Abstract
Tuberculosis remains a leading cause of death worldwide, despite the availability of effective chemotherapy and a vaccine. Bacillus Calmette–Guérin (BCG), the tuberculosis vaccine, is an attenuated mutant of Mycobacterium bovis that was isolated after serial subcultures, yet the functional basis for this attenuation has never been elucidated. A single region (RD1), which is absent in all BCG substrains, was deleted from virulent M. bovis and Mycobacterium tuberculosis strains, and the resulting ΔRD1 mutants were significantly attenuated for virulence in both immunocompromised and immunocompetent mice. The M. tuberculosis ΔRD1 mutants were also shown to protect mice against aerosol challenge, in a similar manner to BCG. Interestingly, the ΔRD1 mutants failed to cause cytolysis of pneumocytes, a phenotype that had been previously used to distinguish virulent M. tuberculosis from BCG. A specific transposon mutation, which disrupts the Rv3874 Rv3875 (cfp-10 esat-6) operon of RD1, also caused loss of the cytolytic phenotype in both pneumocytes and macrophages. This mutation resulted in the attenuation of virulence in mice, as the result of reduced tissue invasiveness. Moreover, specific deletion of each transcriptional unit of RD1 revealed that three independent transcriptional units are required for virulence, two of which are involved in the secretion of ESAT-6 (6-kDa early secretory antigenic target). We conclude that the primary attenuating mechanism of bacillus Calmette–Guérin is the loss of cytolytic activity mediated by secreted ESAT-6, which results in reduced tissue invasiveness.
Bacillus Calmette–Guérin (BCG) was first isolated from Mycobacterium bovis after serial subculturing in ox bile medium (1, 2), when Drs. Calmette and Guérin set out to test the hypothesis that a bovine tubercle bacillus could transmit pulmonary tuberculosis after oral administration (1, 3, 4). However, unexpectedly after the 39th passage, the strain was unable to kill experimental animals (1, 2), and showed no reversion to virulence even after the authors had performed over 200 passages (3), which is consistent with the attenuating mutation being a deletion mutation. In proceeding studies, BCG was determined to be able to protect animals receiving a lethal challenge of virulent tubercle bacilli (5), and in 1921 was first used as an anti-tuberculous vaccine (6). Presently, an estimated 3 billion doses have been used to vaccinate the human population against tuberculosis, yet the mechanism that causes the attenuation of BCG remains unknown.
Mahairas et al. (6) first compared the genomic sequences of BCG and M. bovis, by using subtractive hybridization, and found that there were three regions of difference (designated RD1, RD2, and RD3) present in the genome of M. bovis, but missing in BCG. Behr et al. (7), and others (8), later identified 16 large deletions, including RD1-RD3, which were present in the Mycobacterium tuberculosis genome but absent in BCG. Eleven of these 16 deletions were unique to M. bovis whereas the remaining 5 deletions were unique to BCG. One of these 5 deletions, designated RD1 (9,454 bp), was absent from all of the BCG substrains currently used as tuberculosis vaccines worldwide, and it was concluded that the deletion of RD1 occurred very early during the development of BCG, probably before 1921 (9). Therefore, it is reasonable to hypothesize that RD1 was the primary attenuating mutation, which resulted in the generation of BCG from M. bovis. An attempt to restore virulence to BCG with an RD1-complementing clone was only partially successful (10) although the deletion of the esat-6 gene from M. bovis resulted in an attenuated phenotype in guinea pigs (11). A recent study has also determined that deletion of RD1 from M. tuberculosis results in attenuation of virulence (12), but there has been no definitive study looking at the mechanism of attenuation or at the function of each of the genes that comprise RD1.
Materials and Methods
Mice. BALB/c and C57BL/6 mice were purchased from Charles River Breeding Laboratories. Severe combined immunodeficient (SCID) (BALB/c background) mice were purchased from the National Cancer Institute. Aerosol infections at a low dose of 50 colony-forming units (cfu) per mouse and high dose of 500 cfu per mouse were carried out as described in ref. 13. I.v. injections in the tail vein (at a dose of 2 × 106 cfu) and plating were carried out as described in ref. 14. At least 10 mice were used for survival studies, and at least 3 mice per time point for all other studies. For pathology, half of the organ was plated and half was fixed in either 10% formalin (for light microscopy) or 2.5% glutaraldehyde, in 0.1 M sodium cacodylate buffer (for electron microscopy).
Construction of the RD1 Mutants. The unmarked deletion mutant of M. tuberculosis H37Rv, mc24002, was generated by transformation using a sacB counter selection, as described in ref. 15. Specifically, the plasmid pJH506 was created by cloning UFS (upstream flanking sequence) and DFS (downstream flanking sequence; see Fig. 1A) into pJH12 [a pMV261-derived Mycobacterial shuttle plasmid in which UFS and DFS flanked a green fluorescent protein gene (GFPuv, CLONTECH) controlled by the Mycobacterium leprae promoter of 18 kDa]. pJH508 was created by cloning UFS-DFS-GFPuv (from pJH506) into pyUB657. After transformation into mycobacteria, selection was carried out using hygromycin, followed by 3% sucrose. Southern analysis (16), using UFS- or DFS-specific probes, was performed to confirm the RD1 deletion. The mycobacteriophage-based method of specialized transduction, which utilizes conditionally replicating shuttle phasmids, was also used to construct RD1 mutants, by using UFS and DFS as above, along with the individual gene constructs (Rv3871, Rv3872/3, Rv3874/5, and Rv3876/7), as described in ref. 17. Again, Southern analysis was used to confirm deletion.
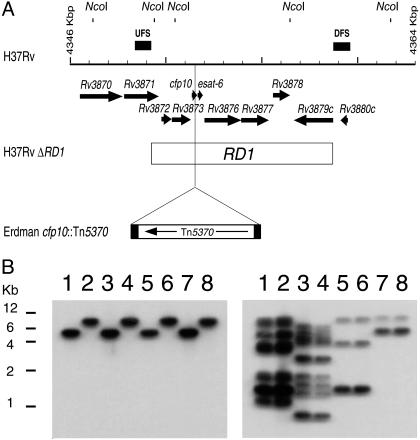
Fig. 1.
Generation of mutants in the RD1 region of M. tuberculosis and M. bovis. (A) Schematic of M. tuberculosis H37Rv RD1 region showing predicted NcoI sites. Arrows at the top represent the genes in this region. UFSs and DFSs used to generate the knockout are indicated as filled bars above the grid line. Each increment in the grid line represents 1 kbp. The RD1 sequence deleted from M. bovis BCG is represented by an open bar spanning from Rv3871 to Rv3879c. The site of the insertion of transposon Tn5370 is also indicated. (B) Southern analysis of the NcoI-digested genomic DNA isolated from the wild type and the ΔRD1 mutants generated by using specialized transduction in M. tuberculosis and M. bovis. Lane 1, M. tuberculosis H37Rv; lane 2, M. tuberculosis H37Rv ΔRD1; lane 3, M. tuberculosis Erdman; lane 4, M. tuberculosis Erdman ΔRD1; lane 5, M. tuberculosis CDC1551; lane 6, M. tuberculosis CDC1551 ΔRD1; lane 7, M. bovis Ravenel; and lane 8, M. bovis Ravenel ΔRD1. The probe used in the Southern analysis was either DFS (Left), demonstrating the deletion of RD1, or IS6110-specific (Right). The IS6110 probe is used to characterize the four different strains (described in ref. 31).
Construction and Screening of the M. tuberculosis Erdman Transposon-Generated Mutant Library. A transposon library of M. tuberculosis Erdman (of ≈6,460 clones) was constructed as described (18) by using the hygromycin-resistant Tn5370 transposon. Screening of the library was carried out by infection of A549 cells in 96-well plates (without the addition of gentamicin), which were then screened for reduced lactate dehydrogenase (LDH) release (described in the infection experiment methodology). Identification of the transposon insertion sites was carried out by sequencing, as described in ref. 18.
Complementation of Mutants. Complementation analyses were performed with the cosmid 2F9, which contains the entire RD1 region (Rv3860–Rv3885c) in the integration proficient vector pYUB412 (constructed by F. Bange, Medical School, Hannover, Germany, identified and provided by S. T. Cole Institut Pasteur, Paris). After transformation of the mutant strains with the constructs, as described in ref. 19, Southern analyses was carried out to determine whether integration was successful.
Infection Experiments. A549 cells [an alveolar epithelial cell line, American type Culture Collection (ATCC)] were infected at a multiplicity of infection of 10:1, as described in detail in ref. 20. Briefly, cells were seeded at 5 × 105 cells/ml, and, after infection with freshly sonicated bacilli for 3 h, any extracellular bacilli were washed off and gentamicin (Sigma) was added to the medium. LDH release was quantified at specified time points by using the cytotoxicity detection kit (Roche Diagnostics), according to the manufacturer's instructions. The maximum and background controls used were 0.1% Triton X-100-treated cells (maximum) and uninfected cells (background). Values were calculated by using the following equation: % LDH release = (sample-background)/(maximum-background). The levels of both apoptosis and necrosis were determined by elucidating the concentration of histone-associated DNA fragments (in the cell lysate and supernatant, respectively), which were quantified by using the cell death detection ELISA (Roche Diagnostics), again according to the manufacturer's instructions. The necrosis positive control was 0.1% Triton X-100, whereas 10 μM campothecin (Sigma) was added for the apoptosis positive control. Five mM exogenous glycine was added to the infected monolayers for the glycine protection assay, as described in ref. 21. Bone marrow-derived macrophages were obtained from the femurs of BALB/c mice and were seeded at 2 × 105 cells/ml, after lysis of red blood cells with 3% acetic acid/PBS and the removal of nonadherent cells.
Artificial Membrane Studies. Artificial planal bi-layers were constructed at room temperature by using the lipid diphytanoylphosphatidylcholine and the Mueller brush technique, as described in ref. 22. Voltages of 20–50 mV were applied, and the conductance across the bilayer was measured on the addition of 1 μg of purified protein.
Preparation of Culture Filtrates and Whole Cell Extracts. The culture filtrate and whole cell lysates of the bacterial strains were isolated as described in ref. 16. Western blot analyses were performed by using anti-ESAT-6 mouse monoclonal primary antibody HYB76-8 at a 1:100 dilution and a sheep anti-mouse Ig secondary antibody (Amersham Pharmacia) at 1:10,000.
Results
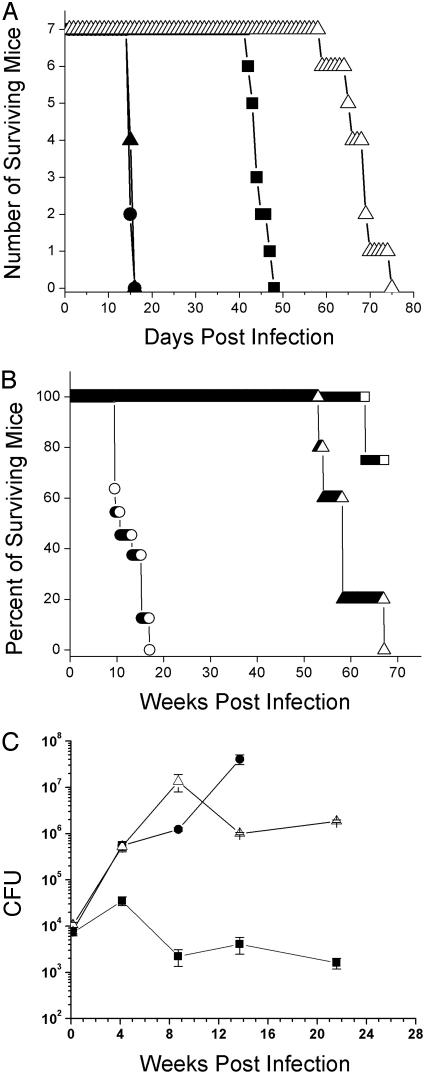
RD1 Deletions of M. bovis and M. tuberculosis Are Attenuated for Virulence in Mice. To test whether RD1 (Fig. 1 A) was essential for virulence in M. bovis and M. tuberculosis, RD1 was deleted from the M. tuberculosis strains H37Rv, Erdman, and CDC1551, and from M. bovis Ravenel (Fig. 1B). The deletion of RD1 resulted in the attenuation of virulence of M. tuberculosis H37Rv ΔRD1 in a SCID mouse model of infection, which was restored on complementation with RD1 (Fig. 2A). Similar attenuating phenotypes were observed for the RD1 deletion mutants made in M. tuberculosis Erdman and CDC1551, and in M. bovis Ravenel (data not shown). The M. tuberculosis H37Rv ΔRD1 mutant was also highly attenuated in immunocompetent BALB/c mice (Fig. 2B), but the pattern of in vivo growth remained similar to wild-type, in contrast to BCG (Fig. 2C).
Fig. 2.
Deletion of RD1 in M. tuberculosis confers an attenuation of virulence in vivo. (A) Survival experiment using SCID mice i.v. infected with 2 × 106 cfu per mouse. Infection was carried out as described in Materials and Methods. Strains used were M. tuberculosis H37Rv (•), M. tuberculosis ΔRD1 (▪), M. tuberculosis ΔRD1 (pYUB412::Rv3860–Rv3885c)(▴), and M. bovis BCG (▵). (B) Survival experiment with BALB/c mice i.v. infected with M. tuberculosis H37Rv (○), M. tuberculosis ΔRD1 (▵), and M. bovis BCG (□), at a dose of 2 × 106 cfu (C) Growth kinetics in the lungs of BALB/c mice i.v. infected with M. bovis BCG (▪), M. tuberculosis H37Rv (•), and M. tuberculosis H37Rv ΔRD1 (▵). The infecting dose per mouse was 2 × 106 cfu. Data represent the mean of cfus from three mice per time point.
Protection of BCG and ΔRD1 Against Challenge with Virulent M. tuberculosis. One of the hallmark characteristics of BCG is its ability to provide protection against aerosolized challenge with virulent M. tuberculosis. At 3 and 8 months after vaccination with either BCG or ΔRD1 mutant, mice were challenged with 50–200 cfus of the acriflavin-resistant strain of M. tuberculosis Erdman, by the aerosol route. Twenty-eight days after the challenge, the mice were killed, and the bacterial burden in the lungs and spleens was determined (see Table 1). In mice challenged after 8 months, both the BCG-vaccinated and M. tuberculosis ΔRD1-vaccinated mice exhibited >1 log protection in the lung, with similar cfu values. Protection was seen to a similar extent in mice challenged 3 months after vaccination (see Table 1). The M. tuberculosis ΔRD1 mutant also protected against hematogenous spread at both 3- and 8-month challenges (see Table 1). Thus, M. tuberculosis ΔRD1 exhibits long-term immunogenicity similar to BCG.
Table 1. Vaccination with M. tuberculosis H37Rv ΔRD1 protects as well as M. bovis BCG (day 28 postvaccination).
| 3-month challenge (log10 cfu)
|
8-month challenge (log10 cfu)
|
|||
|---|---|---|---|---|
| Vaccination strain | Lung | Spleen | Lung | Spleen |
| 4.77 ± 0.06 | 3.57 ± 0.21 | 6.61 ± 0.13 | 5.26 ± 0.11 | |
| BCG | 3.96 ± 0.20** | 2.18 ± 0.18*** | 5.07 ± 0.10*** | 3.85 ± 0.17** |
| H37RvΔRD1 | 3.97 ± 0.39** | 2.12 ± 0.12*** | 5.11 ± 0.14*** | 4.00 ± 0.33** |
, P < 0.01
, P < 0.001
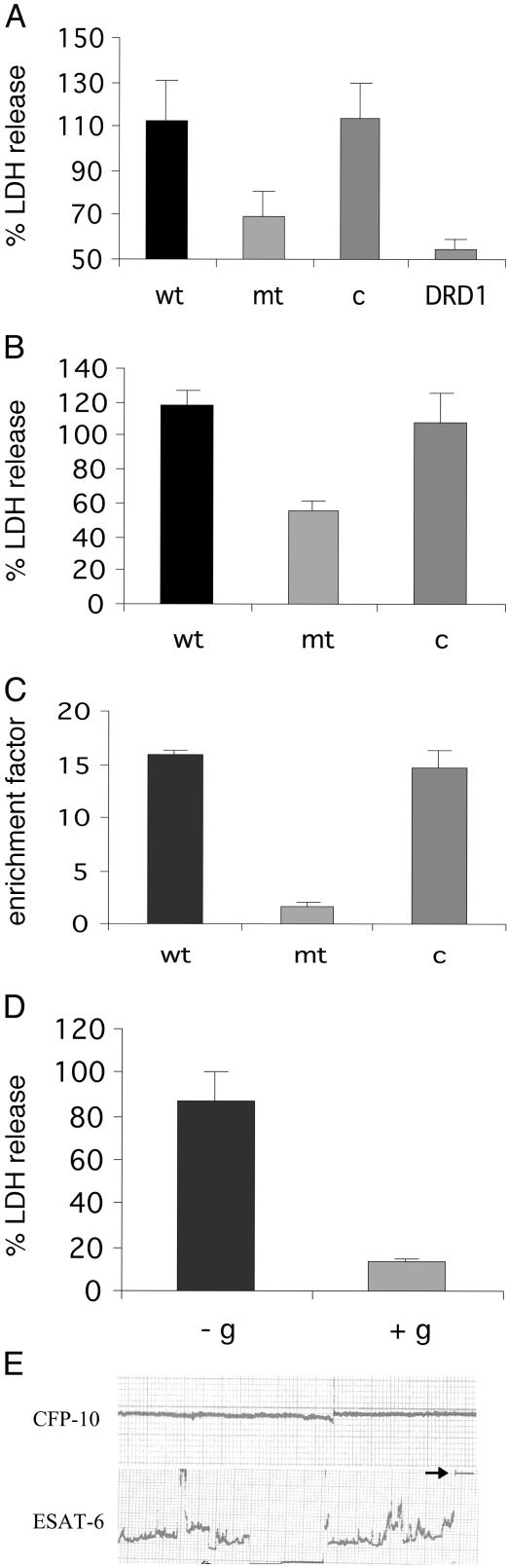
The cfp-10 esat-6 Operon of M. tuberculosis Is Required for Host Cell Lysis. Because virulent M. tuberculosis, but not BCG, was previously reported to induced lysis of alveolar epithelial cells, we screened for mutants that were defective in the ability to induce cytolysis of lung epithelial cells (20, 23). Approximately 700 clones of a transposon-generated library of M. tuberculosis Erdman mutants were screened for reduced cytolysis of lung epithelial cells, by measuring LDH release. This screen resulted in the identification of several mutants, one of which, mc24513, mapped to the cfp-10 (Rv3874) gene, which is contained within the RD1 region (Fig. 1). The mutant phenotype of reduced LDH release was then confirmed in triplicate in A549 cells, along with the parental and complemented strain controls (Fig. 3A). This phenotype was also observed in primary macrophages, but at 7 days postinfection (Fig. 3B). It was also noted that both the broth grown and intracellular growth rates of the mutant were not significantly different to those of the parent strain (data not shown). Previous studies have demonstrated that the cfp-10 (Rv3874) gene and the esat-6 (Rv3875) gene are coordinately expressed as an operon (24). Therefore, a transposon insertion in cfp-10 is likely to have a polar effect and prevent the expression of both genes. Indeed, our studies showed that, as expected, the cfp-10 mutant failed to express ESAT-6 (6-kDa early secretory antigenic target), as evidenced by Western analysis (data not shown).
Fig. 3.
The cfp-10 esat-6 (Rv3874/Rv3875) operon of RD1 is required for host cell lysis. (A) LDH release assay of infected lung epithelial cells, at 72 h postinfection. Cells were infected at a multiplicity of infection of 10:1, and supernatants were analyzed for LDH release. wt, M. tuberculosis Erdman + pYUB412 (empty vector); mt, mc24513 = cfp10::Tn5370 + pYUB412; c, mc24513 + pYUB412::Rv3860–Rv3885c; DRD1, M. tuberculosis Erdman ΔRD1. Values are means ± SD of triplicate measurements. (B) LDH release assay of bone marrow-derived macrophages at day 7 postinfection. Labels wt, mt, and c are as in A. Values are means ± SD of triplicate measurements. (C) Necrosis-determining assay, measuring histone-associated DNA fragments in the supernatant. Labels wt, mt, and c are as in A and B. (D) The M. tuberculosis necrosis phenotype is inhibited by 5 mM exogenous glycine (+g), compared with the control, in which there is no exogenous glycine (-g). Values are means ± SD of triplicate measurements. (E) ESAT-6, but not CFP-10, is sufficient to induce disruption, leading to total destruction (see arrowhead) of an artificial lipid bilayer. Artificial planal bilayers were constructed by using the lipid diphytanoylphosphatidylcholine as described in ref. 28. Voltages of 20–50 mV were applied, and the conductance across the bilayer was measured on the addition of 1 μg of each protein. The addition of ESAT-6 and CFP-10 in combination resulted in similar disruption to ESAT-6 alone. These studies were carried out in triplicate. For further details, see Materials and Methods.
To further elucidate the mode of cell death induced by the wild-type strain and reduced in the mutant strain, the concentration of histone-associated DNA fragments was determined, both in the supernatant (associated with necrosis, in which the cells lyse) and in the cell lysate (associated with apoptosis) in lung epithelial cells. These results confirmed that necrosis was reduced in the mutant when compared with the parental and complemented strains (Fig. 3C), and that there was no difference noted in the levels of apoptosis (data not shown). To further define the mechanism of wild-type-induced cytolysis, glycine was added to the supernatant of the infected monolayer. The addition of exogenous glycine has been reported to protect the cells against lethal ion fluxes across the plasma membrane (21) and was observed to reduce the amount of cytolysis from ≈87% to ≈13% (Fig. 3D). This finding suggests that wild-type-induced cytolysis is mediated by lethal ion fluxes across the plasma membrane. That the effector protein causing lysis and death of the cells was ESAT-6 was demonstrated by experiments using an artificial planal bi-layer, constructed by using the lipid diphytanoylphosphatidylcholine, as described in ref. 22 (Fig. 3E). In these studies it was determined that the ESAT-6 protein, either alone or in combination with culture filtrate protein 10 (CFP-10), resulted in major disruptions in conductance, eventually resulting in total destruction of the artificial membrane. It is also interesting to note that the purified ESAT-6 protein, again either alone or in combination with CFP-10, formed structures similar to those seen in amyloidogenic protein solutions before amyloid fibrils are formed (data not shown), and that some amyloidogenic proteins exhibit pore-forming properties similar to those of bacterial toxins (25).
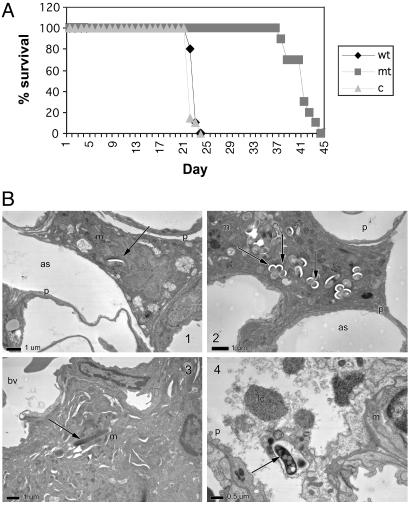
The cfp-10 esat-6 Operon of RD1 Is Required for Virulence. The disruption of the cfp-10 esat-6 operon in M. tuberculosis also resulted in severe attenuation of virulence in an i.v. SCID mouse model of infection, which was comparable to that observed with the RD1 deletion mutants (Fig. 4A). Three weeks after aerosol challenge of immunocompetent BALB/c mice, the mutant-infected mice exhibited rare mild foci composed of very low numbers of mononuclear inflammatory cells, with no lesions detected in some lobes of the lung. Low numbers of infiltrating macrophages were present, predominantly in the alveolar spaces of lungs from mice infected with the mutant strain. In comparison, the wild-type-infected mice showed scattered foci of mild pneumonitis, with focal infiltration of low numbers of macrophages, accompanied by infiltration of epitheloid cells. The cfu counts in the lung, for both the mutant and wild-type-infected mice, were similar (data not shown). Electron microscopy revealed that the mutant, Rv3874::Tn5370, bacilli were located intracellularly within intact macrophages in the alveolar spaces, with some macrophages containing large numbers of bacilli (Fig. 4 B1 and B2). In direct contrast, the wild-type bacilli were typically located in macrophages in the interstitium of the interalveolar wall (Fig. 4 B3 and B4), with some macrophages showing evidence of lysis (Fig. 4B4). Furthermore, ultrastructural studies of the M. tuberculosis cfp-10 esat-6 mutant and wild-type-infected human alveolar epithelial cells (in vitro) revealed that fewer cells were infected in the case of the mutant bacilli (≈20% more cells were infected by wild-type bacilli), and that there were approximately ten times higher numbers of bacilli per cell in the mutant treatment, compared with wild-type (see Table 2). Together, these data suggest that the lack of the cfp-10 esat-6 operon in the mutant results in intracellular containment of the organism and is associated with a lack of cell lysis and tissue invasiveness, and with the attenuation of lung damage. Rv3871 and Rv3876/Rv3877 Are Required for Virulence and for the Secretion of ESAT-6. To test whether Rv3874 and Rv3875 were the only genes of RD1 required for virulence, we generated deletions of the other transcriptional units in RD1 by specialized transduction and observed that the deletion of Rv3871 and Rv3876/77, as well as Rv3874/75, resulted in the same attenuated phenotype as the ΔRD1 or Rv3874::Tn5370 mutants in the SCID mouse model (Fig. 5A).
Fig. 4.
The loss of cfp-10/esat-6 (Rv3874/Rv3875) confers attenuation of virulence. (A) Survival in SCID mice i.v. infected with 2 × 106 cfu of M. tuberculosis Erdman + pYUB412 (wt), mc24513 = cfp10::Tn5370 + pYUB412 (mt), and mc24513 + pYUB412::Rv3860–Rv3885c (c). (B) Electron micrographs of early lesions after high-dose aerosol infection of BALB/c mice with M. tuberculosis Erdman wild type + pYUB412, compared with mc24513 + pYUB412, at 3 weeks postinfection (×5,000). (B1) Mutant-infected macrophage in the alveolar airspace. This result was not observed in the wild-type-infected mice at this time point. (B2) A macrophage in the lumen of the airspace with large numbers of intracellular mutant bacilli. (B3) Wild-type bacillus residing within a macrophage, located interstitially. This result was not observed in mutant-infected mice at this time point. (B4) Cell within the alveolar space with ingested wild-type bacillus is lysed, resulting in the release of the organism. This result was not observed in mice infected with the mutant bacilli. as, alveolar space; bv, blood vessel; lc, lytic cell; m, macrophage; p, type I pneumocyte. Arrows denote mycobacteria.
Table 2. Ultrastructural studies of the behaviour of the M. tuberculosis cfp-10 esat-6 mutant in alveolar epithelial cell culture (each value is representative of 62 4-grid squares).
| mc24513 | Wild type | |
|---|---|---|
| % cells infected (24 h) | 3 | 2 |
| % cells infected (72 h) | 32 | 53 |
| % intracellular bacteria | 67 | 41 |
| % extracellular bacteria | 33 | 59 |
| Average number of bacilli per infected cell | 42 ± 17*** | 4 ± 4*** |
, P < 0.001
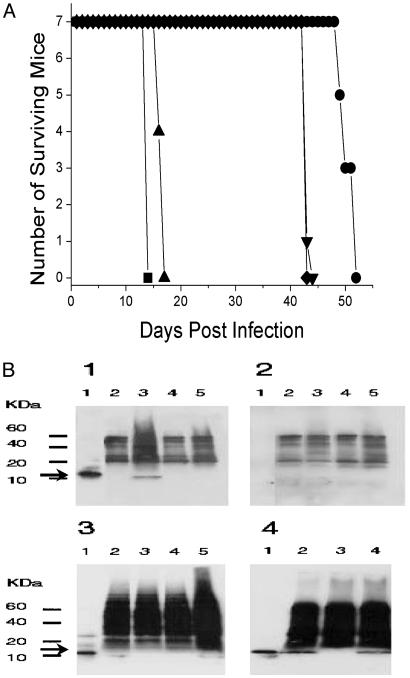
Fig. 5.
The genes Rv3871 and Rv3876/Rv3877 of RD1 are required for virulence and for the secretion of ESAT-6. (A) Survival time of SCID mice infected i.v. with 2 × 106 cfu of M. tuberculosis H37Rv (▪), ΔRv3871 (•), ΔRv3872/3 (▴), ΔRv3874/5 (▾), and ΔRv3876/7 (⋄). (B) Western analyses of the whole cell extracts and culture filtrates. Arrow indicates the ESAT-6-specific band. (B1) Culture filtrates probed with both anti-ESAT-6 primary antibody HYB76-8 and goat anti-mouse Ig secondary antibody. (B2) Culture filtrates probed only with secondary antibody. (B3) Whole-cell lysates probed with both primary and secondary antibodies. In B1–B3, lanes are as follows: lane 1, purified ESAT-6 protein (50 ng); lane 2, ΔRv3871; lane 3, ΔRv3872/3; lane 4, ΔRv3874/5; and lane 5, ΔRv3876/7.(B4) Western analysis of culture filtrates of M. tuberculosis H37Rv, H37Rv ΔRD1, and complemented strain H37Rv ΔRD1 (pYUB412::Rv3860–Rv3885c), reacted with both primary and secondary antibodies. Lane 1, purified ESAT-6 protein (50 ng); lane 2, H37Rv; lane 3, H37Rv ΔRD1; and lane 4, H37Rv ΔRD1 (pYUB412::Rv3860–Rv3885c).
Based on homology to other protein translocation systems (26, 27), we hypothesized that both Rv3871 and Rv3876/77 were involved in secretion of CFP-10 and ESAT-6. Western analyses of the whole cell extracts and culture filtrate proteins (Fig. 5B) clearly demonstrate that both the ΔRv3871 and ΔRv3876/77 mutants can synthesize ESAT-6, but that they are defective in their abilities to secrete ESAT-6 to the culture filtrate. Thus, all of the attenuated mutants of RD1 (ΔRv3871, ΔRv8874/5, and ΔRv3876/77) are defective in the synthesis or secretion of the effector protein ESAT-6.
Discussion
The mechanism of attenuation of BCG had previously not been completely defined. By using a combination of targeted deletion mutagenesis, virulence assays, and complementation analysis, we have been able to unambiguously prove that the RD1 region is required for virulence for M. tuberculosis, and by analogy for M. bovis. The attenuating function resulting from the RD1 deletion was not readily apparent; however, screening for mutants of M. tuberculosis, which failed to cause lysis of alveolar epithelial cells in vitro, revealed this function. The induction of host cell lysis by the Rv3874- and Rv3875-encoded proteins CFP-10 and ESAT-6, respectively, was demonstrated by the reduction in LDH release from alveolar epithelial cells infected with mutant bacilli, when compared with the parent and complemented strains. Moreover, the observed disruption of an artificial lipid bilayer by the ESAT-6 protein defines ESAT-6 as the putative effector of cytolysis. Histopathological and ultrastructural analyses demonstrated a significant decrease in the severity of lung disease in mutant-infected mice, which was associated with reduced invasiveness both in the murine lung and in alveolar epithelial cells in vitro. This evidence is consistent with the defect in cytolytic activity. Interestingly, in prior studies using a model of the alveolar barrier, cytolysis of infected cells was shown to disrupt the barrier and allow more efficient translocation of the bacteria (28). Therefore, we propose that the attenuating function of RD1 is mediated by the effector protein ESAT-6, which induces cytolysis of infected pneumocytes and macrophages and leads to increased cell to cell spread and tissue invasiveness.
The attenuation by the RD1 deletion is not solely the result of the loss of the function of the ESAT-6 protein but also results from loss of the secretion apparatus responsible for the extracellular transportation of ESAT-6. It is interesting to note that CFP-10 reportedly associates as a chaperone with ESAT-6 (29), which is consistent with a secretion apparatus, and that both of these proteins, which are known to be secreted antigens (30), do not possess typical signal sequences. Indeed, a recent report has hypothesized that CFP-10 and ESAT-6 are part of a family of uniquely secreted proteins that may use a novel secretion pathway (26, 27), and our data support that Rv3871 and Rv3876/Rv3877 are part of this secretion system. We conclude that RD1 encodes at least three attenuating mutations, all of which participate in the synthesis or secretion of the effector protein ESAT-6. Furthermore, the secreted ESAT-6 protein mediates cytolysis of the infected cell, resulting in increased egress of the bacilli from the cell and an associated increase in tissue invasiveness. Further studies are warranted to determine the cellular localization, the in vivo expression patterns, and any additional functions mediated by the CFP-10 and ESAT-6 secreted proteins. In addition, whereas our studies show that protection after vaccination with the RD1 mutant is not greater than BCG, this may reflect a limitation of the mouse model of infection and warrants further studies in other protection models. This knowledge may lead to the development of novel therapeutic strategies designed to limit tuberculous pathogenesis.
Note. After submission of this manuscript, Pym et al. (10) reported that esat-6-flanking genes seem to encode an ESAT-6-specific secretory apparatus.
Acknowledgments
We dedicate this manuscript to Frank M. Collins, our long-time collaborator. We thank J. Chan, V. Sambandamurthy, S. Lee, R. Hingley-Wilson, and B. Haynes for helpful comments; S. T. Cole for providing the cosmid 2F9; Alan Finkelstein for the artificial membrane studies; J. Kriakov for providing the temperature-sensitive mycobacteriophage phAE159; Dee Dao for providing BALB/c bone marrow-derived macrophages; Peter Andersen for the CFP-10 and ESAT-6 antibodies; and X. Wang, Marcin I. Apostol, and Leslie Gunther (Albert Einstein Analytical Imaging Facility) for technical assistance. We also thank K. Dobos, E. Spotts-Whitney, L. Ofielu, and the staff from the Albert Einstein Cancer Center histo-technology and comparative pathology facility for their technical expertise. This work was funded by National Institutes of Health (NIH) Grants AI26170 and GM62410, awards to C.H.K. by the Emory Medical Care Foundation (99001), NIH Grant AI52356, and the American Lung Association (RG-029-N).
Abbreviations: ESAT-6, 6-kDa early secretory antigenic target; BCG, bacillus Calmette–Guérin; SCID, severe combined immunodeficient; UFS, upstream flanking sequence; DFS, downstream flanking sequence; LDH, lactate dehydrogenase; CFP-10, culture filtrate protein 10.
References
- 1.Bloom, B. R. & Fine, P. (1994) in Tuberculosis Pathogenesis, Protection, and Control, ed. Bloom, B. R. (Am. Soc. Microbiol., Washington, DC).
- 2.Calmette, A. & Guérin, C. (1909) C. R. Acad. Sci. 149, 716-718. [Google Scholar]
- 3.Gheorghiu, M. (1996) in Vaccinia, Vaccination, and Vaccinology: Jenner, Pasteur and Their Successors, eds. Plotkin, S. A. & Fantini, B. (Elsevier, Paris), pp. 87-94.
- 4.Calmette, A. & Guérin, C. (1905) Ann. Institut Pasteur 19, 601-618. [Google Scholar]
- 5.Calmette, A. & Guérin, C. (1920) Ann. Inst. Pasteur 34, 553-561. [Google Scholar]
- 6.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178, 1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behr, M. A., Wilson, M. A., Gill, W. P., Salamon, H., Schoolnik, G. K., Rane, S. & Small, P. M. (1999) Science 284, 1520-1523. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, S. V., Eiglmeier, K., Garnier, T., Brosch, R., Parkhill, J., Barrell, B., Cole, S. T. & Hewinson, R. G. (2001) Tuberculosis 81, 157-163. [DOI] [PubMed] [Google Scholar]
- 9.Behr, M. A. & Small, P. M. (1999) Vaccine 17, 915-922. [DOI] [PubMed] [Google Scholar]
- 10.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46, 709-717. [DOI] [PubMed] [Google Scholar]
- 11.Wards, B., de Lisle, G. & Collins, D. (2000) Tuber. Lung Dis. 80, 185-189. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, K. N., Liao, R., Guinn, K. M., Hickey, M. J., Smith, S., Behr, M. A. & Sherman, D. R. (2003) J. Infect. Dis. 187, 117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwebach, J. R., Chen, B., Glatman-Freedman, A., Casadevall, A., McKinney, J. D., Harb, J. L., McGuire, P. J., Barkley, W. E., Bloom, B. R. & Jacobs, W. R., Jr. (2002) Appl. Environ. Microbiol. 68, 4646-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAdam, R. A., Quan, S., Smith, D. A., Bardarov, S., Betts, J. C., Cook, F. C., Hooker, E. U., Lewis, A. P., Woollard, P., Everett, M. J., et al. (2002) Microbiology 148, 2975-2986. [DOI] [PubMed] [Google Scholar]
- 15.Pavelka, M. S., Jr., & Jacobs, W. R., Jr. (1999) J. Bacteriol. 181, 4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 17.Bardarov, S., Bardarov, S., Jr., Pavelka, M. S., Jr., Sambandamurthy, V., Larsen, M., Tufariello, J., Chan, J., Hatfull, G. F. & Jacobs, W. R., Jr. (2002) Microbiology 148, 3007-3017 [DOI] [PubMed] [Google Scholar]
- 18.Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R., Jr. (1999) Nature 402, 79-83. [DOI] [PubMed] [Google Scholar]
- 19.Bardarov, S., Kriakov, J., Carriere, C., Yu, S., Vaamonde, C., McAdam, R. A., Bloom, B. R., Hatfull, G. F. & Jacobs, W. R., Jr. (1997) Proc. Natl. Acad. Sci. USA 94, 10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobos, K., Spotts, E., Quinn, F. & King, C. (2000) Infect. Immun. 68, 6300-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby, J. E., Vogel, J. P., Andrews, H. L. & Isberg, R. R. (1998) Mol. Microbiol. 27, 323-336. [DOI] [PubMed] [Google Scholar]
- 22.Nassi, S., Collier, R. J. & Finkelstein, A. (2002) Biochemistry 41, 1445-1450. [DOI] [PubMed] [Google Scholar]
- 23.McDonough, K. A. & Kress, Y. (1995) Infect. Immun., 63, 4802-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthet, F. X., Rasmussen, P. B., Rosenkrands, I., Andersen, P. & Gicquel, B. (1998) Microbiology 144, 3195-3203. [DOI] [PubMed] [Google Scholar]
- 25.Lashuel, H. A., Hartley, D. M., Balakhaneh, D., Aggarwal, A., Teichberg, S. & Callaway, D. J. (2002) J. Biol. Chem. 277, 42881-42890. [DOI] [PubMed] [Google Scholar]
- 26.Pallen, M. (2002) Trends Microbiol. 10, 209-212. [DOI] [PubMed] [Google Scholar]
- 27.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biology 2, research0044.1–0044.18. [DOI] [PMC free article] [PubMed]
- 28.Bermudez, L. E., Sangari, F. J., Kolonoski, P., Petrofsky, M. & Goodman, J. (2002) Infect. Immun. 70, 140-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renshaw, P., Panagiotidou, P., Whelan, A., Gordon, S., Hewinson, R., Williamson, R. & Carr, M. (2002) J. Biol. Chem. 277, 21598-21603. [DOI] [PubMed] [Google Scholar]
- 30.Sorensen, A., Nagai, S., Houen, G., Andersen, P. & Andersen, A. (1995) Infect Immun 63, 1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., 3rd, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]