Abstract
Microarray data reported elsewhere indicated that herpes simplex virus 1 induces the up-regulation of nuclear factor κB (NF-κB)-regulated genes, including that of its inhibitor, IκBα, consistent with the reports that wild-type virus induces the activation of NF-κB. In this report we show that activation of NF-κB in infected cells is linked to the activation of protein kinase R (PKR). Specifically: (i) PKR is activated in infected cells although the effects of the activated enzyme on protein synthesis are negated by the viral gene γ134.5, which encodes a protein phosphatase 1α accessory factor that enables the dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. NF-κB is activated in wild-type murine embryonic fibroblasts but not in related PKR-null cells. (ii) In cells infected with a replication-competent Δγ134.5 mutant (R5104), but carrying a US11 gene expressed early in infection, eukaryotic translation initiation factor 2α is not phosphorylated, and in in vitro assays, PKR bound to the US11 protein is not phosphorylated on subsequent addition of double-stranded RNA. Here we report that this mutant does not activate PKR, has no effect on the accumulation of IκBα, and does not cause the translocation of NF-κB in infected cells. (iii) One hypothesis advanced for the activation of NF-κB is that it blocks apoptosis induced by viral gene products. The replication-competent R5104 mutant does not induce the programmed cell's death. We conclude that in herpes simplex virus 1-infected cells, activation of NF-κB depends on activation of PKR and that NF-κB is not required to block apoptosis in productively infected cells.
In the course of profiling cellular RNAs induced after herpes simplex virus 1 (HSV-1) infection of quiescent human foreskin fibroblasts, we noted that a significant number of the upregulated RNAs are derived from cellular genes normally activated by nuclear factor κB (NF-κB) transcriptional factors (1). Activation of NF-κB in cells infected with HSV-1 has been described (2, 3), but the consequences of the activation of NF-κB in the course of HSV-1 infection are unclear. Thus, of those tested, most of the NF-κB-dependent RNAs transported into the cytoplasm were rapidly degraded or were untranslatable because of the presence of introns (ref. 4; A. Esclatine, B.T., and B.R., unpublished data). This report addresses the mechanism and role of the activation of NF-κB in infected cells.
Among cellular transcriptional factors, NF-κB plays a very dominant, significant role. In essence, NF-κB activates and regulates a wide range of genes involved in virtually all aspects of innate and adaptive immune responses. The list of genes regulated by NF-κB includes cytokines (IL-1, IL-2, IL-6, IL-12, tumor necrosis factor α, granulocyte–macrophage colony-stimulating factor, etc.), chemokines (IL-8, MIP-1α, MCP-1, RANTES, etc.), adhesion molecules (ICAM-1, VCAM-1, etc.), inducible effector enzymes (iNOS, COX-2, etc.), regulators of apoptosis (c-IAP-1, c-IAP-2, Bfl1, Bcl-XL Fas-ligand, c-myc, etc.), serum amyloid A, β-defensin, MHC molecules, IFN-β, etc. (reviewed in ref. 5). In unstimulated cells, NF-κB exists as homo- or heterodimers complexed with regulatory proteins IκBα or IκBβ. Current evidence suggests that NF-κB–IκBα shuttles between the cytoplasm and nucleus, but that its default localization is in the cytoplasm (reviewed in ref. 6). Activation of NF-κB appears to require two events (6). The first is the phosphorylation of IκBα by IκB kinase (IKK) complex. IKK is activated by a growing list of kinases, including protein kinase R (PKR), DNA-dependent protein kinase, and PKC ζ (6). Phosphorylated IκBα then becomes the target of degradation by the ubiquitin proteasomal pathway. The second involves the phosphorylation of the p65 component of NF-κB and its translocation to the nucleus. A rich and ever-expanding literature on the structure and activation of NF-κB and the consequences of its activation attests to the importance of NF-κB in the mediation of innate and immune responses. NF-κB is activated in a number of viral infections (reviewed in ref. 7). The events that lead to the activation of NF-κB and the contribution of this activation to the pathogenesis of HSV-1 infections remain unknown.
As noted above, phosphorylation of IκBα by IKK is an essential event in the activation of NF-κB. Of particular interest to us were several reports that IKK is activated by PKR (8). The significance of these reports stems from studies related to the activation of PKR in HSV-1-infected cells. Specifically, after the onset of viral DNA synthesis at ≈3 h after infection, the infected cells begin to accumulate a large amount of complementary viral RNAs, sequences derived from at least 50% of the viral DNA (9, 10). In HSV-1-infected cells, activation of PKR depends on viral DNA synthesis, and in cells infected with a mutant lacking γ134.5, the α subunit of eukaryotic translation initiation factor 2 (eIF-2α) is phosphorylated and all protein synthesis comes to a halt. The γ134.5 protein blocks the effect of activated PKR by recruiting the protein phosphatase 1α to dephosphorylated eIF-2α. In effect, wild-type HSV-1 circumvents, but does not affect, the accumulation of activated PKR. It is of interest to note and relevant to this report that one mutant, which was selected for continued protein synthesis in the absence of the γ134.5 gene, exhibited a mutation that enables a late gene, US11, to be expressed early (11–13). The US11 protein is not essential for viral replication. The protein is packaged in virions and binds RNA in a sequence- and conformation-dependent manner (14, 15). Subsequent studies revealed that the US11 protein made early in infection blocks the activation of PKR (16). The relevance of these observations to the activation of NF-κB emerged from a recent report showing that poly(rI)·poly(rC) induced PKR and NF-κB. In cells lacking the PKR genes (PKR-/-), NF-κB was not induced (8).
In this report we show that wild-type HSV-1 does not induce NF-κBin PKR-/- cells or in cells infected with a mutant in which the US11 protein is expressed immediately after infection. We also report that at least one function ascribed to the activation of NF-κB in infected cells, namely to preclude apoptosis, does not appear to be correct because the consequences of apoptosis were not apparent in productively infected cells in which NF-κB was not activated.
Materials and Methods
Cells and Viruses. The SK-N-SH cells obtained from American Type Culture Collection were propagated in DMEM supplemented with 10% newborn calf serum. PKR-/- and PKR+/+ mouse embryo fibroblasts kindly provided by B. R. G. Williams (Cleveland Clinic Foundation, Cleveland) were propagated in DMEM supplemented with 10% FCS. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (17). The Δα27 mutant virus, 27LacZ (18), and the d120 mutant lacking both copies of the α4 gene (19) were the kind gifts of S. J. Silverstein (Columbia University, New York) and Neal A. DeLuca (University of Pittsburgh, Pittsburgh), respectively. The ΔUL41 mutant virus, R2621, was reported elsewhere (20). The recombinant virus R7023 lacking US8–US12 genes was described elsewhere (21). Recombinant virus R5104 derived from R7023 also lacks the γ134.5 gene but contains an insertion of the US11 ORF driven by the α47 gene into the UL23 gene encoding thymidine kinase. In cells infected with this mutant, the US11 protein is made earlier rather than later after infection (13). Cell monolayers were exposed to 10 plaque-forming units of the indicated viruses per cell for 1 h at 37°C.
Total RNA Isolation and Electrophoretic Separation. Total RNA was extracted with the aid of TRIzol Reagent (Life Technologies, Rockville, MD) according to the manufacturer's instructions. DNase treatment (Life Technologies), phenol/chloroform extraction, and ethanol precipitation (Fisher Scientific) were carried out to remove possible DNA contamination. For Northern blot analysis, 10 μg of total RNA was loaded onto a denaturing formaldehyde gel and probed with a random hexanucleotide-primed 32P-labeled specific probe after transfer onto a nylon membrane. For IκBα mRNA detection, the PCR-amplified fragment containing the entire IκBα coding sequence was used as the probe. Prehybridization and hybridization were performed with the ULTRAhyb buffer (Ambion, Austin, TX) supplemented with 200 μg/ml of denatured salmon sperm DNA (Stratagene). The membrane was prehybridized for 2 h at 42°C and then overnight after the addition of the 32P-labeled probe. The membrane was washed as suggested by the ULTRAhyb manufacturer and exposed to the film for signal detection.
Preparation of Whole Cell Extract and Cytoplasmic and Nuclear Fractions. Cells were collected by scraping directly into the medium, rinsed once with cold PBS, transferred to a 1.5-ml Eppendorf tube, and lysed in RIPA buffer [PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM sodium orthovanadate, 5 mM EDTA, and protease inhibitor mixture (Complete Protease mixture, Roche Diagnostics)]. Samples were kept on ice for 1 h, and insoluble material was pelleted in an Eppendorf 5415C microfuge at 14,000 rpm for 10 min at 4°C. The fractionation of infected cells into nuclear and cytoplasmic extracts was performed as follows. Cells were collected by scraping directly into the medium, rinsed once with cold PBS, transferred to a 1.5-ml Eppendorf tube, and lysed by gentle inversion in hypotonic lysis buffer (10 mM Hepes, pH 7.5/10 mM KCl/3 mM MgCl2/0.05% Nonidet P-40/1 mM EDTA/1 mM DTT/10 mM NaF/10 mM β-glycerophosphate/0.1 mM sodium orthovanadate/protease inhibitor mixture). Samples were incubated in ice for 30 min prior to centrifugation at 500 × g for 5 min at 4°C. The supernatant fluids (cytoplasmic extract) were transferred into fresh tubes. The nuclear pellets were rinsed twice in hypotonic lysis buffer containing increased amounts of Nonidet P-40 (0.1%) and lysed with buffer containing 50 mM Hepes (pH 7.9), 250 mM KCl, 1% Nonidet P-40, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, and protease inhibitor mixture. The samples were frozen and thawed three times and incubated in ice for 30 min. Insoluble material was pelleted in an Eppendorf 5415C microfuge at 14,000 rpm for 10 min at 4°C.
Immunoblots. Approximately 50 μg of cell proteins from whole extracts or subcellular fractions was separated on a denaturing 10% polyacrylamide gel and electrically transferred to a nitrocellulose membrane at 300 mA (constant) for 4 h in Tris/glycine/methanol buffer, pH 8.3, at 4°C. The membranes were blocked for 2 h with5% nonfat dry milk in PBS and reacted with the appropriate primary antibody overnight at 4°C, rinsed, and then exposed to a secondary antibody, conjugated with either alkaline phosphatase or peroxidase, at room temperature for 1 h. The antibodies were diluted in PBS containing 1% BSA and 0.05% Tween 20. All rinses were done in PBS containing 0.05% Tween 20. To develop alkaline phosphatase-conjugated secondary antibodies, the immunoblots were washed with alkaline phosphatase buffer (100 mM Tris·HCl, pH 9.5/100 mM NaCl/5 mM MgCl2), followed by reaction in alkaline phosphatase buffer containing 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. To develop peroxidase-conjugated secondary antibodies, the immunoblots were reacted with enhanced chemiluminescence (ECL) Western blotting detection reagents, according to the manufacturer's instructions (Amersham Biosciences). The HSV-1 proteins were detected with the anti-US11 monoclonal antibody (22), anti-ICP4 (clone 1112), and anti-ICP27 reported elsewhere (23). The polyclonal rabbit anti-mouse IκBα, the monoclonal mouse anti-human IKKα, and the mouse antihuman NF-κB p65 were purchased from BD Biosciences.
PKR in Vitro Kinase Assay. SK-N-SH cells were infected with 10 plaque-forming units of HSV-1(F) or the R5104 mutant virus per cell. The cells were harvested at 4 or 6 h after infection and lysed in immunoprecipitation lysis buffer (50 mM Tris·HCl, pH 7.6/150 mM NaCl/10% glycerol/1% Nonidet P-40/5 mM EDTA/1 mM DTT/100 mM NaF/20 mM β-glycerophosphate/0.1 mM sodium orthovanadate/protease inhibitor mixture). The supernatant containing 100 μg of proteins was brought up in an equal volume of lysis buffer. Lysates were precleared with preimmune serum for 2 h, and 50 μl of a 50% slurry of protein A conjugated to agarose was added. Samples were centrifuged, and the supernatant was transferred to new tubes. Monoclonal antibody to mouse PKR (Santa Cruz Biotechnology) was added to the samples and incubated at 4°C for 16 h, and immunocomplexes were collected by the addition of 20 μl of 50% protein-A slurry. The bound PKR was washed three times with immunoprecipitation buffer and three times with incomplete DBGA buffer (10 mM Tris·HCl, pH 7.6/50 mM KCl/2 mM magnesium acetate/20% glycerol/7 mM mercaptoethanol) (24) before incubation (20 min at 30°C) in 40 μl of DBGA buffer containing 1 μM ATP, 10 μCi (1 Ci = 37 GBq) of [γ-32P]ATP, and 1.5 μg of histone H1. Reactions were terminated by addition of SDS gel-loading buffer and heated to 95°C for 5 min. The samples were resolved by polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed by autoradiography. Quantification of 32P phosphorylation of the substrates was performed with the aid of a Molecular Dynamics PhosphorImager (Storm 860).
IKKα in Vitro Kinase Assay. PKR-/- and PKR+/+ cells were infected with 10 plaque-forming units of HSV-1(F) per cell. The cells were harvested at 6 h after infection and lysed in RIPA buffer. The supernatant containing 75 μg of proteins was treated as described for PKR kinase assay prior to incubation with a monoclonal antibody to human-IKKα at 4°C for 16 h, and immunocomplexes were collected by the addition of 20 μl of 50% protein-A slurry. The bound IKKα was washed three times with RIPA buffer and three times with incomplete IKK buffer (20 mM Hepes, pH 7.5/20 mM β-glycerophosphate/10 mM MgCl2/100 mM NaCl/0.1 mM sodium orthovanadate/2 mM DTT/10 μg of aprotinin per ml) (25) before incubation (30 min at 30°C) in 40 μl of complete IKK buffer [incomplete IKK buffer containing 10 μM ATP, 10 μCi of [γ-32P]ATP, 1 μg of GST-IκBα (amino acids 1–317) (Santa Cruz Biotechnology)]. The samples were resolved as described above.
DNA Fragmentation Assay. Mock- or HSV-1-infected SK-N-SH cells were collected at 12 h after infection, rinsed in PBS, lysed in a solution containing 10 mM Tris·HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100 for 15 min on ice, digested with RNase A (0.1 mg/ml) at 37°C for 1 h, and centrifuged at 12,000 rpm for 25 min in a microcentrifuge to pellet chromosomal DNA. The supernatant fluids were digested with proteinase K (1 mg/ml) at 50°C for 2 h in the presence of 1% SDS, followed by phenol/chloroform extraction and ethanol precipitation. The samples were subjected to electrophoresis on a 1.5% agarose gel.
Results
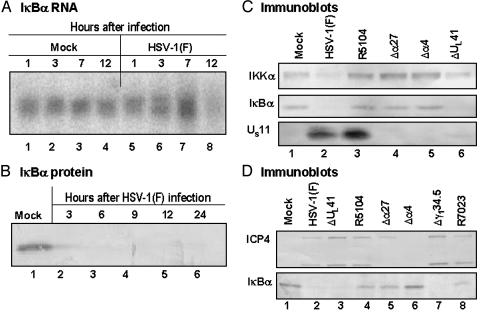
IκBα, the Inhibitor of NF-κB, Is Not Degraded in Cells Infected with the R5104 Mutant Encoding a US11 Gene Expressed Early in Infection. Studies based on microchip analyses have shown that several genes reported to be NF-κB targets were found to be upregulated in human foreskin fibroblasts after infection with HSV-1 (1). This observation was concordant with the report that HSV-1 induces a persistent NF-κB nuclear translocation (2) as a result of the activation of the IKK complex and consequent ubiquitin-dependent degradation of IκBα early in infection (3). Among the various NF-κB-regulated genes whose expression is up-regulated in human foreskin fibroblasts is IκBα, the NF-κB inhibitor. The induction of IκBα RNA in HSV-1-infected human foreskin fibroblasts detected in microarray analyses (1) was verified by Northern blot analysis on total RNA samples extracted at different times from SK-N-SH cells infected with 10 plaque-forming units of HSV-1(F) per cell (Fig. 1A). As expected and shown in Fig. 1B, the IκBα protein was present in mock-infected cells (lane 1) but was barely detectable in HSV-1-infected cells harvested at 3 h post infection (lane 2), and completely disappeared starting in the 6-h sample (lanes 3–6). Because it has been reported that two α proteins, the infected cell proteins 4 and 27 (ICP4 and ICP27), were required for the NF-κB nuclear translocation (2), SK-N-SH cells were infected with a panel of different mutants that fail to express functional forms of these proteins (Δα27, 27LacZ) and ICP4 (Δα4, d120) or late viral proteins such as vhs (ΔUL41), US11 (R7023), or ICP34.5 (Δγ134.5). In addition, the cells were infected with the recombinant virus R5104. As noted in the Introduction and in Materials and Methods, the salient feature of this mutant is that the US11 ORF is driven by the α47 promoter and as a consequence the US11 protein is made early rather than late in infection (13). Cells were harvested at 8 h after infection with wild-type or mutant viruses and processed as described in Materials and Methods. Equal amounts of proteins from whole cell extracts were electrophoretically separated on a denaturing 10% polyacrylamide gel, transferred to a nitrocellulose sheet, and reacted with antibodies directed to cellular proteins IκBα and IKKα and viral proteins US11 and ICP4 (Fig. 1 C and D). IκBα degradation was observed in cells infected with wild-type HSV-1(F) and ΔUL41 mutant virus (Fig. 1 C and D, lanes 2 and 6 and lanes 2 and 3, respectively) as well as in cells infected by Δγ134.5 mutant virus (Fig. 1D, lane 7). As expected based on previous reports, IκBα remained stable in cells infected with either Δα27 (Fig. 1 C, lane 4, and D, lane 5) or Δα4 (Fig. 1 C, lane 5, and D, lane 6). Interestingly, IκBα was not degraded in cells infected by R5104 (Fig. 1 C, lane 3, and D, lane 4). Small amounts of IκBα were also detectable in cells infected by R7023 mutantvirus at 8 h after infection, but these disappeared at later times after infection (data not shown), suggesting that IκBα degradation in cells infected by this mutant lagged behind that observed in wild-type virus-infected cells. It is noteworthy that the total amount of IKKα appeared to decrease in cells infected by wild-type HSV-1(F) and ΔUL41 mutant virus (Fig. 1C, lanes 2 and 6) coincidently with the degradation of IκBα. The molecular reasons of this observation are unclear.
Fig. 1.
Accumulation of IκBα RNA and protein in SK-N-SH cells after HSV-1 infection. (A) SK-N-SH cells were mock-infected or infected with HSV-1(F). Total RNA was extracted from cells harvested at the indicated times after mock infection (lanes 1 and 4) or exposure to HSV-1(F) (lanes 5–8) and processed as described in the text. Ten micrograms of total RNA was loaded onto a denaturing formaldehyde gel and probed with a 32P-labeled fragment containing the entire coding sequence of IκBα. (B) SK-N-SH cells mock-infected or infected with HSV-1(F) were harvested at 3, 6, 9, 12, and 24 h after infection (lanes 2–6) and processed as described in Materials and Methods. The electrophoretically separated proteins were reacted with anti-IκBα antibody. (C and D) SK-N-SH cells were mock-infected or infected with HSV-1(F) or indicated mutant viruses. The cells were harvested at 8 h after infection and processed as described in Materials and Methods. The electrophoretically separated proteins were reacted with antibodies against IκBα, IKKα, or viral proteins US11 or ICP4.
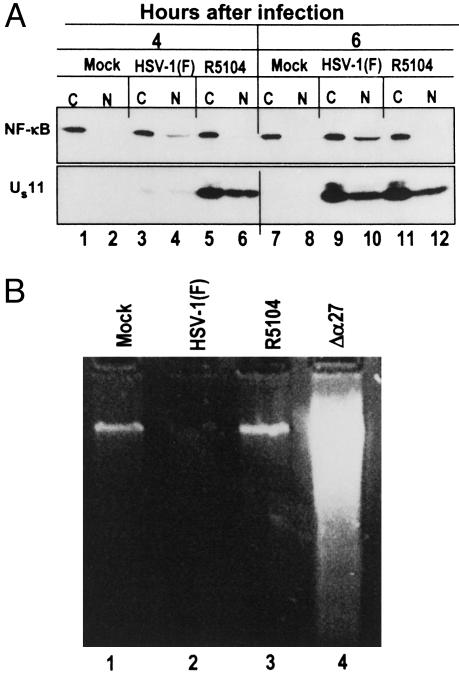
NF-κB Is Retained in the Cytoplasmic Compartment in Cells Infected with the R5104 Mutant. IκBα degradation results in the release of NF-κB with consequent nuclear translocation (6). To test whether the NF-κB is retained in the cytoplasm of cells infected by the R5104 recombinant virus, replicate SK-N-SH cultures were mock-infected or exposed to HSV-1(F) or R5104. The cells were harvested at 4 and 6 h and fractionated into nuclear and cytoplasmic fractions as described in Materials and Methods. Equal amounts of proteins were loaded on a denaturing 10% polyacrylamide gel, transferred to a nitrocellulose sheet, and probed with anti-NF-κB antibody directed against the p65 subunit. The results shown in Fig. 2A were as follows: As expected, in lysates of mock-infected cells, the p65 subunit of NF-κB was detected in the cytoplasmic fraction at both time points tested (Fig. 2 A, lanes 1 and 7). In the HSV-1(F)-infected cells, a low amount of NF-κB appeared in the nuclear fraction at 4 h after infection (Fig. 2 A, lane 4), and the amount increased by 6 h (Fig. 2 A, lane 10). As predicted by the absence of IκBα degradation, NF-κB was retained in the cytoplasm of cells infected with the R5104 recombinant virus (Fig. 2 A, lanes 6 and 12). Concordant with these results, the US11 protein accumulated in R5104-infected cells at early times after infection (Fig. 2 A, lanes 5 and 6).
Fig. 2.
Cytoplasmic localization of NF-κB in SK-N-SH cells does not result in apoptosis. (A) Localization of NF-κB. SK-N-SH cells were mock-infected (lanes 1, 2, 7, and 8) or infected with HSV-1(F) (lanes 3, 4, 9, and 10) or R5104 (lanes 5, 6, 11, and 12). Cytoplasmic (C) and nuclear (N) fractions were prepared from cells harvested at 4 h (lanes 1–6) and 6 h (lanes 7–12) after infection or mock infection and processed as described in Materials and Methods. Electrophoretically separated proteins were reacted with anti-NF-κB or anti-US11 antibodies. (B) Degradation of cellular DNA in infected cells. SK-N-SH cells were mock-infected (lane 1) or infected with HSV-1(F) (lane 2), R5104 (lane 3), or Δα27 mutant viruses (lane 4), and a DNA fragmentation assay was performed on cells collected at 12 h after infection as described in Materials and Methods.
Retention of NF-κB in the Cytoplasm Does Not Lead to Apoptosis in Cells Infected with the R5104 Mutant. Because it has been recently reported that the nuclear translocation of NF-κB early in infection is necessary to prevent apoptosis in wild-type HSV-1-infected HEp-2 cells (26), it seemed appropriate to determine whether the retention of NF-κB in the cytoplasmic fraction of R5104-infected cells led to induction of apoptosis. Replicate cultures of SK-N-SH cells were mock- or HSV-1(F)- or R5104-infected, and a DNA fragmentation assay was performed on cells collected at 12 h after infection as described in Materials and Methods. Infection with Δα27 mutant virus was carried out as a control of the DNA fragmentation assay. As shown in Fig. 2B, lane 3, no DNA fragmentation was detected in cells infected by the R5104 virus. Caspase-3 assays (not shown) yielded results consistent with the results of the experiment described above. These results indicate that failure of NF-κB activation in productively infected cells by replication-competent viruses does not necessarily result in the activation of proapoptosis pathways.
Activation of IKK and Degradation of IκBα Requires Activated PKR. One hypothesis that could explain the results presented above that is consistent with the mechanism by which NF-κB is activated is that in HSV-1-infected cells, NF-κB is activated as a consequence of events initiated by the activation of PKR. To test this hypothesis two series of experiments were done.
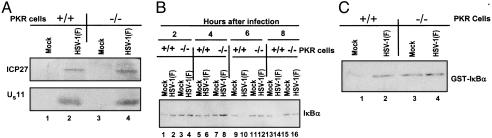
In the first, replicate cultures of murine PKR+/+ and PKR-/- cells were mock- or HSV-1(F)-infected and tested for IKK activity as described in Materials and Methods. Activation of IKK, measured as increased phosphorylation of GST-IκBα in an in vitro kinase assay, was clearly observed in infected PKR+/+ cells (Fig. 3C, lane 2), whereas the level of IκBα phosphorylation in infected PKR-/- cells after HSV-1-infection remained unchanged as compared with the level present in mock-infected cells (Fig. 3C, lanes 3 and 4). Because activation of IKK results in the degradation of IκB complex, whole cell extracts from the two cell lines were tested for IκBα degradation at different times after HSV-1-infection. The levels of IκBα decreased only in PKR+/+ cells at 4 h (Fig. 3B, lane 6) and disappeared completely by 6 h after infection (Fig. 3B, lane 10). Immunoblot assays on lysates of cells harvested at 7 h after infection (Fig. 3A) indicate that the infected PKR+/+ and PKR-/- cells expressed α and late γ2 proteins with equal efficiency.
Fig. 3.
IKKα activity in murine PKR-/- and PKR+/+ cells after HSV-1 infection. (A) Electrophoretically separated proteins from lysates of PKR-/- or PKR+/+ cells harvested at 7 h after infection with HSV-1(F) were reacted with anti-ICP27- or anti-US11-specific antibodies. (B) Electrophoretically separated proteins from lysates of PKR-/- or PKR+/+ cells harvested at 2, 4, 6, or 8 h after infection with HSV-1(F) were reacted with anti-IκBα antibody. (C) Autoradiographic images of GST-IκBα reacted with IKKα immunoprecipitated from PKR-/- or PKR+/+ cells 6 h after infection with HSV-1(F). The reaction conditions were as described in Materials and Methods.
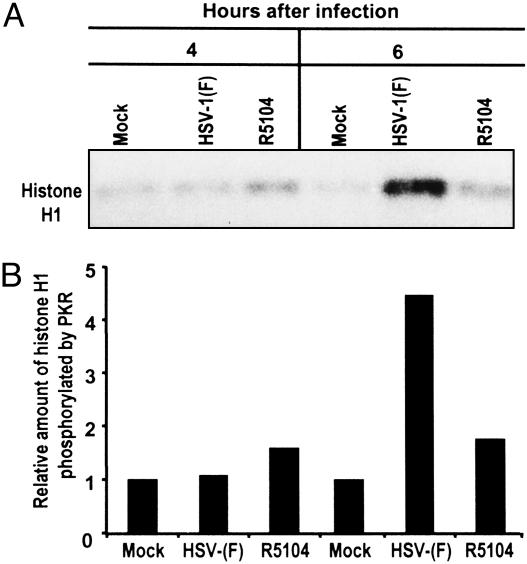
The second series of experiments was based on three observations. First, in cells infected with mutant viruses lacking the γ134.5 gene, but containing a US11 ORF driven by an α promoter, eIF-2 was not phosphorylated. Second, in in vitro assays the US11 added to cultured cells before activation of PKR bound the enzyme and precluded its phosphorylation (16). In the experiments described above, IκBα was not phosphorylated and NF-κB was not translocated to the nucleus in cells infected with this mutant. On the basis of these results, it could be expected that PKR is not activated in cells infected with R5104. To test this hypothesis, SK-N-SH cells were mock- or HSV-1(F)- or R5104-infected, and whole cell extracts were analyzed at 4 and 6 h after infection for PKR activity by immune complex kinase assay as described in Materials and Methods. The level of phosphorylated histone H1 was measured with the aid of Molecular Dynamics PhosphorImager analysis. As shown in Fig. 4, PKR was activated at 6 h after infection in wild-type virus-infected cells but not to a significant level in cells infected with R5104 mutant.
Fig. 4.
PKR activation in SK-N-SH cells after HSV-1 infection. (A) PKR was immunoprecipitated from whole cell extracts prepared from SK-N-SH cells harvested at 4 or 6 h after mock infection or infection with HSV-1(F) or R51204 as described in Materials and Methods. The immune complex was subjected to an in vitro kinase assay in the presence of histone H1, analyzed on a denaturing 10% polyacrylamide gel, electrically transferred to a nitrocellulose sheet, and subsequently subjected to autoradiography to detect substrate phosphorylation. (B) Quantification of the amount of phosphorylated substrate relative to mock-infected cells obtained with the aid of the Molecular Dynamics Storm 860 PhosphorImager.
The results of these two series of experiments indicate that in HSV-1-infected cells activation of NF-κB results from activation of PKR and that expression of US11 early in infection prevents activation of both PKR and NF-κB.
Discussion
The key conclusion of the studies presented in this report is that in cells infected with HSV-1 activation of NF-κB correlates with activation of PKR. Two lines of evidence support this conclusion. First, the wild-type virus failed to induce NF-κB in murine cells from which the PKR genes were knocked out. In these cells the virus expressed both early and late genes in a manner indistinguishable from that expressed in the wild-type parent cells. Second, studies on the R5104 mutant also indicated that this virus failed to activate NF-κB in SK-N-SH cells. As noted in the results, US11 ORF expressed early in infection precludes the phosphorylation of eIF-2α (13). In in vitro assays, US11 protein added to mixtures before the activation of PKR bound the enzyme and precluded its phosphorylation (16). The key feature of the R5104, a replication-competent mutant, is the absence of the γ134.5 gene and insertion of the US11 ORF driven by an α promoter. Analyses of cells infected with this mutant showed that IκBα was not phosphorylated, NF-κB was not translocated to the nucleus, and the activity of PKR, although slightly elevated relative to that of mock-infected cells, was considerably lower than those of wild-type virus-infected cells. In essence, as illustrated in the model depicted in Fig. 5, the results suggest that PKR activates IKK, and this enzyme in turn phosphorylates IκBα, which is subjected to degradation. On the basis of earlier studies and those presented in this report we may also conclude that the US11 protein made early in infection blocks the activation of PKR and consequently also blocks the activation of NF-κB. We also conclude that, although in HSV-1-infected cell's activation of NF-κB is a consequence of the activation of PKR, NF-κB does not appear to play a significant role in the growth of replication-competent viruses in cells in culture. These observations raise several questions regarding both the activation of NF-κB and its relevance to viral replication. Specifically:
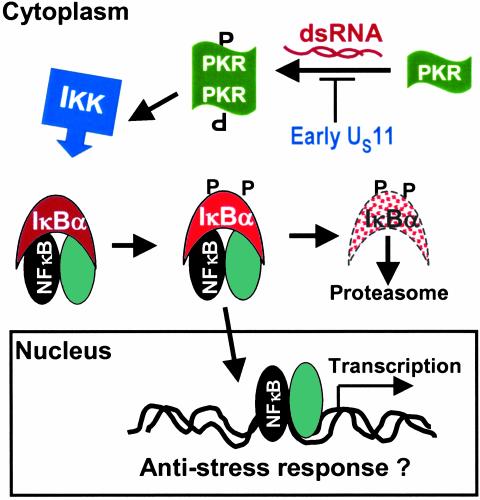
Fig. 5.
A schematic representation of a model of the activation of NF-κB in cells infected with a wild-type virus or R5104 mutant. In mutant virus-infected cells, the US11 protein made early blocks phosphorylation (P) and dimerization of PKR. As a consequence, NF-κB is not activated. In wild-type virus-infected cells, PKR is activated and the cascade of events schematically illustrated in this model leads to activation of NF-κB. dsRNA, double-stranded RNA.
The observation that NF-κB is not activated in cells infected with the replication-defective Δα4 and Δα27 mutants is consistent with the mechanism by which NF-κB appears to be activated in HSV-1-infected cells. Specifically, viral DNA synthesis and expression of late viral proteins require functional ICP4 and ICP27, the products of α4 and α27 genes, respectively. These functions are also required for the activation of PKR in infected cells (27). It would therefore be expected that in cells abortively infected with these mutants, NF-κB would not be activated.
As noted above, in wild-type virus-infected cells PKR is activated after the onset of DNA synthesis. The kinase remains active even though its major manifestation in the infected cells, the phosphorylation of eIF-2α, is negated by the dephosphorylation of the protein by the eIF-2α–protein phosphatase 1α complex. Activation of PKR is the bane of all viruses encoding complementary RNAs, and as a consequence they have evolved ways to block the effects of PKR on eIF-2α and the subsequent shutoff of the protein's synthesis. It is of interest that of the members of the herpesvirus family sequenced to date, only three (HSV-1, HSV-2, and the simian B virus) encode homologs of the γ134.5 gene. Yet all herpesviruses are affected by the activation of PKR. The observation that the carboxyl-terminal amino acid sequences of the γ134.5 gene are homologous to the corresponding domain of GADD34 suggests that it was acquired in the course of the coevolution of herpesviruses with primates and that the new gene replaced an ancestral gene also designed to block the effects of activated PKR. An obvious candidate for an ancestral gene supplanted by the evolution of the γ134.5 gene is the US11, which is expressed early in infection. Irrespective of whether US11 expressed early was the ancestral gene, the questions arises why HSV-1 has evolved a gene that does not block PKR activity and, by extension, does block the activation of NF-κB in favor of a gene that does both. What advantages, if any, accrue HSV-1 by the activation of NF-κB?
Goodkin et al. (26) have suggested that HSV-1 activates NF-κB to preclude apoptosis. The argument is based to a large extent on the observation that mutants lacking all or specific domains of the α27 gene encoding ICP27 fail to induce NF-κB and cause cells to go into apoptosis. These conclusions suffer from two defects. First, viral DNA synthesis is affected by the absence of ICP27, and proteins dependent on viral DNA synthesis that may also be responsible for blocking apoptosis induced by gene products made earlier in infection may not accumulate. The situation is similar to that observed in cells infected with mutants lacking the α4 gene. Cells infected with the d120 mutant lacking the genes encoding ICP4 undergo apoptosis. In this instance apoptosis is blocked by the viral protein kinase US3, whose synthesis depends on ICP4. As noted in this report, d120 does not induce NF-κB, and transduction of d120-infected cells with the US3 gene blocks apoptosis but does not overcome the failure of the d120 mutant to replicate. The second defect apparent from this report is the evidence that the replication-competent virus R5104 fails to activate NF-κB but does not cause the cells to undergo apoptosis.
One possible clue to the selection of the γ134.5 gene emerged from studies designed to verify and establish the significance of cellular RNAs up-regulated after infection with wild-type HSV-1. Of the several RNAs known to be activated by NF-κB, several of the ones tested (ref. 4; A. Esclatine, B.T., and B.R., unpublished data) were not expressed because the cytoplasmic RNA contained introns or were rapidly degraded. Thus, there exists the possibility that HSV-1 is unaffected by the activated NF-κB because the activated cellular genes are not expressed. Further studies on the fate of RNAs activated by NF-κB will be necessary to determine whether the effect is a general one or whether some activated genes are expressed and play a role in the evolution of viral infection in animal hosts.
Acknowledgments
We thank Sunil Advani and Audrey Esclatine for helpful discussions and B. G. R. Williams, S. J. Silverstein, and N. A. DeLuca for invaluable reagents. These studies were aided by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860) of the Public Health Service. T.R.L. was supported by a fellowship of the China Scholarship Council.
Abbreviations: HSV-1, herpes simplex virus 1; PKR, protein kinase R; IKK, IκB kinase; eIF-2α, eukaryotic translation initiation factor 2α; ICP, infected cell protein.
References
- 1.Taddeo, B., Esclatine, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 17031-17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel, A., Hanson, J., McLean, T. I., Olgiate, J., Hilton, M., Miller, W. E. & Bachenhiemer, S. L. (1998) Virology 247, 212-222. [DOI] [PubMed] [Google Scholar]
- 3.Amici, C., Belardo, G., Rossi, A. & Santoro, M. G. (2001) J. Biol. Chem. 276, 28759-28766. [DOI] [PubMed] [Google Scholar]
- 4.Taddeo, B., Esclatine, A., Zhang, W. & Roizman, B. (2003) J. Virol. 77, 6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pahl, H. L. (1999) Oncogene 18, 6853-6866. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, S. & Karin, M. (2002) Cell 109, S81-S96. [DOI] [PubMed] [Google Scholar]
- 7.Hiscott, J., Kwon, H. & Genin, P. (2001) J. Clin. Invest. 107, 143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamanian-Daryoush, M., Mogensen, T. H., DiDonato, J. A. & Williams, B. R. G. (2000) Mol. Cell. Biol. 20, 1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozac, M. & Roizman, B. (1975) J. Virol. 15, 36-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquemont, B. & Roizman, B. (1975) J. Virol. 15, 707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohr, I. & Gluzman, Y. (1996) EMBO J. 15, 4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 12.He, B., Chou, J., Brandimarti, R., Mohr, I., Gluzman, Y. & Roizman, B. (1997) J. Virol. 71, 6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassady, K. A., Gross, M. & Roizman, B. (1998) J. Virol. 72, 7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roller, R. J. & Roizman, B. (1990) J. Virol. 64, 3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sciortino, M. T., Taddeo, B., Poon, A. P. W., Mastino, A. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassady, K. A., Gross, M. & Roizman, B. (1998) J. Virol. 72, 8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ejercito, P. M., Kieff, E. D. & Roizman, B. (1968) J. Gen. Virol. 2, 357-364. [DOI] [PubMed] [Google Scholar]
- 18.Soliman, T. M., Sandri-Goldin, R. M. & Silverstein, S. J. (1997) J. Virol. 71, 9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLuca, N. A., McCarthy, A. & Schaffer, P. A. (1985) J. Virol. 56, 558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poon, A. P. W. & Roizman, B. (1997) Virology 229, 98-105. [DOI] [PubMed] [Google Scholar]
- 21.Longnecker, R. & Roizman, B. (1986) J. Virol. 65, 583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roller, R. J. & Roizman, B. (1992) J. Virol. 66, 3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackermann, M., Braun, D. K., Pereira, L. & Roizman, B. (1984) J. Virol. 52, 108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamanian-Daryoush, M., Der, S. D. & Williams, B. R. G. (1999) Oncogene 18, 315-326. [DOI] [PubMed] [Google Scholar]
- 25.DiDonato, J. A., Hayakawa, M., Rothwarf, D. M., Zandi, E. & Karin, M. (1997) Nature 388, 548-554. [DOI] [PubMed] [Google Scholar]
- 26.Goodkin, M. L., Ting, A. T. & Blaho, J. A. (2003) J. Virol. 77, 7261-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou, J., Chen, J. J., Gross, M. & Roizman, B. (1995) Proc. Natl. Acad. Sci. USA 92, 10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]