Abstract
Protein kinase C (PKC) modulates the function of the capsaicin receptor transient receptor potential vanilloid 1 (TRPV1). This modulation manifests as increased current when the channel is activated by capsaicin. In addition, studies have suggested that phosphorylation by PKC might directly gate the channel, because PKC-activating phorbol esters induce TRPV1 currents in the absence of applied ligands. To test whether PKC both modulates and gates the TRPV1 function by direct phosphorylation, we used direct sequencing to determine the major sites of PKC phosphorylation on TRPV1 intracellular domains. We then tested the ability of the PKC-activating phorbol 12-myristate 13-acetate (PMA) to potentiate capsaicin-induced currents and to directly gate TRPV1. We found that mutation of S800 to alanine significantly reduced the PMA-induced enhancement of capsaicin-evoked currents and the direct activation of TRPV1 by PMA. Mutation of S502 to alanine reduced PMA enhancement of capsaicin-evoked currents, but had no effect on direct activation of TRPV1 by PMA. Conversely, mutation of T704 to alanine had no effect on PMA enhancement of capsaicin-evoked currents but dramatically reduced direct activation of TRPV1 by PMA. These results, combined with pharmacological studies showing that inactive phorbol esters also weakly activate TRPV1, suggest that PKC-mediated phosphorylation modulates TRPV1 but does not directly gate the channel. Rather, currents induced by phorbol esters result from the combination of a weak direct ligand-like activation of TRPV1 and the phosphorylation-induced enhancement of the TRPV1 function. Furthermore, modulation of the TRPV1 function by PKC appears to involve distinct phosphorylation sites depending on the mechanism of channel activation.
Protein kinase C (PKC) in peripheral sensory afferents plays a prominent role in hypersensitivity to thermal and mechanical stimuli after tissue injury. PKC sensitizes heat responses and potentiates peptide release from cultured dorsal root ganglion neurons (1, 2) and sensitizes nociceptive afferent neurons to thermal and mechanical stimuli in intact peripheral nerve preparations (3, 4). Diabetic neuropathic hyperalgesia and epinephrine-induced hyperalgesia are attenuated by PKC inhibitors in vivo (5, 6). Recently, several studies have focused on the role of the PKCε isoform. Specific blockade of PKCε diminishes PKC-mediated enhancement of heat currents in sensory neurons and epinephrine-induced hypersensitivity in vivo (7, 8). PKCε knockout mice exhibit reduced hyperalgesia after intracutaneous injection of epinephrine and nerve growth factor (8). Whereas a role of PKC in peripheral sensitization is well established, PKC-mediated phosphorylation and modulation of specific substrates during peripheral sensitization is not fully understood.
Transient receptor potential vanilloid 1 [TRPV1; formerly known as vanilloid receptor 1 (VR1)] is an attractive PKC effector in peripheral nociceptors. TRPV1 was cloned as a capsaicin receptor and is a ligand-gated ion channel, which is also activated by heat, protons, leukotrienes, and anandamide (9–12). TRPV1 is specifically localized to small-diameter, primarily nociceptive sensory neurons and integrates noxious thermal and chemical stimuli analogous to polymodal nociceptors (9, 10). In response to these stimuli, TRPV1 depolarizes sensory neurons and either directly or indirectly initiates peptide release from afferent terminals (13). These properties suggest that TRPV1 plays a critical role in noxious thermal transduction and in neurogenic components of inflammation. Consistent with this, TRPV1 knock-out mice exhibit reduced detection of strong thermal stimuli and reduced inflammatory thermal hypersensitivity (14, 15).
Given the important roles of PKC and TRPV1 in inflammatory hypersensitivity, TRPV1 may act as a PKC substrate after tissue injury. In cultured dorsal root ganglion neurons, PKC potentiates heat and capsaicin responses (1, 7, 16–19). In heterologous expression systems, studies have shown that PKC activation enhances TRPV1 function (16, 18, 20, 21). A recent study has suggested that PKC-activating phorbol esters may directly bind to TRPV1 (22), and a number of studies have suggested that PKC-mediated phosphorylation may cause TRPV1 activation (16, 18, 23). In this study, we show that PKC activation results in direct phosphorylation of TRPV1. By using in vitro phosphorylation and protein sequencing techniques, we identified several potential PKC phosphorylation sites on TRPV1 intracellular domains. Functional studies suggest that activation of TRPV1 by phorbol esters is phosphorylation-independent but modulated by PKC-mediated phosphorylation. Further, we show that distinct phosphorylation sites are involved in the modulation of TRPV1 currents activated by capsaicin and phorbol esters.
Materials and Methods
32P Metabolic Labeling and Immunoprecipitation. COS7 cells were transfected with pcDNA3 TRPV1 by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol 12–24 h after plating onto six-well plates. Cells were rinsed with phosphate-free Eagle's minimal essential medium with Earle's salts (P-EMEM; Sigma) and incubated with P-EMEM containing 250 μCi/ml (1 Ci = 37 GBq) [32P]orthophosphate for 4 h. Phorbol 12-myristate 13-acetate (PMA; Biomol, Plymouth Meeting, PA) was added to the medium at 100 nM and incubated for 15 min, after which 100 nM PMA dissolved in standard electrophysiology external solution (see Electrophysiology) was applied. Cells were lysed in 500 μl of lysis buffer (50 mM NaCl/50 mM NaF/25 mM sodium phosphate, pH 7.4/2.5 mM EDTA/1% Triton X-100/10 mM sodium pyrophosphate/2 mM sodium orthovanadate/0.1 mM PMSF/10 μg/ml leupeptin/5 μg/ml aprotinin/10 μg/ml pepstatin/1 μM microcystin-LR), and the resulting lysate was centrifuged at 20,800 × g in a microcentrifuge for 15 min at 4°C. After preclearing with 25 μl of protein A-Sepharose (Pierce), 2–3 μg of affinity-purified rabbit anti-TRPV1 C-terminal peptide antibody (raised against the epitope CLKPEDAEVFKDSMVPGEK) was added to the lysate and incubated overnight at 4°C. Twenty-five microliters of protein A-Sepharose was incubated with the lysate for 3 h to precipitate antigen–antibody complexes. Beads were rinsed three times with 1 ml of lysis buffer, twice with high ionic strength buffer (lysis buffer but with 500 mM NaCl), and twice with low ionic strength buffer (lysis buffer without NaCl and with sodium phosphate replaced by 20 mM Tris–Cl, pH 6.8). Two percent SDS sample buffer was added to the beads and heated for 10 min at 40°C. Samples were electrophoresed on SDS/8% polyacrylamide gels. Radiolabeled TRPV1 was detected by phosphorimaging (Packard Instruments).
Fusion Protein Production. N- and C-terminal cytoplasmic fragments (amino acids 1–432 and 686–838, respectively) of TRPV1 were cloned into pGEX-4T1 by using PCR with XhoI ends. These constructs were transformed into BL21-RP Escherichia coli bacteria (Stratagene), and fusion protein production was induced with 0.1 mM isopropyl β-d-thiogalactoside for 4 h at room temperature. Cells were converted to spheroplasts with lysozyme (24) and lysed by sonication. The resulting lysate was clarified by centrifugation, and fusion proteins were purified with GSTrap FF columns essentially according to the manufacturer's instructions (Amersham Biosciences). After elution with 10 mM reduced glutathione, fusion proteins were concentrated to 0.5–4 mg/ml by ultrafiltration and centrifuged at 100,000 × g for 60 min to remove protein aggregates and debris.
In Vitro Phosphorylation. For stoichiometry estimates, reaction mixtures consisted of 25 μM first intracellular loop peptide (CLQRRPSLKSLF) or 1.8 μM fusion protein (≈0.1 mg/ml), 50 mM Tris–Cl at pH 7.5, 1 mM NaEDTA, 12 mM magnesium acetate, 0.25 mM NaATP, 1 μCi/μl [γ-32P]ATP (≈6,000–8,000 cpm/pmol ATP), and 0.8 ng/μl catalytically active PKC (Calbiochem; Biomol). Reaction mixtures were incubated at 30°C for various durations and stopped by spotting onto P81 phosphocellulose paper (peptide) or by adding an equal volume of 2× Laemmli sample buffer (fusion proteins). For kinetic analysis, reaction mixtures were assembled in a similar fashion, but fusion protein reaction mixtures contained 0.16 ng/μl activated PKC and peptide reactions contained 0.1 ng/μl activated PKC. Reactions were incubated at 30°C and stopped after 5 min. 32P incorporation was linear at time points <10 min.
P81 paper squares were washed four times with large volumes of 75 mM phosphoric acid, rinsed quickly with methanol, air-dried, and placed into 10 ml of scintillation fluid. Fusion proteins were electrophoresed on SDS/PAGE gels and quantified by using Coomassie blue staining and nearby BSA standards. 32P incorporation was determined by scintillation counting of blotted peptides or phosphorimaging and densitometry of fusion protein bands (Packard Instruments).
Phosphorylation Site Determination. In-gel digests, HPLC, and Edman sequencing were conducted essentially as described (25). For electrospray ionization mass spectroscopy (API 3000 LC/MS/MS System, PE Sciex, Thornhill, ON, Canada), peptides from in-gel digests were desalted by using a C18 ZipTip column (Millipore). Phosphopeptides were initially delineated by using a -79 precursor ion scan and sequenced with tandem mass spectrometry.
Mutagenesis. TRPV1 mutants were created by using primer extension by Pfu DNA polymerase (Stratagene) and DpnI to digest the wild-type template as described (26). The entire ORF of all constructs was sequenced.
Electrophysiology. COS7 cells were maintained in DMEM plus 10% FBS at 37°C, under 5% CO2/95% O2, and transfected with pCDNA3 TRPV1 or various mutants with pEGFP (9:1) by using Lipofectamine 2000 according to the manufacturer's instructions. Whole-cell patch-clamp recordings were conducted by using standard techniques with an Axopatch 200B amplifier and PCLAMP 8 software at room temperature ≈24–36 h after transfection. The external solution consisted of (in mM) 140 NaCl, 5 KCl, 2 MgCl2, 5 EGTA, 10 Hepes, and 10 glucose at pH 7.4; whereas the pipette solution contained (in mM) 140 CsCl, 5 EGTA, and 10 Hepes at pH 7.4. Cells were voltage-clamped at -60 mV. Dose–response curves shown in Fig. 4 C–E were obtained from CHO-K1 cells maintained in Ham's F12 medium with other aspects of culture maintenance and transfection as above.
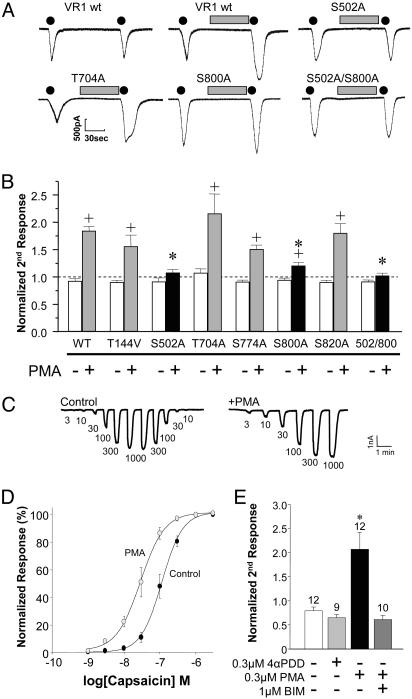
Fig. 4.
S502 and S800 mediate PKC modulation of capsaicin-evoked currents. (A) Representative current responses to consecutive applications of 20 nM capsaicin (black dots; 10 s) without or with an intervening 1-min application of PMA (100 nM, gray bars) in transiently transfected COS7 cells. (B) Ratio of the second to the initial peak capsaicin response (mean ± SEM) is plotted for wild-type TRPV1 and TRPV1 PKC site mutants in the absence (-) or presence (+) of an intervening PMA application. None of the mutants show a significant difference compared with wild type without PMA (ANOVA, P = 0.20). Only S502A, S800A, and the double mutant S502A/S800A show a decrease in PMA potentiation compared with wild type (ANOVA followed by post hoc Dunnett's test; *, P < 0.05), whereas all of the constructs, except S502A and S502/800A, exhibit some form of PMA potentiation (-PMA versus +PMA, unpaired t test; +, P < 0.05). (C and D) Dose–response relationships for capsaicin are shown under control conditions (•) and after a 3-min pretreatment with the PKC activator, PMA (300 nM, ○). Data are plotted as a percentage of the maximal response elicited by capsaicin (n = 6). C shows representative traces of two cells showing the currents induced by various doses of capsaicin (indicated below the traces, in nM). (E) PMA enhancement of capsaicin-evoked currents is PKC-mediated. Two successive applications of 30 nM capsaicin were performed. The second response is potentiated by the PKC-activating phorbol ester PMA but not by the inactive phorbol ester 4α-phorbol 12,13-didecanoate (4αPDD; 300 nM each). The PMA modulation of capsaicin responses was blocked by the PKC inhibitor bisindolylmaleimide (BIM; 1 μM).
Calcium Imaging. COS7 cells grown on poly-d-lysine-coated coverslips were transfected 18 h before imaging with TRPV1 and enhanced GFP (9:1). Transfected cells were loaded just before imaging with 250 μl of 10 mM fura 2-AM (Molecular Probes) from a 1 M stock prepared with 20% Pluronic F-127 (Molecular Probes) in dimethyl sulfoxide (Sigma) diluted in Hanks' balanced salt solution (Invitrogen) with 10 mM Hepes, pH 7.3, for 1 h at room temperature then rinsed three times with Hanks' balanced salt solution plus Hepes. Cells were then equilibrated for 30 min. Imaging was carried out by using a Hamamatsu Orca cooled charge-coupled device camera on an Olympus IX70 inverted microscope with an Olympus UPlanF1 ×10 lens using SIMPLEPCI software (Compix Imaging Systems, Cranberry Township, PA). Coverslips containing transfected cells were continuously perfused at a rate of 3–4ml/min. GFP-positive cells were defined as transfected cells and imaged at room temperature by using ratio imaging with a 380/357 filter set. After acquisition of 2 min of baseline, cells were perfused with 1 μM 4α-PMA (Biomol) for 1 min, followed by a 5-min rinse and then by a 1-min application of 1 μM PMA. Ruthenium red (ammoniated ruthenium oxychloride, 10 μM; Sigma), when used, was present in the bath and phorbol ester-containing solutions. Ratio measurements were taken for each individual cell with SIMPLEPCI and the peak responses to 4α-PMA and PMA were determined. These data were analyzed with MICROCAL ORIGIN Version 5.0 (Microcal Software, Northampton, MA).
Results
To determine whether PKC directly phosphorylates TRPV1, we immunoprecipitated TRPV1 from metabolically 32P-labeled COS7 cells transfected with TRPV1 to examine TRPV1 phosphate incorporation. TRPV1 32P incorporation significantly increased after a brief incubation with a phorbol ester, PMA (15 min, 100 nM), suggesting that PKC directly phosphorylates TRPV1 in cultured cells.
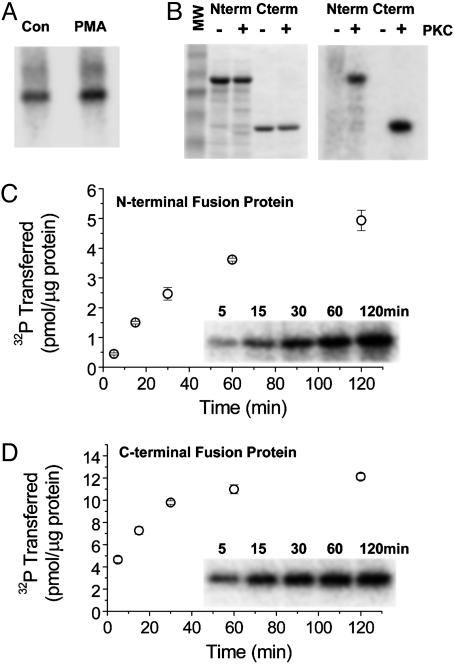
Given that TRPV1 may act as a PKC substrate, we in vitro phosphorylated engineered TRPV1 cytoplasmic domains with PKC to identify candidate phosphorylation sites. Bacterially produced GST fusion proteins of the large N and C termini (Fig. 1B) were incubated with and without activated PKC, and phosphorimaging revealed that both proteins acted as in vitro substrates (Fig. 1C). With our reaction conditions, the N-terminal fusion protein showed a stoichiometry of ≈40% (Fig. 1C), whereas the C-terminal protein exhibited a saturating stoichiometry of ≈55% (Fig. 1D).
Fig. 1.
TRPV1 acts as a PKC substrate in cultured cells and in vitro.(A) Bands represent TRPV1 immunoprecipitated from transiently transfected, metabolically 32P-labeled COS7 cells. PMA treatment (right lane) increases TRPV1 32P incorporation compared with control (left lane). (B) Coomassie blue staining of GST N-terminal (Nterm) and C-terminal (Cterm) TRPV1 fusion proteins without (-) or with (+) added PKC (Left) and the corresponding phosphorimage (Right) showing 32P incorporation into fusion proteins only with added PKC. Reactions were incubated at 30°C for 1 h. (C) Time course of phosphorylation for GST TRPV1 N-terminal fusion protein. Phosphorylation stoichiometry is at least 5 pmol/μg fusion protein or ≈40%. (D) GST TRPV1 C-terminal fusion protein phosphorylation time course showing saturating stoichiometry of ≈12 pmol/μg fusion protein or ≈55%.
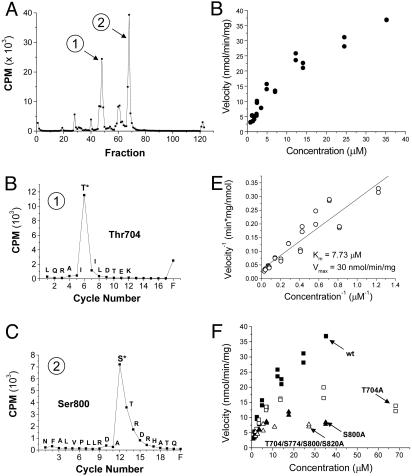
We used classic HPLC peptide mapping and Edman sequencing to delineate PKC in vitro phosphorylation sites located on the engineered cytoplasmic domains. Cerenkov counting of the GST C-terminal fusion protein, which was digested with endoproteinase Lys-C and fractionated by HPLC, revealed two major phosphopeptides (Fig. 2A). Sequencing of these fractions coupled with scintillation counting of the sequencing cycles revealed T704 and S800 as in vitro PKC phosphorylation sites (Fig. 2 B and C). In addition, electrospray ionization mass spectroscopy of peptides from an in-gel trypsin digest of the C-terminal fusion protein delineated S774 and S820 as minor PKC phosphorylation sites (data not shown).
Fig. 2.
T704 and S800 identified as in vitro PKC phosphorylation sites on the TRPV1 C terminus. (A) Cerenkov counting of HPLC fractions from an in-gel Lys-C digest of PKC-phosphorylated TRPV1 C-terminal fusion protein reveals two major phosphopeptides. (B) Edman sequencing and scintillation counting of the sequencing cycles from the first phosphopeptide fraction delineates T704 as a phosphorylated site. (C) Scintillation counting of the Edman sequencing cycles from the second phosphopeptide fraction reveals S800 as an in vitro PKC phosphorylation site. (D) PKC phosphorylation velocity of the TRPV1 C-terminal fusion protein plotted against protein concentration, showing a relatively strong PKC substrate. (E) Lineweaver–Burk plot (inverse velocity versus inverse concentration) and linear regression delineates a Km of 7.73 μM and Vmax of 30 nmol·min-1·mg-1. (F) Kinetic analysis of various mutants plotted alongside wild-type kinetics from D. The decrement observed in the S800 to alanine mutant is similar to that seen with a mutant fusion protein with all identified PKC phosphorylation sites (T704, S774, S800, and S820) converted to alanine, suggesting that S800 is the predominant substrate.
To determine the relative importance of the C-terminal phosphorylation sites, we analyzed the phosphorylation kinetics of the wild-type and various mutant fusion proteins. Wild-type C-terminal TRPV1 fusion protein acted as a relatively strong substrate with a Km of 7.73 μM and a Vmax of 30 nmol·min-1·μg-1 (Fig. 2 D and E). Significant deficits in reaction velocity occurred with S800 mutated to alanine (S800A) and T704A mutants (Fig. 2F). However, little difference in phosphorylation kinetics was found between the single S800A mutant and the multiply mutated fusion protein with T704A, S774A, S800A, and S820A mutations (Fig. 2F). These data suggest that S800 is the most efficacious or rate-limiting substrate amongst the four identified PKC phosphorylation sites on the TRPV1 C-terminal fusion protein.
In-gel digestion with Lys-C followed by HPLC of the N-terminal fusion protein revealed a single major phosphopeptide peak (Fig. 3A). Edman sequencing and scintillation counting delineated T144 as the in vitro PKC phosphorylation site (Fig. 3B). Mutating T144 to alanine eliminates the major phosphorylation peak observed in the wild-type fusion protein, whereas a small peptide in the HPLC flow-through exhibits increased phosphorylation (Fig. 3C). This indicates that the major phosphorylation site was correctly identified as T144. The increased phosphorylation in the flow-through may be compensatory and may explain the relatively high phosphorylation stoichiometry of the T144A mutant (data not shown).
Fig. 3.
Identification of phosphorylation sites on the TRPV1 N-terminal fusion protein and first intracellular loop. (A) Cerenkov counting of the HPLC fractions after an in-gel endoproteinase Lys-C digestion of the PKC-phosphorylated N-terminal fusion protein reveals one major phosphopeptide. (B) Edman sequencing correlated with scintillation counting pinpoints T144 as the phosphorylated site. (C) Cerenkov counting of the HPLC fractions for the T144A mutant TRPV1 N-terminal fusion protein. The major phosphopeptide is eliminated, indicating correct identification of the major site in the wild-type protein, but the radioactivity peak in the flow-through (fraction 83) is dramatically increased. (D) Time course of 32P transfer to TRPV1 first intracellular loop peptide indicates a saturating stoichiometry of ≈50%. (E) Edman sequencing correlated with scintillation counting clearly demarcates S502 as the phosphorylated site rather than S505. (F) Phosphorylation kinetics of the loop peptide with Lineweaver–Burk analysis (Inset) reveals a highly efficacious PKC substrate with a Km of 2.77 μM and Vmax of 7.26 μmol·min-1·mg-1.
The first intracellular loop contains two serines, and a corresponding peptide exhibited a phosphorylation stoichiometry of ≈50% with a Km of 2.77 μM and Vmax of 7.26 μmol·min-1·mg-1 (Fig. 3 D and E). Edman sequencing and scintillation counting of the phosphorylated peptide confirmed S502 as the major phosphorylation site rather than nearby S505 (Fig. 3F).
Taken together, the in vitro phosphorylation data identified T144, S502, T704, and S800 as major PKC phosphorylation sites most likely to possess functional value. The other sites may represent minor in vitro phosphorylation sites with potential functional significance.
We constructed alanine or valine mutants of these sites in TRPV1 and first examined PKC modulation of capsaicin-evoked currents in these mutants by using whole-cell patch-clamp techniques. As shown by other studies (16, 18, 20, 23), a short treatment of TRPV1-expressing cells with phorbol ester (PMA, 100 nM, 1 min) potentiates nondesensitizing TRPV1 currents ≈2-fold (Fig. 4 A and B). When all of the inactivating phosphorylation site mutants were treated with PMA in a similar fashion, only the S502A and S800A mutants exhibited diminished PMA potentiation (Fig. 4 A and B). Whereas the S800A mutant continues to exhibit some PMA potentiation, the S502A mutant shows little, if any, PMA potentiation. Mutating both S502 and S800 to alanine eliminates PMA potentiation similar to mutating S502 alone (Fig. 4B). These data suggest that PKC directly phosphorylates TRPV1 on S502 and S800 to enhance nondesensitizing capsaicin-evoked currents. Essentially identical results were obtained when capsaicin-evoked currents were recorded in the presence of external Ca2+ and under modestly desensitizing conditions (data not shown). As has been previously reported for human TRPV1 expressed in Xenopus oocytes (23), we found that activation of PKC by PMA led to a leftward shift in the capsaicin dose–response curve for activation of rat TRPV1 expressed in mammalian cells (Fig. 4 C and D). This effect of PMA was clearly mediated by PKC activation because an inactive phorbol ester, 4α-phorbol 12,13-didecanoate, did not enhance capsaicin-evoked currents (Fig. 4E). Furthermore, the enhancement of capsaicin-evoked currents by PMA was blocked by the PKC inhibitor bisindolylmaleimide (1 μM; Fig. 4E) or by inclusion of the PKC-inhibiting peptide PKC19–31 in the recording pipet (data not shown).
Previous studies have shown that TRPV1 can be directly activated by PKC-activating phorbol esters (16, 18, 23). In TRPV1-transfected cells, we observed inward currents in response to application of PMA in only 6 of 13 cells (data not shown). Because of the low percentage of cells that responded to phorbol ester application, we chose to measure phorbol ester activation of TRPV1 by using Ca2+ imaging, with which we can measure the responses of many individual cells simultaneously. We have previously shown that native TRPV1 responses in dorsal root ganglion neurons can be faithfully recorded by using calcium imaging (19). We found that cells transfected with TRPV1 responded to application of PMA with a rise in intracellular Ca2+ (Fig. 5). Ca2+ responses to PMA were completely blocked by the TRPV1 antagonist ruthenium red (10 μM) and were not observed in untransfected cells or cells transfected with GFP alone (Fig. 5A). The activation of TRPV1 by PMA was largely caused by PKC activation, because the response to PMA was dramatically reduced by preincubation with the PKC inhibitor Ro31-8220 (Fig. 5B). Interestingly, when the non-PKC-activating PMA analogue 4α-PMA was applied, responses were also observed. Although responses to 4α-PMA were much smaller than those observed in response to PMA, they were statistically indistinguishable from responses to PMA in the presence of Ro31-8220. By using whole-cell recording, we also observed responses to 4α-PMA in three of nine cells tested. Again, the average response to 4α-PMA was smaller than that seen with PMA; the average PMA response was 11.8 + 4.6 pA/pF (n = 6), whereas responses to 4α-PMA averaged 4.7 + 1.3 pA/pF (n = 3). Another inactive analogue of PMA, 4α-phorbol, did not induce Ca2+ influx in TRPV1-transfected cells (not shown). Whereas 4α-phorbol is frequently used as a negative control analogue for phorbol esters, this compound lacks the fatty acid group seen in PKC-activating phorbol esters. 4α-PMA most closely approximates the structure of the PKC-activating PMA. Our data suggest that the fatty acid moiety is necessary for the PKC-independent activation of TRPV1 by phorbol esters.
Fig. 5.
PKC-dependent and -independent effects of phorbol esters on TRPV1. (A) Averaged traces of wild-type TRPV1 responses to 4α-PMA (1 μM) and PMA (1 μM) under control conditions or in the presence of Ro31-8220 (1 μM, present in all perfusion solutions) or of ruthenium red (10 μM). Lower trace shows the response of vector-transfected cells to the same concentrations of 4α-PMA and PMA. (B) Population data showing the average peak responses to 4α-PMA and PMA under control conditions and in the presence of Ro31-8220 [n = 189 (control) and 145 (Ro31-8220)]. (C) Mutation of T704 and S800 to alanine residues reduces the response of TRPV1 to PMA to the level of the 4α-PMA response, whereas mutation of S502A statistically increases the responses to both 4α-PMA and PMA (n = 103–233 cells). *, P < 0.05. Asterisks above the bars indicate a significant difference between the 4α-PMA and PMA response for a given construct. Asterisks within the bars indicate a significant difference for the response to 4α-PMA or PMA compared with the response of wild-type TRPV1 to the same compound. (D) Capsaicin responses of wild-type and TRPV1 PKC-phosphorylation site mutants are not statistically different (n = 38–161). A wild-type control was done on each experimental day to eliminate variability resulting from day-to-day changes in expression. The responses of each mutant were then normalized to the size of the wild-type 4α-PMA response within each day's experiment. Statistical analyses were performed by using a one-way ANOVA followed by a Tukey's post hoc test.
It is important to note that Chuang et al. (22) have shown that PMA decreases binding of [3H]resiniferatoxin to TRPV1, suggesting that PMA directly interacts with TRPV1. Taken together with our data, this finding suggests that there may be two components involved in the activation of TRPV1 by phorbol esters: (i) a direct ligand-like action that accounts for our finding that 4α-PMA can activate TRPV1 and (ii) a second phosphorylation-dependent enhancement of TRPV1 currents that accounts for a PMA response that is much larger than the response elicited by 4α-PMA. To test these possibilities, we performed a series of studies examining the activation of TRPV1 PKC phosphorylation site mutants by 4α-PMA and PMA. We found, rather surprisingly, that the enhanced response to PMA compared with 4α-PMA was not dependent on the same amino acid residues that mediate enhancement of capsaicin-evoked responses by PKC (Fig. 5C). Specifically, the S800A mutant, which blocked PMA-induced enhancement of capsaicin responses, also eliminated the relative enhancement of the response to PMA compared with 4α-PMA. However, the S502A mutant, which also abrogated almost all PMA-induced potentiation of capsaicin responses, did not eliminate the relative enhancement of the response to PMA compared with 4α-PMA. Furthermore, the T704A mutant, which had no effect on the modulation of capsaicin responses by PMA, also completely eliminated the relative enhancement of the response to PMA compared with 4α-PMA. These findings suggest that PKC regulates the activation of TRPV1 differentially depending on the ligand used for activation. The responses to both 4α-PMA and PMA in the S502A mutant were significantly larger than those for wild-type TRPV1 or any of the other mutants (Fig. 5C), although the Ca2+ responses to capsaicin for S502A did not differ from wild type or the other mutants (Fig. 5D). The reason for this discrepancy is not clear.
Discussion
PKC modulation of TRPV1 function has recently become an active, yet confusing, field with several proposed mechanisms. Premkumar and Ahern (16) reported that PMA not only potentiates capsaicin-activated TRPV1 currents but also activates TRPV1 itself in TRPV1-expressing Xenopus oocytes. Neither of these effects was replicated by the inactive 4α-phorbol, and both effects were inhibited by a PKC inhibitor. However, subsequent studies have disputed and extended some of the initial findings. First, direct activation of TRPV1 by phorbol esters is quite variable. One group reported direct activation in transiently transfected HEK293 cells, but not in stably transfected cells (18). Second, direct activation of TRPV1 appears to have a higher EC50 than phorbol ester-mediated potentiation of TRPV1-mediated currents. Whereas 100 nM phorbol 12,13-dibutyrate or 200 nM PMA maximally potentiates TRPV1 currents, at least 1 μM phorbol 12,13-dibutyrate or PMA appears to be required to observe direct activation (data not shown) (16, 18, 20, 23). Third, phorbol esters have been found to directly displace resiniferatoxin binding to TRPV1 (22), further obfuscating the issue of whether phorbol ester-mediated activation of TRPV1 involves phosphorylation by PKC or an agonist-like action. Fourth, phorbol ester potentiation of capsaicin-evoked currents is variable. We and others report a relatively ephemeral 2-fold potentiation (18), whereas others obtain extremely robust 5- to 10-fold potentiation of capsaicin-evoked currents (16, 20, 21, 23). Many of the disparities may simply reflect different expression systems, culture conditions, and drug application paradigms. For instance, it has been reported recently that expression of the α isoform of PKC correlates with the ability of phorbol esters to directly activate TRPV1 (27). Other studies clearly implicate an important role for PKCε in modulation of TRPV1 (21). Thus, it is possible that expression systems vary in PKCα and PKCε expression, leading to variable phorbol ester activation of TRPV1. Ultimately a more important question is whether PKCα and PKCε play different roles in modulating sensory transduction by TRPV1.
However, at the crux of the matter lies the issue of whether phorbol esters activate and/or potentiate TRPV1 through direct binding or by activation of PKC or both. Whereas phorbol ester binding to TRPV1 appears to support an agonist or allosteric modulator model, experiments involving PKC catalytic inhibitors that compete with ATP or substrate, rather than diacylglycerol, point to phosphorylation. The data here show that phosphorylation contributes to phorbol ester potentiation of TRPV1 currents because phorbol esters increase 32P incorporation into TRPV1, and mutations at S502 and S800 eliminate phorbol ester potentiation of capsaicin-evoked currents. The findings with the mutants agree with previous reports (21). However, the previous findings do not address phorbol ester activation of TRPV1, nor do they eliminate the possibility that both phorbol ester binding and PKC-mediated phosphorylation are required to activate and/or potentiate TRPV1 currents.
Our studies using PKC phosphorylation site mutants may help clarify the multiple mechanisms involved in the activation of TRPV1 by phorbol esters. First, we show that TRPV1 can be activated by 4α-PMA, which does not activate PKC, suggesting that phorbol esters can gate TRPV1 either directly or through some other PKC-independent mechanism. Second, our data argue that PKC-mediated phosphorylation of TRPV1 enhances currents activated by PMA by means of an overlapping mechanism distinct from that involving phosphorylation-dependent enhancement of capsaicin-evoked currents. We found that the enhancement of currents gated by either PMA or capsaicin was eliminated when S800 was mutated, whereas a mutation at S502 reduced PKC potentiation of capsaicin-evoked currents but not PMA-evoked currents. Similarly, a mutation at T704 dramatically reduces the PKC-dependent enhancement of PMA-mediated activation of TRPV1 but has no effect on PKC-dependent enhancement of capsaicin-evoked currents. The reason for this distinction is not clear, but likely involves different mechanisms of channel-gating induced by phorbol esters and capsaicin. In general, the results suggest a model in which phosphorylation of S800 by PKC is a necessary step for modulation of TRPV1 function by PKC, and phosphorylation of S502 or T704 is additionally required, depending on the mechanism of channel activation. Whereas this model is the best fit for our data, it is important to keep in mind that TRPV1 is a polymodal receptor, and, although under our conditions we saw no evidence for direct gating of the channel by PKC-mediated phosphorylation, it is possible that at higher temperatures or under slightly different recording conditions phosphorylation of TRPV1 by PKC could directly lead to channel gating.
Acknowledgments
We thank Dr. Richard Cook and the Baylor College of Medicine Protein Core Facility for assistance with Edman sequencing and mass spectroscopy, and Dr. David Julius for kindly providing rat TRPV1 cDNA. This work was supported by grants from the National Institutes of Health (R01MH60230 and R01NS42595 to R.W.G., F32MH65766 to K.S.G., and R01NS18788 and P01NS39420 to G.S.O.). G.B. is a McNair Scholar of the Baylor College of Medicine Medical Scientist Training Program.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TRPV1, transient receptor potential vanilloid 1; PMA, phorbol 12-myristate 13-acetate.
References
- 1.Cesare, P. & McNaughton, P. (1996) Proc. Natl. Acad. Sci. USA 93, 15435-15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, L. A. & Vasko, M. R. (1996) J. Neurochem. 67, 72-80. [DOI] [PubMed] [Google Scholar]
- 3.Leng, S., Mizumura, K., Koda, H. & Kumazawa, T. (1996) Neurosci. Lett. 206, 13-16. [DOI] [PubMed] [Google Scholar]
- 4.Schepelmann, K., Messlinger, K. & Schmidt, R. F. (1993) Exp. Brain Res. 92, 391-398. [DOI] [PubMed] [Google Scholar]
- 5.Ahlgren, S. C. & Levine, J. D. (1994) J. Neurophysiol. 72, 684-692. [DOI] [PubMed] [Google Scholar]
- 6.Khasar, S. G., McCarter, G. & Levine, J. D. (1999) J. Neurophysiol. 81, 1104-1112. [DOI] [PubMed] [Google Scholar]
- 7.Cesare, P., Dekker, L. V., Sardini, A., Parker, P. J. & McNaughton, P. A. (1999) Neuron 23, 617-624. [DOI] [PubMed] [Google Scholar]
- 8.Khasar, S. G., Lin, Y. H., Martin, A., Dadgar, J., McMahon, T., Wang, D., Hundle, B., Aley, K. O., Isenberg, W., McCarter, G., et al. (1999) Neuron 24, 253-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D. & Julius, D. (1997) Nature 389, 816-824. [DOI] [PubMed] [Google Scholar]
- 10.Tominaga, M., Caterina, M. J., Malmberg, A. B., Rosen, T. A., Gilbert, H., Skinner, K., Raumann, B. E., Basbaum, A. I. & Julius, D. (1998) Neuron 21, 531-543. [DOI] [PubMed] [Google Scholar]
- 11.Hwang, S. W., Cho, H., Kwak, J., Lee, S. Y., Kang, C. J., Jung, J., Cho, S., Min, K. H., Suh, Y. G., Kim, D. & Oh, U. (2000) Proc. Natl. Acad. Sci. USA 97, 6155-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zygmunt, P. M., Petersson, J., Andersson, D. A., Chuang, H., Sørgård, M., Di Marzo, V., Julius, D. & Högestätt, E. D. (1999) Nature 400, 452-457. [DOI] [PubMed] [Google Scholar]
- 13.Szallasi, A. & Blumberg, P. M. (1999) Pharmacol. Rev. 51, 159-212. [PubMed] [Google Scholar]
- 14.Caterina, M. J., Leffler, A., Malmberg, A. B., Martin, W. J., Trafton, J., Petersen-Zeitz, K. R., Koltzenburg, M., Basbaum, A. I. & Julius, D. (2000) Science 288, 306-313. [DOI] [PubMed] [Google Scholar]
- 15.Davis, J. B., Gray, J., Gunthorpe, M. J., Hatcher, J. P., Davey, P. T., Overend, P., Harries, M. H., Latcham, J., Clapham, C., Atkinson, K., et al. (2000) Nature 405, 183-187. [DOI] [PubMed] [Google Scholar]
- 16.Premkumar, L. S. & Ahern, G. P. (2000) Nature 408, 985-990. [DOI] [PubMed] [Google Scholar]
- 17.Zhou, Y., Zhou, Z. & Zhao, Z. (2001) Neuropharmacology 41, 601-608. [DOI] [PubMed] [Google Scholar]
- 18.Vellani, V., Mapplebeck, S., Moriondo, A., Davis, J. B. & McNaughton, P. A. (2001) J. Physiol. 534, 813-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, H. J., Bhave, G. & Gereau, R. W. (2002) J. Neurosci. 22, 7444-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tominaga, M., Wada, M. & Masu, M. (2001) Proc. Natl. Acad. Sci. USA 98, 6951-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Numazaki, M., Tominaga, T., Toyooka, H. & Tominaga, M. (2002) J. Biol. Chem. 277, 13375-13378. [DOI] [PubMed] [Google Scholar]
- 22.Chuang, H. H., Prescott, E. D., Kong, H., Shields, S., Jordt, S. E., Basbaum, A. I., Chao, M. V. & Julius, D. (2001) Nature 411, 957-962. [DOI] [PubMed] [Google Scholar]
- 23.Crandall, M., Kwash, J., Yu, W. & White, G. (2002) Pain 98, 109-117. [DOI] [PubMed] [Google Scholar]
- 24.Witholt, B., Boekhout, M., Brock, M., Kingma, J., Heerikhuizen, H. V. & Leij, L. D. (1976) Anal. Biochem. 74, 160-170. [DOI] [PubMed] [Google Scholar]
- 25.Bhave, G., Zhu, W., Wang, H., Brasier, D. J., Oxford, G. S. & Gereau, R. W. (2002) Neuron 35, 721-731. [DOI] [PubMed] [Google Scholar]
- 26.Gereau, R. W. & Heinemann, S. F. (1998) Neuron 20, 143-151. [DOI] [PubMed] [Google Scholar]
- 27.Olah, Z., Karai, L. & Iadarola, M. J. (2002) J. Biol. Chem. 277, 35752-35759. [DOI] [PubMed] [Google Scholar]