Abstract
The release of enveloped viruses from infected cells often requires a virally encoded activity, termed a late-budding domain (L domain), encoded by essential PTAP, PPXY, or YPDL sequence motifs. PTAP-type L domains recruit one of three endosomal sorting complexes required for transport (ESCRT-I). However, subsequent events in viral budding are poorly defined, and neither YPDL nor PPXY-type L domains require ESCRT-I. Here, we show that ESCRT-I and other class E vacuolar protein sorting (VPS) factors are linked by a complex series of protein–protein interactions. In particular, interactions between ESCRT-I and ESCRT-III are bridged by AIP-1/ALIX, a mammalian orthologue of the yeast class E VPS factor, Bro1. Expression of certain ESCRT-III components as fusion proteins induces a late budding defect that afflicts all three L-domain types, suggesting that ESCRT-III integrity is required in a general manner. Notably, the prototype YPDL-type L domain encoded by equine infectious anemia virus (EIAV) acts by recruiting AIP-1/ALIX and expression of a truncated form of AIP-1/ALIX or small interfering RNA-induced AIP-1/ALIX depletion specifically inhibits EIAV YPDL-type L-domain function. Overall, these findings indicate that L domains subvert a subset of class E VPS factors to mediate viral budding, some of which are required for each of the L-domain types, whereas others apparently act as adaptors to physically link specific L-domain types to the class E VPS machinery.
Many enveloped viruses encode a late-budding domain (L domain), whose function is essential for particle release (1–8). The currently defined viral L domains contain at least one of three sequence motifs: PT/SAP, PPXY, or YPDL. L domains act by recruiting host factors during egress, and a variety of findings indicate that PTAP-type L domains, encoded by HIV-1 and Ebola virus, recruit Tsg101 (9–12), a component of endosomal sorting complex required for transport-I (ESCRT-I) (13).
ESCRT-I normally functions in the sorting of cargo into vesicles that bud into the lumen of multivesicular bodies (MVBs) (14, 15). Tsg101 is the mammalian orthologue of a yeast protein, Vps23p, which, along with ≈16 other proteins, is required for MVB formation (16). Genetic ablation of any of these factors results in the formation of a class E compartment, an aberrant, multilamellar prevacuolar endosome that lacks luminal vesicles. A subset of yeast class E vacuolar protein sorting (VPS) factors assemble into complexes, including ESCRT-I (which includes Vps23p), ESCRT-II, and ESCRT-III (13, 17, 18). A current model for cargo sorting and vesicle formation invokes the sequential recruitment of ESCRT-I, -II and -III, and budding concurrent with disassembly of ESCRT components, catalyzed by an AAA-ATPase, Vps4p (17, 18). Homologues of ESCRT components exist in mammals, but with the exception of the ESCRT-I components Tsg101 and VPS28 (14, 15), their role in mammalian MVB formation is poorly defined. Indeed, Eap45, Eap30, and Eap20, mammalian homologues of the yeast ESCRT-II factors Vps36p, Vps22p, and Vps25p, respectively, have been studied only in the context of transcription regulation (14, 15). Mammalian homologues of ESCRT-III components, the charged MVB proteins (CHMPs) have not been studied in detail, but CHMP1A and CHMP2A bind to mammalian VPS4 (19).
The equivalent, and otherwise unique, topology of enveloped viral budding and MVB vesicle budding suggests that the two processes might share a common mechanism (20). Catalytically inactive mutants of VPS4 induce the formation of class E compartment-like structures in mammalian cells (11, 21–23). Moreover, the activity of PTAP, YPDL, and PPXY L domains encoded by HIV-1 p6, equine infectious anemia virus (EIAV) p9, and murine leukemia virus (MLV) p12, respectively, is inhibited by VPS4 mutants (11, 21, 24). Conversely, most dominant negative forms of Tsg101 and small interfering RNA (siRNA)-mediated Tsg101 depletion specifically inhibit PTAP-type L-domain function (11, 12, 21, 25). Thus, current evidence suggest that all viral L domains function by overlapping VPS4-dependent mechanisms, but that PTAP-type L domains are unique in requiring ESCRT-I for viral budding.
At least two important questions are raised by these findings. First, how does ESCRT-I access the machinery (currently thought to be ESCRT-III) that mediates vesicle/viral budding and fission? Second, how do non-PTAP-type L domains obviate the requirement for ESCRT-I? To address these issues, we constructed a protein–protein interaction map of human class E VPS factors, and screened these proteins for interaction with non-PTAP-type L domains. We find that human class E VPS factors participate in a complex network of interactions that bridge ESCRT-I, -II, and -III, and other class E VPS factors. Moreover, a mammalian orthologue of the yeast VPS factor, Bro1 (AIP-1/ALIX) (26, 27), interacts with both the ESCRT-I and ESCRT-III complexes, binds to, and is a necessary cofactor for, a prototype YPDL-type L domain.
Materials and Methods
Mammalian Class E VPS Factor Expression Plasmids. Plasmids expressing Tsg101, VPS28, and VPS4 were described (10, 21). Human cDNAs encoding orthologues of the following yeast class E VPS genes: Hrs, HBP/STAM (AB012611), AIP-1/ALIX (AF349951), Eap45 (AF151903), Eap30 (AF156102), Eap20 (BC006282), CHMP1A (AF281063), CHMP1B (AF281064), CHMP2A (AF042384), CHMP2B (AF151842), CHMP3 (AF151907), CHMP4A (CAC14088), CHMP4B (AF161483), CHMP4C (BC014321), CHMP5 (AF132968), and CHMP6 (BC010108) (GenBank accession nos. in parentheses) were amplified by PCR from Jurkat, H9, or HeLa cell cDNA by using primers directed to the 5′and 3′ ends of the coding sequences. The primers incorporated EcoRI or XhoI sites that were used to insert the cDNAs into pGBKT7 (Clontech) and pVP16 (10) for yeast two-hybrid assays, and into pCR3.1/HA, pCR3.1/YFP, and pCAGGS/GST for mammalian expression of epitope, yellow fluorescent protein (YFP)-, or GST-tagged proteins.
Yeast Two-Hybrid Assays. Yeast Y190 cells were transformed with pGBKT7 and pVP16 derivatives and interactions measured by β-galactosidase reporter activity, as described (10).
Coprecipitation Assays. Pairs of GST- and YFP- or HA-epitope-tagged proteins were coexpressed in 293T cells. Forty-eight hours later, cells were lysed in buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/5 mM EDTA/5% glycerol/1% Triton X-100, and a protease inhibitor mixture), and clarified lysates were incubated with glutathione-Sepharose beads. The beads were washed three times with buffer containing 0.1% Triton X-100. Lysates and glutathione-bound proteins were analyzed by SDS/PAGE and Western blot with α-HA or α-GFP monoclonal antibodies.
Assays of L-Domain Function. A previously described complementation assay was used to measure L-domain function (10, 21). 293T cells were cotransfected with 300 ng of an HIV-1 proviral plasmid (NLδp6) that lacks the p6 L domain, and 200 ng of a plasmid expressing a truncated HIV-1 Gag protein (Gagδp6) fused to either the p6 domain of HIV-1 Gag, the p9 domain of EIAV, or a minimal Tsg101-binding motif (PEPTAPPEES). Alternatively, EIAV particles were generated by using a GFP vector and packaging system (28). In some experiments, 200 ng of a YFP fusion protein expression plasmid or 20 pmol of siRNA duplexes (Dharmacon, Lafayette, CO) were included in the transfection mixture. Virion production in supernatants was measured 48 h later by using an infection assay with P4/R5 (HeLa-CD4/CCR5 LTR-lacZ) indicator cells for HIV-1, or CRFK cells for EIAV, as described (10, 28). Alternatively, particle yield was determined by ultracentrifugation through 20% sucrose, and by Western blot analysis, as described (10). Western blot signal intensities were quantitated by using nih image.
Results
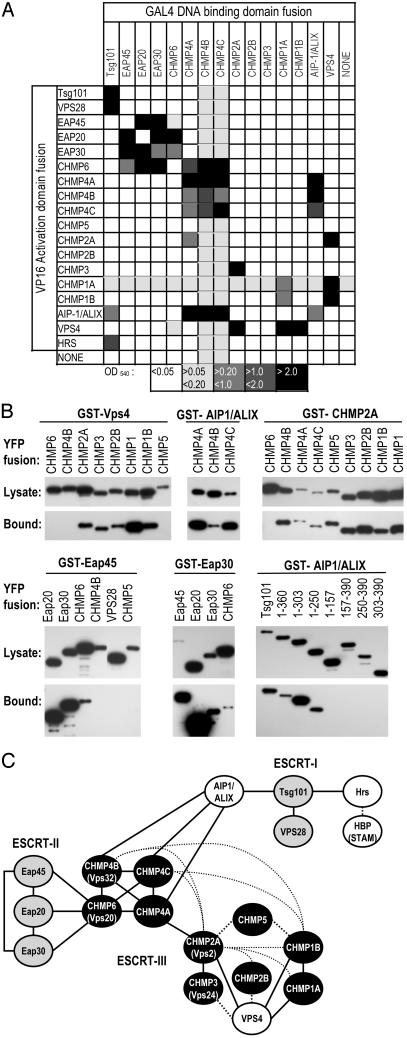
Protein–Protein Interactions Among Mammalian Class E VPS Factors. To better understand how ESCRT-I and other class E VPS proteins mediate L-domain function, we constructed a protein–protein interaction map incorporating each of the human orthologues of the yeast class E VPS factors. Interactions were scored initially by using yeast two-hybrid assays, and 24 positive interactions between heterologous proteins were identified (Fig. 1A, and Table 1 and Fig. 6, which are published as supporting information on the PNAS web site, www.pnas.org). Some class E VPS proteins also exhibited homomultimerizing activity. A GST fusion protein coprecipitation approach was used as a confirmatory assay to validate many of the interactions, and examples of this analysis are shown in Fig. 1B. Each of the YFP and GST fusion proteins was expressed in an intact form in mammalian cells, but low-level expression of several GSTCHMP proteins precluded confirmation of some of the interactions. Nonetheless, 17 of the 24 positive interactions in yeast were tested by using the coprecipitation assay, and of these interactions, all were confirmed. In addition, some additional interactions among the CHMP proteins that were not detected in the yeast two-hybrid assay were detected by coprecipitation (Fig. 1 B and C). It is possible that these interactions were bridged by endogenous human CHMP proteins, or that their failure to be detected in the yeast two-hybrid test was due to low affinity or interference by the fused GAL4 or VP16 domains. In general, however, there was good concordance between the yeast two-hybrid and coprecipitation assays. The results of yeast two-hybrid and coprecipitation assays are interpreted in the form of a protein–protein interaction map in Fig. 1C, and its features are described below.
Fig. 1.
Interactions between mammalian class E VPS proteins. (A) Matrix showing interactions between VPS proteins in the yeast two-hybrid assay. Shading indicates the level of β-galactosidase activity in optical density (OD) units as indicated by the key. The GAL4-fusions of VPS28, CHMP5, and Hrs were strong transcriptional activators in the absence of a coexpressed VP16-fusion protein and were excluded. A more detailed version of these data are given in Table 1. (B) Examples of coprecipitation assays to evaluate class E VPS protein interactions. GST fusion proteins were coexpressed with one of a series of YFP fusion proteins, as indicated. Samples of clarified cell lysate and glutathione-Sepharose bead-bound proteins were analyzed by Western blot with an α-GFP antibody. In each case, the full-length ORF was fused to YFP, except in the Lower Right, where a series of truncated Tsg101 proteins were also used. (C) Interpretation of the yeast two-hybrid assay and coprecipitation results in the form of a protein–protein interaction map incorporating components of ESCRT-I, ESCRT-II (gray shading), ESCRT-III (black shading), and additional human homologues of yeast class E genes. Positive interactions scored by yeast two-hybrid assay are indicated by solid lines. Interactions observed in the GST fusion coprecipitation assay, but not in the yeast two-hybrid assay, are indicated by dotted lines.
ESCRT-I. It has been previously shown that Tsg101 binds to VPS28 (15, 21). We found that Tsg101 also bound to two other class E VPS factors, AIP-1/ALIX and Hrs (Fig. 1 A), which are not components of any ESCRT complex, but are homologous to the yeast class E VPS factors, Bro1 and Vps27p, respectively (16, 26, 27, 29). That Tsg101 and Hrs bind to each other is consistent with a recent report (30) implicating Tsg101–Hrs interaction in receptor down-regulation. The interaction between Tsg101 and AIP-1/ALIX was mediated by the N-terminal UBC-like and proline-rich Tsg101 domains; residues 1–250 were both necessary and sufficient to bind AIP-1/ALIX (Fig. 1B). No other VPS protein that was tested bound to Tsg101, and VPS28 did not bind to any other VPS protein, except for Tsg101.
ESCRT-II. Each of the putative ESCRT-II components Eap45, Eap30, and Eap20 bound to each other, and to the ESCRT-III protein, CHMP6, but not to any other class E VPS protein. In particular, we found no evidence for direct interactions between ESCRT-I and ESCRT-II.
ESCRT-III. The majority of CHMP proteins that comprise ESCRTIII fell into two groups, which correlated with groupings based on sequence homology (see Table 1). One group contained members who bound to VPS4, including the previously described VPS4-binding proteins, CHMP1A and CHMP2A, whereas a second group contained proteins that bound promiscuously to each other and homomultimerized, but did not bind to VPS4. The VPS4-binding group included CHMP2A and CHMP3, whereas the second group included CHMP4B and CHMP6, whose orthologues (Vps2p, Vps24p, Vps32p/Snf7, and Vps20p, respectively) comprise two distinct subcomplexes of ESCRT-III in yeast, which are similarly distinguished by their ability to bind Vps4p (18). Thus, the functional anatomy of ESCRT-III appears to be conserved in yeast and humans. The two mammalian ESCRT-III subgroups were linked by multiple interactions between their respective members.
Each of the CHMP proteins that bound to CHMP6 (CHMP4A, -B, and -C) did not bind to ESCRT-I or -II, but instead bound to AIP-1/ALIX. Thus, it appeared that AIP-1/ALIX plays a pivotal role in linking ESCRT-I with ESCRT-III and (indirectly) ESCRT-II.
The only other reported interaction involving mammalian class E VPS factors is that of Hrs binding to HBP/STAM (31), which for technical reasons, we did not test. Overall, whereas the precise stoichiometry of the mammalian class E VPS complex remains to be defined, it is clear that the components can be linked by a complex series of interactions that are potentially important for viral budding.
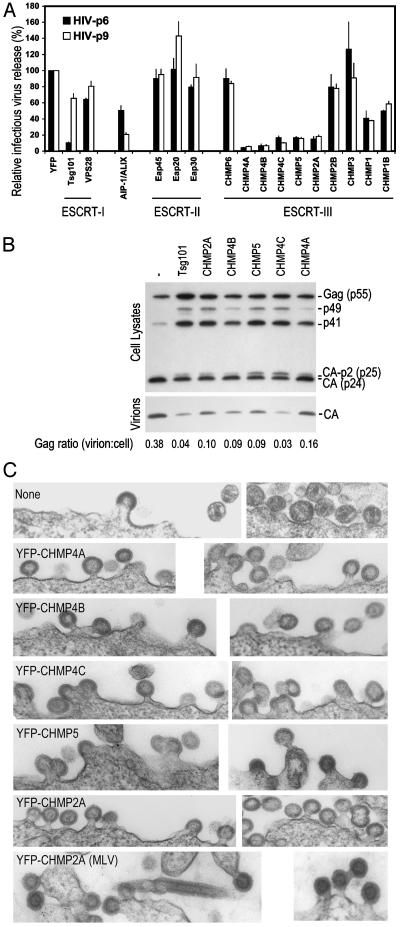
YFP-CHMP Fusion Protein Overexpression Blocks Retroviral L-Domain Function. To address whether the class E VPS factors that link ESCRT-I and VPS4 play a role in retroviral budding, in a general or L-domain-specific manner, we used a described strategy (19) whereby the factors were fused to a heterologous protein in an attempt to convert them to dominant-negative forms when overexpressed. To measure effects on virus release, we used a complementation assay in which HIV-1 budding is rendered dependent on the native PTAP-type, ESCRT-I-dependent p6 L-domain, or a heterologous, ESCRT-I-independent, YPDL-type L domain from EIAV (p9) (10, 21). As an example, YFP-Tsg101 overexpression inhibited p6-dependent release of infectious HIV-1 particles, whereas p9-dependent virus production was only marginally affected (Fig. 2A). When this strategy was applied to the other class E VPS factors, YFP-AIP-1/ALIX expression, surprisingly, preferentially inhibited p9-dependent virus production. In contrast, none of the ESCRT-II components had any inhibitory activity, nor did the ESCRT-II-binding component of ESCRT-III, CHMP6. However, several other ESCRT-III components, including each of those that bound AIP-1/ALIX, inhibited infectious HIV-1 release (Fig. 2 A). With the exception of CHMP2A, the VPS4-binding YFP-CHMP proteins were either inactive or only weakly inhibitory, suggesting that inhibition by YFP-CHMP proteins was not simply due to sequestration of VPS4. Notably, both p6 and p9 L-domain activity was equivalently blocked by each of the inhibitory YFP-CHMP fusion proteins. In addition, infectious MLV release showed a virtually identical pattern of susceptibility to inhibition by the various YFP-CHMP fusion proteins (data not shown). Inhibition of virus release by the YFP fusion proteins did not correlate with abundance, and the noninhibitory fusion proteins were expressed at a level that was equal to or higher than to the inhibitory YFP-CHMP proteins (Fig. 1B).
Fig. 2.
Generalized L-domain dysfunction induced by ESCRT-III perturbation. (A) Infectious HIV-1 release mediated by HIV-1 p6 or EIAV p9 L domains in the complementation assay in the presence of YFP class E VPS fusion protein expression. Infectious virion production, measured by β-galactosidase assay after infection of P4/R5 cells, is plotted as a percentage of that obtained in the presence of unfused YFP (371,390 relative light units for HIV-p6 and 79,938 relative light units for HIV-p9). (B) Gag processing and virion release defects induced by inhibitory YFP-CHMP fusion protein expression. (C) Electron microscopic images of late viral budding defects induced by YFP-CHMP fusion proteins. The top six pairs of images show HIV-1 budding in the absence or presence of the inhibitory YFP-CHMP fusion proteins, whereas the bottom pair of images shows MLV budding in the presence of YFP-CHMP2A.
To verify that YFP-CHMP proteins induced an L-domain defect, and not, for example, nonspecific cellular toxicity, we coexpressed the inhibitory YFP-CHMP fusion proteins with a replication-competent HIV-1 construct, and examined cell-associated Gag expression and extracellular particle formation. Defective HIV-1 L-domain function is accompanied by an aberrant and characteristic cell-associated Gag processing phenotype, in which the cleavage intermediates p49 and p25 are increased in abundance (4, 10, 11, 32). As can be seen in Fig. 2B, the YFP-CHMP proteins did not inhibit Gag expression, but did induce precisely these Gag processing and release defects to varying degrees. Electron microscopic examination of HIV-1 producing cells coexpressing YFP-CHMP fusion proteins (Fig. 2C) revealed a phenotype that was consistent with an L-domain defect. In place of the mature extracellular virions, numerous incompletely budded virions were observed (at least 5-fold more frequently) when inhibitory YFP-CHMP fusion proteins were coexpressed. Moreover, occasional chains of virions that were tethered to each other and to cells by membranous stalks were evident. An analysis of MLV producing cells coexpressing CHMP2A revealed a similar defect. Moreover, tubes, rather than spherical virions, were occasionally observed, which was similar to those generated by a PPXY-deleted MLV mutant (8). Overall, the data in Fig. 2 suggest that the integrity of complexes that act downstream of ESCRT-I, ESCRT-III in particular, is required in a general manner for retroviral L-domain function.
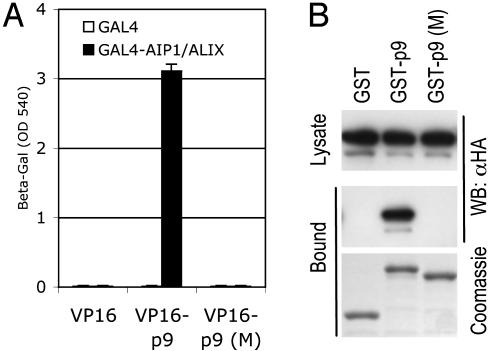
The YPDL-Type EIAV p9 L Domain Binds AIP-1/ALIX. Based on the findings that ESCRT-III and VPS4 are apparently required by non-PTAP-type L domains (refs. 11 and 21 and Fig. 2), and that ESCRT-I is dispensable (11, 21), we reasoned that they might simply bypass ESCRT-I and recruit class E VPS factors that act downstream. We screened the class E VPS factors shown in Fig. 1 for interactions with PPXY and YPDL L domains by using the yeast two-hybrid assay. Whereas none of the proteins tested bound to the PPXY L domains encoded by Rous sarcoma virus (p2) or MLV (p12) (data not shown), we detected a strong and specific interaction between EIAV p9 and AIP-1/ALIX (Fig. 3A). This interaction was reproduced in coprecipitation assays by using GST-p9 as bait, but not by using GST (Fig. 3B) or GST-p12 (data not shown). The YPDL motif in EIAV p9 is critical for L-domain activity (5), and substitution of these residues completely abolished the ability of p9 to bind AIP-1/ALIX in both assays (Fig. 3 A and B), suggesting that the interaction could be functionally important.
Fig. 3.
YPDL-dependent EIAV p9 binding to AIP-1/ALIX. (A) Yeast two-hybrid analysis. β-galactosidase reporter levels in yeast expressing GAL4 or GAL4-AIP-1/ALIX and VP16 or VP16-EIAV p9 were plotted. The p9 (M) mutant contained a four-residue substitution (YPDL to AAAA). (B) GST fusion proteins were expressed in 293T cells along with HA-tagged AIP-1/ALIX. Samples of clarified cell lysate and glutathione-bound proteins were analyzed by Western blot. Equivalent loading of the GST fusion proteins was verified by Coomassie blue staining of the glutathione-bound fraction.
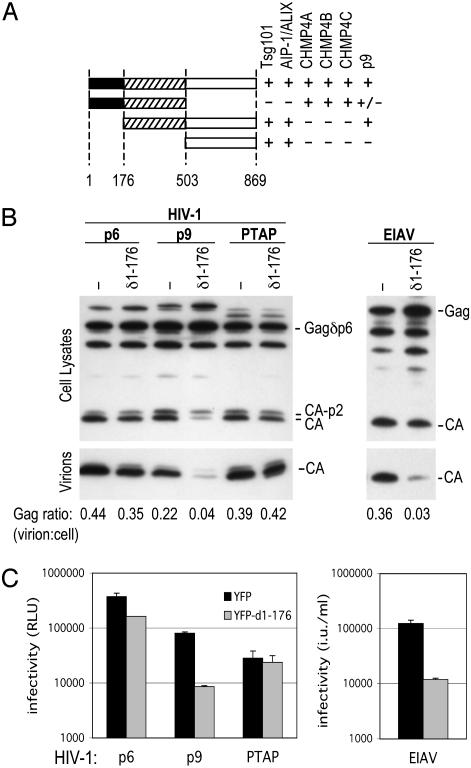
Specific Inhibition of YPDL-Type L-Domain Function by Dominant-Negative AIP-1/ALIX and siRNAs. To address whether AIP-1/ALIX was required for EIAV L-domain function, we attempted to derive more potent and specific dominant-negative forms of the protein. First, we mapped the domains of AIP-1/ALIX that mediate interaction with Tsg101 and with ESCRT-III by using yeast two-hybrid assays. As shown in Fig. 4A, this analysis revealed that N-terminal and central domains were necessary and sufficient for interaction with CHMP4A, -B, and -C, whereas a C-terminal domain was sufficient for interaction with Tsg101, and for AIP-1/ALIX multimerization. Central and C-terminal domains were necessary and sufficient for binding to EIAV p9. Thus, in principle, AIP-1/ALIX could simultaneously bind to ESCRT-I, and to ESCRT-III, and act as a bridging factor to connect either or both the HIV-1 and EIAV L domains to downstream factors required for virus budding. Of the truncated forms of AIP-1/ALIX, a protein lacking the N-terminal domain (δ1–176), which did not interact with ESCRT-III (Fig. 4A), was a potent and selective inhibitor of p9-mediated HIV-1 budding. As shown in Fig. 4 B and C, expression of this protein inhibited EIAV p9-dependent HIV-1 budding by 5- or 10-fold, as measured by Western blot or infectivity assays, respectively. In contrast, a minimal Tsg101-binding L domain from HIV-1 (PEPTAPPEES), was completely insensitive to δ1–176AIP-1/ALIX. Interestingly, the full-length HIV-1 p6 L-domain function was marginally inhibited by δ1–176AIP-1/ALIX. We and others have recently shown that the HIV-1 p6 protein contains a bipartite L domain (20, 38), and since PTAP function was resistant to δ1–176AIP-1/ALIX, we speculate that it inhibits the second PTAP-independent function encoded within p6. As was the case for p9-dependent HIV-1 budding, the egress of authentic EIAV virions was inhibited ≈10-fold by δ1–176AIP-1/ALIX, as measured by Western blot or infectivity assays (Fig. 4 B and C).
Fig. 4.
A truncated form of AIP-1/ALIX that lacks ESCRT-III-binding activity is a specific inhibitor of a YPDL-type L domain. (A) AIP-1/ALIX domain organization. The indicated truncated versions of AIP-1/ALIX were tested for interaction with Tsg101, AIP-1/ALIX, CHMP4A, -B, -C, and EIAV p9 in the yeast two-hybrid assay. The ± ascribed to p9 interaction with AIP-1/ALIX residues 1–503 indicates a weak positive (β-galactosidase reporter levels ≈10% of those obtained by using the full-length or 503–869 residue fragment). (B) Western blot analysis of cell lysates and pelleted virions after transfection of 293T cells with a p6-deleted HIV-1 proviral plasmid, along with complementing Gagδp6-p6, Gagδp6-p9, and Gagδp6-PTAP expression vectors (Left). Alternatively, EIAV vector plasmids were transfected (Right). YFP or YFP-δ1–176 AIP-1/ALIX was coexpressed, as indicated. (C) Results are the same as for B, except that HIV-1 in culture supernatants was quantified by infection of P4/R5 indicator cells and β-galactosidase activity measurement, given in relative light units (RLU), or infectious EIAV virions were measured by titration on CRFK cells given in infectious units (iu)/ml of supernatant.
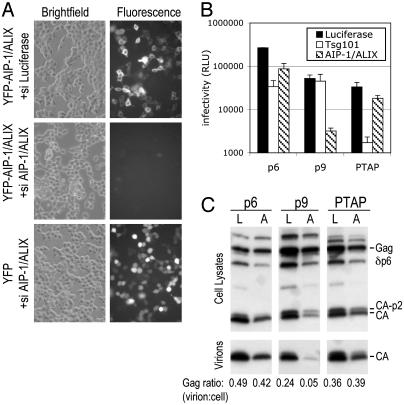
We also used the complementation assay to test whether EIAV p9- or PTAP-dependent HIV-1 budding could be inhibited by siRNA-mediated depletion of AIP-1/ALIX. As shown in Fig. 5A, AIP-1/ALIX-targeted siRNAs effectively suppressed YFP-AIP-1/ALIX, but not YFP expression. PTAP-dependent infectious HIV-1 production was only marginally affected (≈2-fold) by AIP-1/ALIX depletion, whereas p9-dependent virion egress was inhibited by 17-fold (Fig. 5B). Conversely, PTAP-mediated budding was profoundly inhibited (20-fold) by Tsg101 depletion by using described siRNA duplexes (11), whereas p9-mediated budding was unaffected. Thus, these data suggest that even though Tsg101 and AIP-1/ALIX interact, they function largely independent of each other to mediate the function of their cognate viral L domains. As was the case with δ1–176AIP-1/ALIX, HIV-1 p6 activity was marginally more sensitive to AIP-1/ALIX siRNA than was PTAP. Moreover, p6 was somewhat less sensitive than was PTAP to Tsg101 siRNAs (Fig. 5B). These findings are consistent with the notion of a bipartite late domain in HIV-1 p6.
Fig. 5.
Effects of siRNA-mediated AIP-1/ALIX depletion on YPDL and PTAP-type L-domain function. (A) Validation of AIP-1/ALIX siRNAs. The 293T cells transfected with YFP or YFP-AIP-1/ALIX expression plasmids in the presence of siRNAs directed against luciferase (control) or AIP-1/ALIX, as indicated. Bright-field and fluorescent images of the same field, acquired 24 h after transfection, are shown. (B) Inhibition of virus release by Tsg101- and AIP-1/ALIX-specific siRNAs. Gagδp6-p6-, Gagδpb-p9-, and Gagδpd-PTAP-complemented HIV-1 was generated as in Fig. 4, but, in this case, luciferase (control)-, Tsg101-, or AIP-1/ALIX-specific siRNAs were cotransfected. Virion production was measured by using P4/R5 cells as in Fig. 4. (C) Western analysis of virion HIV-1 formation mediated by p6, p9, or PTAP in the presence of siRNAs. Lanes L, luciferase control siRNAs; lanes A, AIP-1/ALIX siRNAs.
The interpretation of these results was slightly complicated by the fact that AIP-1/ALIX depletion by using siRNA likely had deleterious effects on cell viability, because a Western blot analysis showed slightly reduced Gag expression at later time points (Fig. 5C). Nonetheless, whereas p9-dependent HIV-1 budding was dramatically inhibited under conditions of AIP-1/ALIX depletion, p6- or PTAP-dependent budding was clearly less affected (Fig. 5C).
Discussion
This study demonstrates that a complex series of protein–protein interactions involving human class E VPS factors occurs, and identifies AIP-1/ALIX as a cofactor for the YPDL-type L-domain encoded by EIAV. Thus, there is a remarkable functional convergence between two nonhomologous sequence motifs, YPDL and PTAP, that use two distinct entry points into the interaction network to exploit VPS factors for viral budding.
Previously, the AP50 subunit of the AP2 complex was proposed to bind the EIAV L domain (33). However, a definite role for AP2 in EIAV budding is yet to be established. It might be due to the fact that interactions between EIAV p9, AP2, and AIP-1/ALIX are important. Indeed, the ability of the EIAV YPDL motif to bind both AP2 and AIP-1/ALIX suggests that AP2-binding YXXL/I sorting motifs in cellular proteins might share the ability to bind AIP-1/ALIX. As such, these motifs might have a dual function in recruiting distinct cellular functions.
EIAV p9 L-domain activity is resistant to dominant-negative forms of Tsg101 (21) and siRNA-mediated Tsg101 depletion (Fig. 5). However, both p6 and p9, as well as MLV PPXY L domains, are similarly blocked by dominant-negative VPS4 (11, 21) and CHMP fusion protein overexpression (Fig. 2). Intuitively, the protein interaction map in Fig. 1 suggests that the EIAV YPDL-type L domain bypasses the requirement for Tsg101 simply by recruiting class E VPS factors at one step downstream in a linear pathway that proceeds from ESCRT-I to ESCRT-III and VPS4 through AIP-1/ALIX. A prediction of this hypothesis is that ESCRT-I-dependent L-domain function should also require AIP-1/ALIX. However, PTAP L-domain activity was dramatically less sensitive than was YPDL L-domain activity to dominant-negative AIP-1/ALIX, and to siRNA-mediated AIP-1/ALIX depletion (Figs. 4 and 5). Moreover, a Tsg101 fragment lacking C-terminal sequences required to bind VPS28 is sufficient to bind AIP-1/ALIX (Fig. 1), but is not able to mediate HIV-1 budding (10, 21). These data suggest that alternative mechanisms of ESCRT-III/VPS4 recruitment by ESCRT-I exist. This confirmation would require additional bridging factors or interactions that were not detected in this analysis. A further distinction between HIV-1 and EIAV L domains is that only the former requires an active proteasome degradation pathway to mediate budding (34–37). How this fact relates to the exploitation of class E VPS factors is unclear at present.
Based on findings in yeast that ESCRT-II overexpression partly suppresses sorting defects that result from ESCRT-I ablation, and that ESCRT-II, but not ESCRT-I, binds to ESCRT-III (17, 18), current models suggest that ESCRT-I, ESCRT-II, and ESCRT-III act sequentially to mediate MVB cargo sorting and vesicle invagination. Whereas the interaction map of human class E VPS proteins shown in Fig. 1 is consistent with previously documented class E VPS factor interactions in yeast, it is more complex, and other aspects of the mammalian interaction network suggest alternative ESCRT-II-independent pathways. Alternatively, ESCRT-I and ESCRT-II could act sequentially in the pathway, but without direct physical contact. However, it is conceivable that the map shown in Fig. 1 underestimates the total complexity of interactions involving class E VPS factors.
Whereas each of the class E VPS genes in yeast are required for budding of vesicles into MVBs (16), some of the mammalian factors (e.g., ESCRT-I and AIP-1/ALIX) appear to be required for viral budding, only in the context of specific viral L-domain types. Thus, only a subset of class E VPS factors, particularly ESCRT-III, seems to be generally required for viral budding. Nonetheless, it is increasingly evident that the budding of MVB vesicles and many enveloped viruses, is functionally analogous (20). As such, further studies of how viral L domains parasitize the class E VPS machinery is likely to illuminate the mechanism by which this cellular process occurs.
Supplementary Material
Acknowledgments
We thank Eleana Sphicas of the Rockefeller Bioimaging Resource Center for electron microscopy, Trinity Zang for technical assistance, and Hua Chen, Kyriacos Mitrophanous, and Stephen Goff for reagents. This work was supported by National Institutes of Health Grants RO1AI52774 and RO1AI50111 and American Foundation for AIDS Research Grant 02865-31 (to P.D.B.). J.M.-S. is the recipient of a postdoctoral fellowship from the Spanish Ministerio de Educacion, Cultura y Deporte. P.D.B. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ESCRT, endosomal sorting complex required for transport; EIAV, equine infectious anemia virus; L domain, late-budding domain; MLV, murine leukemia virus; VPS, vacuolar protein sorting; AIP-1/ALIX, a mammalian orthologue of the yeast class E VPS factor, Bro1; MVB, multivesicular body; CHMP, charged MVB protein; YFP, yellow fluorescent protein; siRNA, small interfering RNA.
References
- 1.Wills, J. W., Cameron, C. E., Wilson, C. B., Xiang, Y., Bennett, R. P. & Leis, J. (1994) J. Virol. 68, 6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yasuda, J. & Hunter, E. (1998) J. Virol. 72, 4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harty, R. N., Brown, M. E., Wang, G., Huibregtse, J. & Hayes, F. P. (2000) Proc. Natl. Acad. Sci. USA 97, 13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlinger, H. G., Dorfman, T., Sodroski, J. G. & Haseltine, W. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puffer, B. A., Parent, L. J., Wills, J. W. & Montelaro, R. C. (1997) J. Virol. 71, 6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven, R. C., Harty, R. N., Paragas, J., Palese, P. & Wills, J. W. (1999) J. Virol. 73, 3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jayakar, H. R., Murti, K. G. & Whitt, M. A. (2000) J. Virol. 74, 9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan, B., Campbell, S., Bacharach, E., Rein, A. & Goff, S. P. (2000) J. Virol. 74, 7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VerPlank, L., Bouamr, F., LaGrassa, T. J., Agresta, B., Kikonyogo, A., Leis, J. & Carter, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2001) Nat. Med. 7, 1313-1319. [DOI] [PubMed] [Google Scholar]
- 11.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55-65. [DOI] [PubMed] [Google Scholar]
- 12.Demirov, D. G., Ono, A., Orenstein, J. M. & Freed, E. O. (2002) Proc. Natl. Acad. Sci. USA 99, 955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzmann, D. J., Babst, M. & Emr, S. D. (2001) Cell 106, 145-155. [DOI] [PubMed] [Google Scholar]
- 14.Babst, M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, 248-258. [DOI] [PubMed] [Google Scholar]
- 15.Bishop, N. & Woodman, P. (2001) J. Biol. Chem. 276, 11735-11742. [DOI] [PubMed] [Google Scholar]
- 16.Katzmann, D. J., Odorizzi, G. & Emr, S. D. (2002) Nat. Rev. Mol. Cell Biol. 3, 893-905. [DOI] [PubMed] [Google Scholar]
- 17.Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B. & Emr, S. D. (2002) Dev. Cell 3, 283-289. [DOI] [PubMed] [Google Scholar]
- 18.Babst, M., Katzmann, D. J., Estepa-Sabal, E. J., Meerloo, T. & Emr, S. D. (2002) Dev. Cell 3, 271-282. [DOI] [PubMed] [Google Scholar]
- 19.Howard, T. L., Stauffer, D. R., Degnin, C. R. & Hollenberg, S. M. (2001) J. Cell Sci. 114, 2395-2404. [DOI] [PubMed] [Google Scholar]
- 20.Pornillos, O., Garrus, J. E. & Sundquist, W. I. (2002) Trends Cell Biol. 12, 569-579. [DOI] [PubMed] [Google Scholar]
- 21.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2003) J. Virol. 77, 4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop, N. & Woodman, P. (2000) Mol. Biol. Cell 11, 227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babst, M., Wendland, B., Estepa, E. J. & Emr, S. D. (1998) EMBO J. 17, 2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanzi, G. O., Piefer, A. J. & Bates, P. (2003) J. Virol. 77, 8440-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goila-Gaur, R., Demirov, D. G., Orenstein, J. M., Ono, A. & Freed, E. O. (2003) J. Virol. 77, 6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odorizzi, G., Katzmann, D. J., Babst, M., Audhya, A. & Emr, S. D. (2003) J. Cell Sci. 116, 1893-1903. [DOI] [PubMed] [Google Scholar]
- 27.Vincent, O., Rainbow, L., Tilburn, J., Arst, H. N., Jr., & Penalva, M. A. (2003) Mol. Cell. Biol. 23, 1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitrophanous, K., Yoon, S., Rohll, J., Patil, D., Wilkes, F., Kim, V., Kingsman, S., Kingsman, A. & Mazarakis, N. (1999) Gene Ther. 6, 1808-1818. [DOI] [PubMed] [Google Scholar]
- 29.Komada, M., Masaki, R., Yamamoto, A. & Kitamura, N. (1997) J. Biol. Chem. 272, 20538-20544. [DOI] [PubMed] [Google Scholar]
- 30.Lu, Q., Hope, L. W., Brasch, M., Reinhard, C. & Cohen, S. N. (2003) Proc. Natl. Acad. Sci. USA 100, 7626-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asao, H., Sasaki, Y., Arita, T., Tanaka, N., Endo, K., Kasai, H., Takeshita, T., Endo, Y., Fujita, T. & Sugamura, K. (1997) J. Biol. Chem. 272, 32785-32791. [DOI] [PubMed] [Google Scholar]
- 32.Huang, M., Orenstein, J. M., Martin, M. A. & Freed, E. O. (1995) J. Virol. 69, 6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puffer, B. A., Watkins, S. C. & Montelaro, R. C. (1998) J. Virol. 72, 10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., Coren, L. V., Sowder, R. C., II, Adams, J., Nagashima, K. & Schubert, U. (2002) J. Virol. 76, 3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patnaik, A., Chau, V., Li, F., Montelaro, R. C. & Wills, J. W. (2002) J. Virol. 76, 2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strack, B., Calistri, A., Accola, M. A., Palu, G. & Gottlinger, H. G. (2000) Proc. Natl. Acad. Sci. USA 97, 13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schubert, U., Ott, D. E., Chertova, E. N., Welker, R., Tessmer, U., Princiotta, M. F., Bennink, J. R., Krausslich, H. G. & Yewdell, J. W. (2000) Proc. Natl. Acad. Sci. USA 97, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Serrano, J. & Bieniasz, P. D. (2003) J. Virol., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.