitopes in the context of HLA-DQ2.5 and HLA-DQ2.2.
| Peptide-binding register, P1—P9
|
Binding/IC50, μM
|
T cell stimulation
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gluten epitope | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | DQ2.5 | DQ2.2 | DQ2.5 | DQ2.2 |
| Glia-α2 | P | Q | P | E | L | P | Y | P | Q | 15 | 130 | + | — |
| Glia-α9 | P | F | P | Q | P | E | L | P | Y | 8 | >500 | + | — |
| Glia-α20 | F | R | P | E | Q | P | Y | P | Q | 30 | >500 | + | — |
| Glu-5 | E | X | P | E | Q | P | Q | Q | F | 100 | >500 | + | — |
| Glia-γ2 | P | Y | P | E | Q | P | E | Q | P | 65 | 350 | + | — |
| Glia-γ1 | P | Q | Q | S | F | P | E | Q | E | 14 | 35 | + | + |
| Glia-γ30 | I | I | Q | P | E | Q | P | A | Q | 10 | 50 | + | + |
| Glt-17 | P | F | S | E | Q | E | Q | P | V | 25 | 12 | + | + |
| MHCIα* | I | E | Q | E | G | P | E | Y | W | 1.6 | 1.6 | ND | ND |
The peptides contain glutamic acid residues at positions that are deamidated by tTG. X = I or L. ND, not done.
The MHCIα(46—63) reference peptide
Next, T cell proliferation assays were performed with T cell clones specific for the gluten peptides presented by HLA-DQ2.5- or HLA-DQ2.2-positive antigen-presenting cells (APC). The results obtained with two of the T cell clones are shown in Fig. 1, and an overview for all T cell clones is given in Table 1. All gluten peptides stimulated the T cell clones in the presence of HLA-DQ2.5-positive APC. In contrast, only those peptides that lack proline at P3 [glia-γ1 (7), glt-17, and glia-γ30] stimulated the T cell clones in the presence of HLA-DQ2.2-positive APC (Table 1 and Fig. 1). Thus, a proline residue at position P3 in gluten peptides has a strong negative influence on binding to HLA-DQ2.2, but not to HLA-DQ2.5, which is reflected in the T cell-stimulatory properties of the peptides.
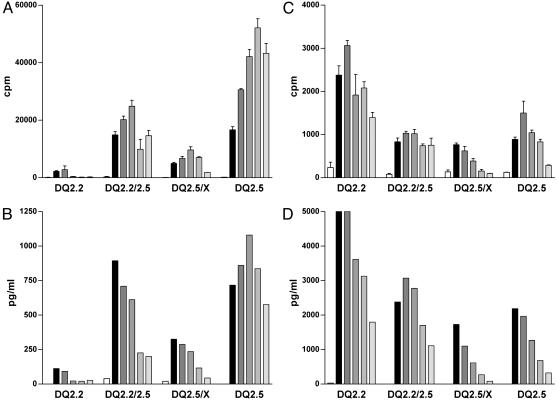
Fig. 1.
Gene dose effect in stimulation of gluten-specific T cell clones. Stimulation of two gluten-specific T cell clones with the gluten epitopes glia-α9(A and B) and glt-17 (C and D) is shown. Five concentrations of the epitopes were tested in the range from 0.2 μg/ml (light-gray bars) to 20 μg/ml (black bars). Peripheral blood mononuclear cells (PBMC) of various HLA-DQ2+ healthy individuals were used as APC (x axis). The epitopes were tested as deamidated peptides. T cell stimulation was determined by measurement of proliferation (A and C) and IFN-γ production (B and D). Production of tumor necrosis factor α, IL-10, IL-5, and IL-4 correlated with the levels of IFN-γ secreted (data not shown). The percentage of HLA class II-expressing cells in the PBMCs was determined by FACS analysis and was similar for each donor (data not shown). These results are representative of three independent experiments using PBMCs from various donors.
HLA-DQ2 Gene Dose Determines the Level of Gluten-Specific T Cell Stimulation. Epidemiological studies have shown that HLA-DQ2-homozygous individuals have the highest risk of developing CD (9-11). We have now investigated the influence of the dose and type of the HLA-DQ2 alleles expressed by APC on gluten-specific T cell responses. T cell clones specific for gluten peptides (22) were stimulated with these peptides presented by APC homozygous or heterozygous for HLA-DQ2.5, HLA-DQ2.2, and unrelated HLA-DQ alleles, and specific proliferation and cytokine secretion were measured. Representative experiments are shown for an α-gliadin and a glutenin epitope (Fig. 1). The α-gliadin epitope induced strong T cell proliferation and IFN-γ production after stimulation in the context of the homozygous HLA-DQ2.5 APC. In contrast, HLA-DQ2.5/nonDQ2.2 heterozygous APC induced weak T cell responses, whereas intermediate stimulation was achieved when HLA-DQ2.5/DQ2.2 heterozygous APC were used (Fig. 1 A and B). As expected, no T cell stimulation was observed with the HLA-DQ2.2-positive APC, because this peptide does not bind to the HLA-DQ2.2 molecule (Table 1). Similar results were obtained with the five gluten peptides that bind preferentially to HLA-DQ2.5 (Table 1 and data not shown).
The glutenin epitope (designated glt-17 in Table 1), which has a high affinity for the HLA-DQ2.2 allele (Table 1), induced maximal T cell stimulation in the context of the HLA-DQ2.2 homozygous APC. This peptide induced intermediate and minimal T cell stimulation in the context of the HLA-DQ2.5/DQ2.2 and HLA-DQ2.5/non-DQ2.2 heterozygous APC, respectively (Fig. 1 C and D). This complementary effect of the non-disease-associated HLA-DQ2.2 allele in combination with the disease-associated HLA-DQ2.5 allele on T cell stimulation was found for all epitopes listed in Table 1 (data not shown).
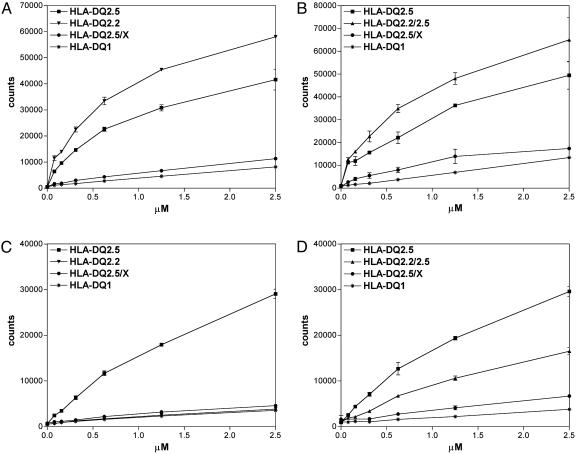
HLA-DQ2 Gene Dose Determines the Level of Gluten Peptide Binding. Subsequently, peptide-binding studies were performed to investigate whether the observed gene dose effect in the T cell proliferation assays originated from a quantitative difference in presentation of the gluten epitopes by the APC. For this HLA-DQ2 molecules isolated from either HLA-homozygous or HLA-heterozygous B cell lines were used. FACS analysis of these B cell lines confirmed similar levels of HLA-DQ expression on the cell surface (data not shown). An MHC class I indicator peptide, known to bind specifically to both HLA-DQ2.5 and HLA-DQ2.2 (6), bound strongly to HLA-DQ isolated from both HLA-DQ2-homozygous cell lines and the HLA-DQ2.5/DQ2.2-heterozygous cell lines (Fig. 2 A and B). Substantially lower binding of this indicator peptide was observed when HLA-DQ molecules from a heterozygous HLA-DQ2.5/non-DQ2.2 cell line were used (Fig. 2 A and B).
Fig. 2.
Peptide binding to a panel of HLA-DQ combinations. Shown is the binding of a concentration range of the indicator peptides MHCIα46-63 (A and B) and glia-α9(C and D) to HLA-DQ molecules isolated from HLA-homozygous and -heterozygous EBV-transformed B cell lines. The results shown are representative of at least three independent experiments.
The glia-α9 peptide bound strongly to the HLA-DQ molecules derived from an HLA-DQ2.5 homozygous cell line (Fig. 2 C and D), but binding to the HLA-DQ molecules from an HLA-DQ2.5/non-DQ2.2 cell line was hardly detectable (Fig. 2 C and D). Intermediate binding to HLA-DQ from an HLA-DQ2.5/DQ2.2-heterozygous cell line was found (Fig. 2D). As expected (Table 1), this peptide did not bind to HLA-DQ2.2 (Fig. 2C).
These results indicate highly significant differences in the number of HLA-DQ2-peptide complexes that can be formed on HLA-DQ2-homozygous vs. -heterozygous cells. This correlates with the observed differences in T cell-stimulatory capacity of HLA-DQ-homozygous and -heterozygous APC.
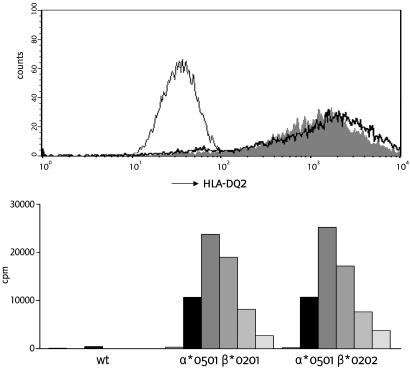
Expression and Function of Cis and Trans HLA-DQ2 Dimers. HLA-DQ α- and β-chains potentially pair in cis and in trans (25). To establish the formation of disease-relevant HLA-DQ2 trans dimers, the DQα1*0501 α-chain was stably introduced into an HLA class II-negative EBV-transformed B cell line together with either the disease-associated DQβ1*0201 β-chain or the non-disease-associated DQβ1*0202 β-chain. FACS analysis indicated equivalent expression of both DQαβ-chain combinations on the cell surface (Fig. 3 Upper). Moreover, both dimers presented the glia-α9 (Fig. 3 Lower) and the glia-γ1 (data not shown) epitopes in an identical fashion to gluten-specific T cell clones. This finding implies that HLA-DQ2.5/2.2 heterozygous individuals can express two HLA-DQ2-dimers that bind and present all characterized gluten epitopes. The other two HLADQ2 dimers that can be formed in these individuals would share identical HLA-DQ-α1 and -β1 domains and therefore display the peptide-binding properties of HLA-DQ2.2.
Fig. 3.
Functional expression of HLA-DQ2.5 in cis and in trans. (Upper) Absence of HLA-DQ2 on an HLA class II negative B cell line (thin line), and presence of HLA-DQ2 on the cell line transduced with HLA-DQα*0501 and DQβ*0201 (solid gray), and HLA-DQα*0501 and DQβ*0202 (black line). (Lower) T cell stimulation by the glia-α9 epitope in the context of the untransduced HLA class II-negative cell line (wt), and transduced cell lines with the HLA-DQ2 molecules in cis (DQα*0501 and DQβ*0201) and in trans (DQα*0501 and DQβ*0202). Five concentrations of the glia-α9 epitope were tested in the range from 0.2 μg/ml (light-gray bars) to 20 μg/ml (black bars).
Discussion
Our present study provides a functional explanation for the correlation between the HLA-DQ2 gene dose and the risk of developing CD (9-11). We demonstrate that the HLA-DQ2 gene dose has a strong quantitative effect on the magnitude of gluten-specific T cell responses (Fig. 1) and that this correlates with the level of (gluten) peptide binding to HLA-DQ2 homozygous and heterozygous APC (Fig. 2). HLA-DQ dimers can originate from αβ-chain combinations in cis and in trans (25). Hence, four αβ-chain combinations can be formed in HLA-DQ heterozygous individuals, whereas in homozygous individuals all HLA-DQ molecules are identical (Table 2). Our results demonstrate that in HLA-DQ2.2/2.5-heterozygous cells two HLA-DQ αβ-chain combinations lead to the expression of a functional HLA-DQ2.5 molecule, whereas the other two combinations would form a functional HLA-DQ2.2 molecule. In contrast, in a heterozygous HLA-DQ2.5/non-DQ2 individual, only one of the four possible combinations represents an HLADQ2.5 molecule (Table 2).
Table 2. Correlation among the HLA-DQ gene dose, the number of potentially presented gluten peptides, and the relative risk for CD.
| HLA-DQ2 genotype | α/β-Chain combinations | Functional expression | Gluten epitopes presented | Haplotype risk of CD (9—11) |
|---|---|---|---|---|
| HLA-DR3 DQ2 | α0501, β0201 | + | All | High |
| HLA-DR3/7 DQ2 | α0501, β0201 | + | All | High |
| α0201, β0202 | + | Few | ||
| α0501, β0202 | + | All | ||
| α0201, β0201* | (+) | (Few) | ||
| HLA-DR3/X DQ2/X | α0501, β0201 | + | All | Low |
| αXXX, βXXX | — | — | ||
| α0501, βXXX | — | — | ||
| αXXX, β0201 | — | — | ||
| HLA-DR7 DQ2 | α0201, β0202 | + | Few | None |
Functional expression and T cell stimulation were not formally determined for this HLA-DQ2.2 transdimer, but this putative transdimer would have HLA-DQα1 and β1 domains identical with those of the HLA-DQ2.2 cis dimer and thus most likely possess identical peptide-binding properties
Furthermore, we show that the non-disease-associated HLADQ2.2 displayed a far more restricted presentation of gluten peptides than HLA-DQ2.5, which is due to a subtle difference in the peptide-binding properties of both molecules. Five of eight tested T cell-stimulatory gluten peptides were found to carry a proline at relative position P3, which had a deleterious effect on binding to HLA-DQ2.2, but not to HLA-DQ2.5 (ref. 6 and Table 1). Other recently characterized gluten epitopes also contain proline residues at P3 (5). In fact, proline is the second most abundant amino acid in gluten, and gluten epitopes cluster in proline-rich regions (20). Consequently, the protein composition of gluten favors the generation of gluten peptides with HLADQ2.5-but not HLA-DQ2.2-binding properties.
The gluten peptides that do bind to HLA-DQ2.2 trigger inflammatory gluten-specific T cell responses that are indistinguishable from those triggered by peptides in the context of HLA-DQ2.5 (7) (Table 1 and Fig. 1). That HLA-DQ2.2 is not associated with CD (9-11) suggests that a limited T cell response to gluten does not necessarily result in disease development.
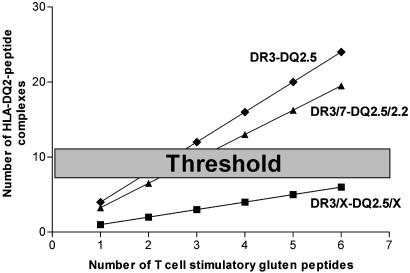
Together, our results indicate that the observed high risk associated with the HLA-DQ2.5 homozygous and HLA-DQ2.5/2.2-heterozygous genotypes, the low risk associated with the HLADQ2.5/X-heterozygous genotype, and the nonrisk associated with the HLA-DQ2.2 genotype (9-11) can be explained by the impact of the number and quality of the HLA-DQ2 molecules on gluten peptide presentation to T cells (Table 2). A model that demonstrates the impact of the HLA-DQ2 gene dose on the formation of HLA-DQ2-gluten peptide complexes is presented in Fig. 4.
Fig. 4.
Model for the impact of the HLA-gene dose on the formation of immunogenic HLA-DQ2-gluten peptide complexes. With an increase in the number of available gluten peptides (x axis), the number of immunogenic HLA-DQ-peptide complexes (y axis) rises the strongest in HLA-DR3-DQ2.5-homozygous individuals, breaking through a hypothetical threshold when relatively few gluten peptides are present. HLA-heterozygous individuals need to recognize a larger group of gluten peptides before they reach the threshold. HLA-DQ2.5/2.2 individuals are in between.
This insight may have important consequences for disease prevention in individuals at risk. Strong evidence shows that early exposure to gluten leads to an increased risk for development of CD. In Sweden, the addition of higher amounts of gluten to infant food led to a 5-fold increase in the occurrence of CD in the 1980s (26). Therefore, it is now common practice that gluten consumption by infants should not commence until after 7 months of age. After this period, however, no restriction in gluten consumption exists. A normal diet at the age of 12 months has been estimated to contain between 6 and 9 g of gluten daily (27), a dose far above the microgram amounts that are sufficient to trigger recall gluten-specific T cell responses (Fig. 1). Such a high gluten intake may be particularly critical to HLA-DQ2.5 homozygous and HLA-DQ2.5/2.2 heterozygous individuals, because they are able to present high amounts of immunogenic gluten peptides (Figs. 1 and 2). We therefore speculate that, for such individuals, a more gradual increase in gluten intake may be called for. This gradual increase may prevent the development of uncontrolled T cell responses to the sudden occurrence of high numbers of immunogenic HLA-DQ2-gluten complexes. This possibility will have to be investigated in future studies.
It is likely, however, that other genetic or environmental factors also contribute to the loss of tolerance. We have never observed gluten-specific T cell responses in small intestinal biopsies of HLA-DQ2-positive patients that do not suffer from CD. The large majority of HLA-DQ2-positive individuals thus appear to be able to regulate gluten-specific T cell responses.
In this respect, it is of interest to note that the expression levels of HLA-DQ are under the influence of cytokines. HLA-DQ expression is regulated by the coactivator CIITA, which is in turn under the influence of IFN-γ (28). Local production of IFN-γ, e.g., as the result of an infection, would thus lead to elevated HLA-DQ expression and, consequently, gluten peptide presentation, which could contribute to the development and/or progression of gluten-specific T cell responses. Clearly, our results indicate that such effects would have the strongest impact in individuals homozygous for HLA-DQ2.5.
In conclusion, this study provides an explanation for the HLA-DQ2 gene dose effect in the development of CD. Similar mechanisms may form the basis for HLA-gene dose effects in other diseases, including type I diabetes, rheumatoid arthritis, and multiple sclerosis (29).
Acknowledgments
We thank R. R. P. de Vries, T. H. M. Ottenhoff, R. Offringa, F. H. J. Claas, C. J. M. Melief, B. O. Roep, and J. van Bergen for critical reading of the manuscript, A. de Ru and P. van Veelen for mass spectrometric analysis, J. W. Drijfhout and W. Benckhuijsen for peptide synthesis, P. van den Elsen for providing the HLA-class II negative EBV-transformed B cell line, G. Nolan for ϕ-NX-A cells, C. Mazzilli for HLA-DQ2-specific mAb, F. Verreck for help with the peptide-binding assay, and E. Hopman for advice. This study was supported by grants from the European Community (BHM4-CT98-3087 and QLK1-2000-00657), the Foundation for Nutritional Research at Leiden University Medical Centre, and the Dutch Organization for Scientific Research (Netherlands Organization for Health Research and Development Grant 912-02-028).
Abbreviations: CD, celiac disease; tTG, tissue transglutaminase; APC, antigen-presenting cells; EBV, Epstein-Barr virus; FACS, fluorescence-activated cell sorter.
References
- 1.van de Wal, Y., Kooy, Y. M. C., Drijfhout, J. W., Amons, R. & Koning, F. (1996) Immunogenetics 44, 246-253. [DOI] [PubMed] [Google Scholar]
- 2.Vartdal, F., Johansen, B. H., Friede, T., Thorpe, C. J., Stevanovic, S., Eriksen, J. E., Sletten, K., Thorsby, E., Rammensee, H. G. & Sollid, L. M. (1996) Eur. J. Immunol. 26, 2764-2772. [DOI] [PubMed] [Google Scholar]
- 3.Molberg, O., McAdam, S. N., Korner, R., Quarsten, H., Kristiansen, C., Madsen, L., Fugger, L., Scott, H., Noren, O., Roepstorff, P., et al. (1998) Nat. Med. 4, 713-717. [DOI] [PubMed] [Google Scholar]
- 4.van de Wal, Y., Kooy, Y., van Veelen, P., Pena, S., Mearin, L., Papadopoulos, G. & Koning, F. (1998) J. Immunol. 161, 1585-1588. [PubMed] [Google Scholar]
- 5.Vader, L. W., De Ru, A., van der Wal, Y., Kooy, Y. M., Benckhuijsen, W., Mearin, M. L., Drijfhout, J. W., van Veelen, P. & Koning, F. (2002) J. Exp. Med. 195, 643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Wal, Y., Kooy, Y. M., Drijfhout, J. W., Amons, R., Papadopoulos, G. K. & Koning, F. (1997) Immunogenetics 46, 484-492. [DOI] [PubMed] [Google Scholar]
- 7.Quarsten, H., Molberg, O., Fugger, L., McAdam, S. N. & Sollid, L. M. (1999) Eur. J. Immunol. 29, 2506-2514. [DOI] [PubMed] [Google Scholar]
- 8.Sollid, L. M., Markussen, G., Ek, J., Gjerde, H., Vartdal, F. & Thorsby, E. (1989) J. Exp. Med. 169, 345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louka, A. S., Nilsson, S., Olsson, M., Talseth, B., Lie, B. A., Ek, J., Gudjonsdottir, A. H., Ascher, H. & Sollid, L. M. (2002) Tissue Antigens 60, 147-154. [DOI] [PubMed] [Google Scholar]
- 10.Mearin, M. L., Biemond, I., Pena, A. S., Polanco, I., Vazquez, C., Schreuder, G. T., de Vries, R. R. & van Rood, J. J. (1983) Gut 24, 532-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ploski, R., Ek, J., Thorsby, E. & Sollid, L. M. (1993) Tissue Antigens 41, 173-177. [DOI] [PubMed] [Google Scholar]
- 12.Spits, H., Borst, J., Giphart, M., Coligan, J., Terhorst, C. & De Vries, J. E. (1984) Eur. J. Immunol. 14, 299-304. [DOI] [PubMed] [Google Scholar]
- 13.Heemskerk, M. H., Blom, B., Nolan, G., Stegmann, A. P., Bakker, A. Q., Weijer, K., Res, P. C. & Spits, H. (1997) J. Exp. Med. 186, 1597-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruggieri, L., Aiuti, A., Salomoni, M., Zappone, E., Ferrari, G. & Bordignon, C. (1997) Hum. Gene Ther. 8, 1611-1623. [DOI] [PubMed] [Google Scholar]
- 15.Kinsella, T. M. & Nolan, G. P. (1996) Hum. Gene Ther. 7, 1405-1413. [DOI] [PubMed] [Google Scholar]
- 16.Heemskerk, M. H., de Paus, R. A., Lurvink, E. G., Koning, F., Mulder, A., Willemze, R., van Rood, J. J. & Falkenburg, J. H. (2001) Proc. Natl. Acad. Sci. USA 98, 6806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, S. G. & Bodmer, J. G. (1989) Immunol. Today 10, 305-312. [DOI] [PubMed] [Google Scholar]
- 18.Anderson, R. P., Degano, P., Godkin, A. J., Jewell, D. P. & Hill, A. V. (2000) Nat. Med. 6, 337-342. [DOI] [PubMed] [Google Scholar]
- 19.Arentz-Hansen, H., Korner, R., Molberg, O., Quarsten, H., Vader, W., Kooy, Y. M., Lundin, K. E., Koning, F., Roepstorff, P., Sollid, L. M., et al. (2000) J. Exp. Med. 191, 603-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arentz-Hansen, H., McAdam, S. N., Molberg, O., Fleckenstein, B., Lundin, K. E., Jorgensen, T. J., Jung, G., Roepstorff, P. & Sollid, L. M. (2002) Gastroenterology 123, 803-809. [DOI] [PubMed] [Google Scholar]
- 21.Sjostrom, H., Lundin, K. E., Molberg, O., Korner, R., McAdam, S. N., Anthonsen, D., Quarsten, H., Noren, O., Roepstorff, P., Thorsby, E., et al. (1998) Scand. J. Immunol. 48, 111-115. [DOI] [PubMed] [Google Scholar]
- 22.Vader, W., Kooy, Y., van Veelen, P., De Ru, A., Harris, D., Benckhuijsen, W., Pena, S., Mearin, L., Drijfhout, J. W. & Koning, F. (2002) Gastroenterology 122, 1729-1737. [DOI] [PubMed] [Google Scholar]
- 23.van de Wal, Y., Kooy, Y. M., van Veelen, P. A., Pena, S. A., Mearin, L. M., Molberg, O., Lundin, K. E., Sollid, L. M., Mutis, T., Benckhuijsen, W. E., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 10050-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Wal, Y., Kooy, Y. M., van Veelen, P., Vader, W., August, S. A., Drijfhout, J. W., Pena, S. A. & Koning, F. (1999) Eur. J. Immunol. 29, 3133-3139. [DOI] [PubMed] [Google Scholar]
- 25.Charron, D. J., Lotteau, V. & Turmel, P. (1984) Nature 312, 157-159. [DOI] [PubMed] [Google Scholar]
- 26.Ivarsson, A., Persson, L. A., Nystrom, L., Ascher, H., Cavell, B., Danielsson, L., Dannaeus, A., Lindberg, T., Lindquist, B., Stenhammar, L., et al. (2000) Acta Paediatr. 89, 165-171. [DOI] [PubMed] [Google Scholar]
- 27.van Overbeek, F. M., Uil-Dieterman, I. G., Mol, I. W., Kohler-Brands, L., Heymans, H. S. & Mulder, C. J. (1997) Eur. J. Gastroenterol. Hepatol. 9, 1097-1099. [DOI] [PubMed] [Google Scholar]
- 28.Ting, J. P. & Trowsdale, J. (2002) Cell 109, Suppl., S21-S33. [DOI] [PubMed] [Google Scholar]
- 29.Thorsby, E. (1997) Hum. Immunol. 53, 1-11. [DOI] [PubMed] [Google Scholar]