Abstract
DNA topoisomerase II (Top2) is the target of some of the most effective anticancer DNA intercalators. To determine the effect of intercalating ligands at defined positions relative to a known DNA cleavage site for human Top2α, we synthesized oligodeoxynucleotides containing single trans-opened benzo[a]pyrene 7,8-diol 9,10-epoxide (DE) deoxyadenosine (dA) adducts of known absolute configuration, placed at specific positions in a duplex sequence containing staggered Top2 cleavage sites on both strands. Because the orientations of the intercalated hydrocarbon are known from NMR solution structures of duplex oligonucleotides containing these dA adducts, a detailed analysis of the relationship between the position of intercalation and trapping of Top2 is possible. Our findings demonstrate that (i) Top2 cleavage complexes are trapped by intercalation of the hydrocarbon at either of the staggered cleavage sites or immediately adjacent to the base pairs flanking the cleavage sites within the stagger; (ii) both concerted and nonconcerted cleavage by both subunits of a Top2 homodimer were detected depending on the position of the benzo[a]pyrene DE dA adduct; and (iii) intercalation immediately outside of the staggered Top2 cleavage site, and to a lesser extent in the middle of the stagger, prevents Top2 from cleaving DNA at this site, consistent with the effect of some intercalators as suppressors of Top2-mediated DNA cleavage. These results identify specific binding sites for intercalators that result in trapping of Top2. Such poisoning of Top2 by bulky polycyclic aromatic hydrocarbon DE adducts constitutes a potential mechanism for their carcinogenic activity.
DNA topoisomerase II (Top2) is a homodimer essential for DNA transactions (relaxation/supercoiling, catenation/decatenation, and possibly recombinations) (1–3). Human cells contain two Top2 enzymes, Top2α and Top2β, that are encoded by two different genes (2). The molecular mechanism by which Top2 catalyzes such DNA transactions is by an ATP-dependent induction of two coordinated nicks in complementary strands of the DNA duplex followed by transfer of another DNA double helix through the broken DNA duplex (4–7). The sites of Top2-mediated cleavage are staggered by four bases on the opposite DNA strands (8–10). These staggered DNA breaks are generated by phosphodiester-bond formation between the catalytically essential tyrosine 805 in each homodimer and the 5′-phosphate termini of the cleaved strands. The enzyme-cleaved DNA intermediate is known as a Top2 “cleavage complex” (9).
Top2 is the primary target of a variety of clinically relevant anticancer drugs such as epipodophyllotoxins [etoposide (VP-16) and teniposide], anthracyclines (doxorubicin, epirubicin, and idarubicin), anthrapyrazoles (mitoxantrone), acridines (amsacrine), and ellipticines (3, 11, 12). About half of current cancer chemotherapy protocols include Top2-targeted drugs (3, 11). These drugs interfere with the normal functioning of the enzyme during the cleavage/religation process. They cause accumulation of Top2 cleavage complexes by inhibiting DNA religation (i.e., by “trapping”) the enzyme homodimers covalently bound to DNA (3, 11, 12), leading eventually to cell death. Position-specific trapping of Top2 can also result from base damage (3, 13). Because crystal structures for Top2 have been determined only in the absence of DNA (10), molecular interactions among drugs, DNA, and the protein are unknown.
Because many clinically used Top2 poisons including anthracyclines, mitoxantrone, acridines, and ellipticines are intercalators, the main objective of this study was to establish the spatial relationship between structurally well defined intercalating ligands and Top2 trapping. Benzo[a]pyrene (BaP) 7,8-diol 9,10-epoxide (DE) adducts at the exocyclic N6-amino group of deoxyadenosine (dA) are well suited to this purpose because they are intercalated into DNA with well defined conformations. BaP, a product of incomplete combustion, is present in tobacco smoke (20–40 ng per cigarette) and has been implicated in the development of smoking-related lung cancer (14). Metabolism of BaP (15) yields DEs, which form mutagenic adducts (16) at the exocyclic amino groups of the purine bases (17) such as the dA adducts in the present study. The resultant mutations provide an attractive mechanism whereby these adducts can initiate cell transformation and cancer. The results to be reported here demonstrate that these adducts also act as potent Top2 poisons. This provides an additional mechanism for their carcinogenic activity, because levels of Top2 poisons that produce insufficient DNA damage to trigger apoptosis are believed to induce cancer via chromosomal translocations (3, 18, 19).
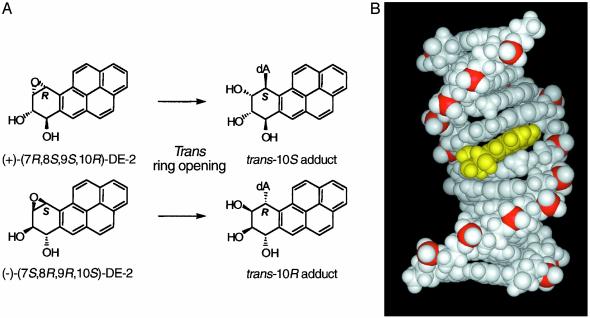
Recent studies from our laboratories established that site-specific BaP DE dA adducts, when incorporated either at the +1 position of the scissile strand or at the -1 position (nonscissile strand) relative to a normal Top1 cleavage site, efficiently trap Top1 cleavage complexes (20), which were readily detectable in the absence of camptothecin. The present study utilizes a similar approach to study the effect of these BaP DE dA adducts on cleavage and religation of duplex DNA by purified human Top2α. The adducts used are derived by trans-opening of the enantiomeric (+)-(7R,8S,9S,10R)- and (-)-(7S,8R,9R,10S)-DEs of 7,8,9,10-tetrahydro BaP (the DE-2 or anti-diastereomer in which the benzylic 7-hydroxyl group and the epoxide oxygen are trans; see Fig. 1A). The (+)-DE-2 enantiomer is the major DE metabolically formed from BaP (15), and unlike (-)-DE-2 is highly carcinogenic (21). The trans-(S) BaP DE dA adduct is derived from the (+)-DE-2 enantiomer, and the trans-(R) adduct is derived from (-)-DE-2 by inversion of configuration at C-10 (Fig. 1 A). These adducts are particularly useful as probes of DNA–enzyme interactions because the trans-(R) dA adducts intercalate toward the 5′ end of the modified strand (22, 23), whereas trans-(S) dA adducts intercalate toward its 3′ end (24, 25) (Figs. 1B and 2B).
Fig. 1.
(A) Structure of the trans-opened 10R and 10SN6-BaP DE-2 dA adducts. Note that trans-opening of the precursor DEs results in inversion of configuration at C-10. (B) View looking into the major groove of a minimized average NMR structure of an 11-mer duplex containing a trans-opened 10R BaP DE-2 dA adduct (yellow) intercalated toward the 5′ end (top) of the adducted strand (P. Pradhan, X. Liu, J.M.S., D.M.J., and H. J. C. Yeh, unpublished results). The phosphate backbone atoms are shown in red.
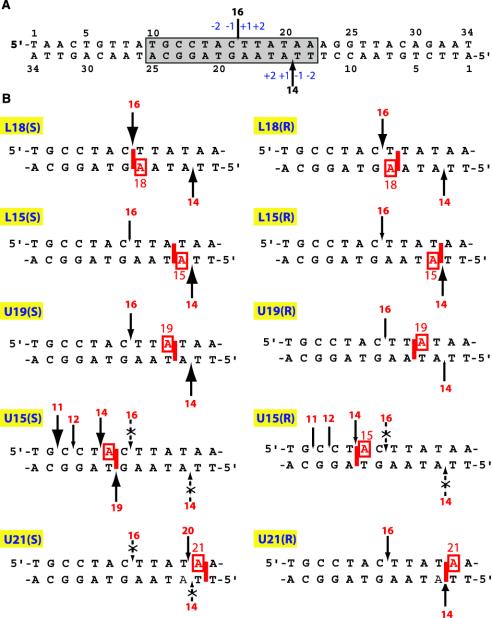
Fig. 2.
(A) Sequence of the oligonucleotide substrate derived from the simian virus 40 nuclear matrix-associated region. The Top2 cleavage site stabilized by VP-16 at position 4265 (34) generates a 16-mer oligonucleotide when the upper strand (U) is 5′-end-labeled and a 14-mer when the lower strand (L) is 5′-end-labeled. The shaded box corresponds to the sequences represented in B. The sizes of the arrows correspond to Top2 cleavage in the absence of VP-16. (B) Summary of the position-specific effects of intercalated BaP DE-2 dA adducts (only the boxed sequence in A is shown). The sizes of the arrows are in proportion to the intensity of Top2-mediated cleavage observed in the presence of the adduct independent of VP-16. Dotted arrows with x symbols denote suppression of Top2-mediated DNA cleavage regardless of the presence or absence of VP-16.
These trans-opened BaP DE dA adducts were incorporated into a 34-bp oligonucleotide (Fig. 2 A) derived from the simian virus 40 nuclear matrix-associated region (26, 27), which contains a single drug-sensitive staggered Top2 cleavage site (28). The oligonucleotide was well suited for these studies because of its relatively high dA content. This sequence allowed placement of BaP DE dA adducts at various positions relative to the known Top2 upper- and lower-strand cleavage sites (9 A/T base pairs of the 13 highlighted base pairs in Fig. 2 A). Our results indicate that depending on its position relative to the cleavage site, an adduct can either block cleavage or efficiently trap the Top2 homodimer by stabilizing the Top2–DNA cleavage complex.
Materials and Methods
Enzymes and Chemicals. Recombinant human Top2α was expressed in yeast and purified as described (29). T4 polynucleotide kinase was purchased from GIBCO/BRL, [γ-32P]ATP was purchased from New England Nuclear, polyacrylamide was purchased from Bio-Rad, and VP-16 and DMSO were purchased from Sigma.
Oligonucleotide Syntheses. Although we previously observed (20) that the 10R and 10S adducted diastereomers of BaP DE adducted oligonucleotides (up to 22-mers) were generally easily separable by HPLC, it seemed possible that the 10R/10S pairs of 34-mers (Fig. 2) in the present study might not separate because their greater length could overwhelm specific effects of the adducts on chromatographic retention. To avoid this potential difficulty, we synthesized each oligonucleotide from the individual, diastereomerically pure 10R or 10S phosphoramidites (see Materials and Methods, Scheme 1, and Figs. 5–7, which are published as supporting information on the PNAS web site, www.pnas.org). A semiautomated method with a manual step as described (30) for addition of the adducted 10R or 10S dA phosphoramidite was used. The oligonucleotides were first purified by reverse-phase HPLC as their 5′-dimethoxytrityl derivatives to separate the full-length adducted oligonucleotides from shorter failure sequences that occurred after coupling of the hydrocarbon adduct near the middle of the 34-mer sequence, and then rechromatographed after removal of the 5′-dimethoxytrityl group (see Table 1, which is published as supporting information on the PNAS web site, for HPLC conditions and retention times). When necessary, oligonucleotides were purified further by polyacrylamide gel electrophoresis. Unmodified oligonucleotides were obtained from MWG Biotech (High Point, NC).
Top2-Mediated DNA Cleavage Assays. Oligonucleotides were 5′-end-labeled with [γ-32P]ATP and T4 kinase (27). Annealing to the complementary strand was performed by heating the reaction mixture to 95°C and overnight cooling to room temperature in 10 mM Tris·HCl (pH 7.8)/100 mM NaCl/1 mM EDTA.
DNA substrates (≈10 pmol per reaction) were incubated with 500 ng of Top2α in the presence or absence of VP-16 (100 μM) for the indicated times at 25°Cin10 μl of reaction buffer (10 mM Tris·HCl, pH 7.5/50 mM KCl/5 mM MgCl2/1 mM ATP /0.2 mM DTT/0.1 mM EDTA/15 μg/ml BSA). Reactions were stopped by adding SDS (final concentration 0.5%). For the EDTA-induced religation experiments, the SDS stop was preceded by adding EDTA (10 mM final concentration) for the indicated times. Before loading the samples for electrophoresis, 3.3 volumes of Maxam–Gilbert loading buffer (98% formamide/0.01 M EDTA/10 mM NaOH/1 mg/ml xylene cyanol/1 mg/ml bromophenol blue) were added to reaction mixtures. Denaturing 16% polyacrylamide gels (7 M urea) were run at 40 V/cm at 50°C for 3 h. Imaging was performed with a PhosphorImager (Molecular Dynamics).
Results
Oligonucleotide Notation. Fig. 2 depicts the duplex oligonucleotides used in this study with a schematic representation for each of the corresponding trans-(R) or trans-(S) BaP DE-2 dA adducts. The experimental results for each of these oligonucleotides are summarized in legend for Fig. 2B. Our general conventions are: (i) the upper and lower strands are referred to as U and L, respectively; (ii) oligonucleotides are named according to the strand modified (U or L) followed by the position of the modified dA (counted from the 5′ end of the modified strand) and the (S) or (R) configuration at C-10 of the adduct; and (iii) an asterisk indicates which of the two strands was labeled and examined. For instance, in Fig. 3A, *U/L18(R) indicates that the substrate is a duplex oligonucleotide with an unmodified upper strand (U) labeled at the 5′ end (asterisk) and annealed to a lower strand bearing a trans-(R) BaP DE-2 dA adduct at position 18. 5′-End labeling of either strand allows detection of a 16-mer from the upper strand or a 14-mer from the lower strand in the presence of VP-16 when cleavage occurs at the normal staggered site (see Fig. 2 A).
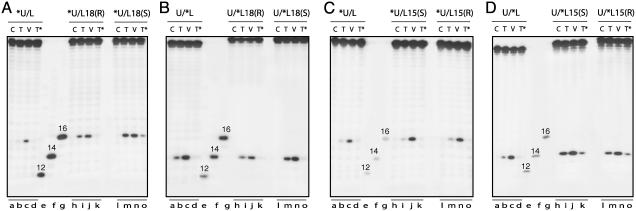
Fig. 3.
Enhancement of Top2-mediated DNA cleavage by trans-opened BaP DE-2 dA adducts intercalated at the cleavage sites. (A and B) Effects of L18 adducts on Top2-mediated DNA cleavage in the upper (A) and lower (B) strand, respectively. (C and D) Effects of L15 adducts on Top2-mediated DNA cleavage on the upper (C) and lower (D) strands, respectively. One strand of the duplex oligonucleotides was labeled at the 5′ terminus as indicated by the asterisk. C, DNA alone; T, +Top2; V, T + VP-16 (C, T, and V were incubated for 30 min at 25°C); T*, Top2 for 30 min as described above followed by an incubation with 10 mM EDTA for an additional 5 min. 5′-End-labeled 12-, 14-, and 16-mer oligonucleotides were used as markers.
Intercalation from the Lower Strand at the Normal Upper- and Lower-Strand Cleavage Sites or Immediately Adjacent to Them Within the Stagger Causes Accumulation of Top2-Mediated DNA Cleavage Complexes. We first examined the effect of lower-strand intercalated adducts at the dA located at positions L18 and L15. L18(S) and L15(R) have adducts intercalated at the upper- and lower-strand cleavage sites, respectively (Fig. 2). The L18(R) and L15(S) adducts are both within the stagger and are intercalated adjacent to their respective cleavage sites. For both the L18(R) and L18(S), Top2-mediated DNA cleavages on the upper strand (generating a 16-mer) accumulated independently of VP-16 (Fig. 3A, compare lanes i and m with lane b). Presence of the adduct at L18 substantially enhanced the neighboring cleavage on the upper strand, whereas less of an effect was observed on the lower-strand cleavage site, 3 bases away from the adducted dA (Fig. 3B). The observed accumulation of cleavage complexes can result from an increased rate of cleavage, a decreased rate of religation, or both. To determine the effect of the adducts on religation, we examined the effect of EDTA, which shifts the equilibrium of Top2–DNA complexes from cleavage to ligation by chelating the magnesium that promotes the forward reaction (31, 32). Persistence of the cleavage complex in the presence of EDTA is diagnostic of Top2 trapping by blockage of religation (32), although the ligation reaction in the presence of EDTA may not be an accurate mimic of the normal religation reaction. Fig. 3A shows that a significant fraction of the upper-strand cleavage product (16-mer) from the L18(S) oligonucleotide (lane o) but not L18(R) (lane k) failed to religate. In contrast, reversal of the lower-strand cleavage (generating a 14-mer; Fig. 3B, lanes k and o) was similar to control (oligo U/*L; Fig. 3B, lane d).
In the case of the L15(S) and L15(R) adducts, Top2-mediated DNA cleavage products accumulated independently of VP-16 in the lower strand (Fig. 3D, compare lanes i and m with lane b). The amount of cleavage product from the upper strand was affected only minimally (Fig. 3C, compare lanes i and m with lane b). Accumulation of the lower-strand cleavage complex was associated with reduced religation (Fig. 3D, compare lanes k and o with lane d), consistent with trapping of the Top2 cleavage complex by both intercalated (R) and (S) BaP DE adducts. Thus, as was observed for the L18 adducts, the intercalated L15 adducts caused accumulation of Top2 cleavage complexes at or immediately adjacent to the position of intercalation. Three of these four adducts, L18(S), L15(S), and L15(R), trapped the cleavage complex as shown by its failure to religate after addition of EDTA. Both L15(R) and U21(R) have the hydrocarbon moiety intercalated at the same lower-strand cleavage site but from opposite strands (Fig. 2B). In contrast to the substantial enhancement of lower-strand cleavage by L15(R) (Fig. 3D), only a weak effect was observed with U21(R) (data not shown).
Intercalation in the Middle of the Top2 Cleavage Stagger Decreases Top2-Mediated Cleavage. The U19(S) diastereomer intercalates adjacent to the lower-strand cleavage site between the same base pairs as the L15(S) adduct but from the upper strand (see Fig. 2B). This adduct caused the Top2-mediated DNA cleavage product to accumulate independently of VP-16 at the upper-strand 16-mer site (Fig. 4A, compare lanes h and b). Cleavage on the lower strand was also enhanced by the U19(S) adduct (Fig. 4B, compare lanes h and b) in a manner consistent with our observations with L15(S).
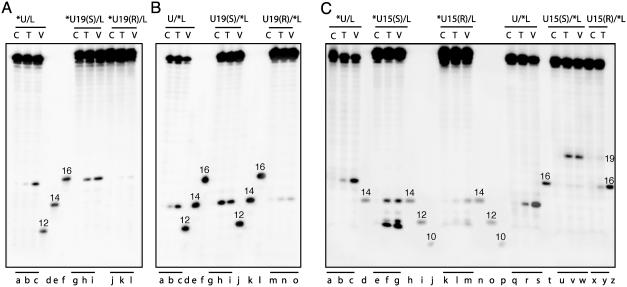
Fig. 4.
Attenuation of Top2-mediated DNA cleavage in the presence of trans-opened BaP DE-2 dA adducts intercalated either in the middle of the preexisting staggered double-stranded cleavage sites [A and B, U19(R)] or immediately outside of the staggered site [C, U15(S) and U15(R)] and induction of new cleavage sites [C, U15(S) and U15(R)]. One strand of each of the duplex oligonucleotides was labeled at the 5′ terminus as indicated by the asterisk. Symbols and markers are the same as described for Fig. 3.
In contrast, the diastereomeric U19(R) adduct intercalates in the center of the stagger equidistant from the upper- and lower-strand cleavage sites (see Fig. 2B). Interestingly, this U19(R) adduct decreased Top2-mediated DNA cleavage. VP-16 had no effect on cleavage in the presence of either U19(R)or(S) adducts (compare lanes l and k with c and b in Fig. 4A and lanes o and n with c and b in Fig. 4B). This result is in contrast to the enhancement of cleavage observed in Fig. 3 for other adducts in the presence of VP-16. Thus, intercalation needs to be at or adjacent to the cleavage site for efficient trapping of Top2-mediated DNA cleavage.
Intercalation Immediately Outside of the Preexisting Staggered Top2 Cleavage Sites Suppresses the Normal Top2 Cleavage Sites and Induces New Cleavages. We also investigated the effects of BaP DE adducts on the upper-strand dA at position U15 (see Fig. 2B). Both the diastereomeric U15(S) and U15(R) adducts completely suppressed Top2-mediated DNA cleavage at the preexisting sites on both strands (Fig. 4C). New cleavages were observed independently of VP-16 particularly for the U15(S) oligonucleotide (Fig. 4C). On the upper strand, three new sites generated 11-, 14- and 12-mers (Fig. 4C, lanes f and g), whereas on the lower strand, a single new site was observed, which generated a 19-mer (lanes v and w). Reversal experiments demonstrated that among these new sites, both the upper-strand 14-mer and the lower-strand 19-mer sites religated slowly (data not shown), consistent with the interpretation that the new cleavage sites caused by the U15(S) adduct are trapped by inhibition of religation. U21(S), which like U15(S) intercalates adjacent to the cleavage site and outside the stagger, suppressed normal cleavage in both strands and induced a small amount of displaced cleavage (20-mer) in the upper strand (data not shown).
Discussion
Previous studies had revealed that different classes of intercalators exhibit a strong bias for the specific bases immediately flanking Top2 cleavage sites (by convention, positions -1 and +1; Fig. 2 A) (33–35). Thus, it was proposed that intercalators as well as VP-16 and VM-26 trap Top2 by stacking with the base pairs (+1 and -1) flanking the cleavage site generated by Top2 (34). As a result, Top2-mediated DNA religation is inhibited. Freudenreich and Kreuzer provided further evidence for the “stacking model” with the phage T4 Top2 by using detailed mutational analysis (36) and drug crosslinking (37). The present study was designed to investigate the effects of covalently bound intercalators introduced site-specifically by chemical synthesis. Thus, the position of intercalation is rigorously defined and has been systematically varied.
Intercalation at the normal cleavage site (between positions -1 and +1) results in trapping of Top2 cleavage complexes. Thus, the present experiments provide the most direct evidence for the model proposed (33) on the basis of observations with noncovalently bound intercalating drugs. This drug-stacking/intercalation model is strikingly similar to the molecular mechanism proposed for trapping of Top1 by camptothecins (38), in which the camptothecins stack with the +1G at the cleavage site. Recently this model was refined by BaP DE intercalation studies (20) and by crystallography of a cleavage complex containing a camptothecin derivative (39). Thus, a common model emerges for both Top1 and Top2 poisons, whereby the drugs inhibit the enzymes by forming complexes comprising the drug and the covalent enzyme–DNA cleavage complex, thereby preventing religation of the DNA and its dissociation from the enzyme.
Trapping of cleavage complexes is generally more effective when the intercalated adduct at the cleavage site is covalently attached to the cleaved strand [compare L15(R) and U21(R) and L15(S) and U19(S); Fig. 2B]. In our present adducts the aromatic moiety intercalates from the major groove while a portion of the cyclohexene ring remains “visible” in the groove at the site of the modified adenine base (Fig. 1B). This suggests a possible role for major groove contacts with the enzyme in recognition of these bulky adducts. This contrasts with Top1, where minor groove contacts with the protein are more important (40–42).
In addition to intercalation at the cleavage site, intercalation within the “stagger” between the +1 and +2 positions from either strand also traps Top2 cleavage complexes (see Fig. 2) both when the adduct is on the +1 base [as with L15(S)] of the cleaved strand and when it is intercalated from the opposite strand between the +1 and +2 positions [L18(R) and U19(S)]. In these constructs, the adduct is located where it could alter the conformation at the position of attachment of the DNA via a 5′-phosphodiester linkage to the enzyme, thereby producing a misalignment with the 3′-hydroxyl group of the cleaved strand and inhibiting religation.
Our finding that intercalation at either side of position -2 (see Fig. 2) suppresses Top2 cleavage suggests that proper contacts between the protein and the DNA outside the 4-bp stagger are also required for the initial cleavage step to occur. Interestingly, suppression of normal cleavage can be accompanied by the appearance of new cleavage sites at or immediately adjacent to the position of intercalation [most notably the 19-mer cleavage of the lower strand and the 14-mer cleavage of the upper strand of U15(S)]. This suggests that one role of poisons that are intercalators may be to induce such “abnormal” cleavages and to trap the resultant cleavage complexes. Some strong intercalators such as ellipticines and anthracyclines (43, 44) are potent suppressors of Top2 cleavage at high concentration, whereas at low concentration they trap the cleavage complex. At high concentrations, these drugs potentially could occupy many sites on the DNA including positions distal to the normal Top2 cleavage sites, thereby preventing Top2 from cleaving DNA.
It was initially proposed that a single intercalated drug molecule was sufficient for trapping both subunits of a Top2 homodimer and forming a concerted DNA double-strand break because the base sequence requirement (-1A for doxorubicin) was often found at only one of the two cleavage sites of a Top2 cleavage complex (33). Some of our experiments provide clear evidence for this possibility. For example, the single U19(S) adduct enhanced both upper-strand cleavage to the 16-mer and lower-strand cleavage to the 14-mer. Even more strikingly, a single (S) BaP dA adduct at U15 induced the accumulation of abnormal upper-strand (11-mer) as well as lower-strand (19-mer) cleavage products, corresponding to displaced cleavage on both strands with the 4-base stagger, which is characteristic of concerted cleavage by a Top2 homodimer (see Fig. 2B). The existence of this stagger suggests that a single intercalated adduct likely traps both subunits of the homodimer. Conversely, concerted suppression of the normal Top2 cleavage site on both strands was also observed by a single BaP dA adduct at U15(S) or (R). However, the extent of cleavage can be asymmetrical and in several cases was significantly greater at or near the intercalation site. This was the case for the L18 and L15 adducts (Fig. 2B). Moreover, in the case of the U15(S) adduct, we even found “uncoupled cleavage” for two relatively weak sites in the upper strand (generating the 12- and 14-mers). Uncoupled (nonconcerted) cleavage of the two DNA strands by the two subunits of a Top2 homodimer was demonstrated recently by Osheroff and coworkers (45) using base mutations of a VP-16-specific sequence. We conclude that a single intercalated ligand can affect both subunits of a Top2 homodimer, although with some selectivity for the cleavage site close to the adduct, demonstrating that both subunits of a Top2 homodimer can function in concert.
In summary, the present study provides structural evidence for Top2 trapping and inhibition by DNA intercalators at specific positions relative to a known Top2 cleavage site. Our observations can be summarized as follows. (i) Intercalation within the stagger at or immediately adjacent to a cleavage site results in the accumulation of cleavage complexes, due at least in part to blocking of religation. (ii) An intercalator at one of the two staggered cleavage sites can induce the accumulation of cleavage complexes derived from both DNA strands, indicative of partial coupling between the reactions of the two Top2 subunits. (iii) Intercalation outside the stagger blocks cleavage at these sites and can induce new cleavages at and adjacent to the position of intercalation. (iv) Intercalation at the center of the stagger, two bases away from both the upper- and lower-strand cleavage sites, reduces cleavage complex formation both in the absence and presence of VP-16. By delineating the relationship between the position of intercalation of an adduct and its ability to trap the Top2 homodimer, our results provide a basis for modeling poisons in Top2 cleavage complexes and potentially for the development of novel drugs to inhibit the enzyme.
Top2 is relatively insensitive to small, covalent DNA lesions such as 8-oxopurines, O6-methyl deoxyguanosine, and N6-methyl dA (≤3-fold enhancement of cleavage). However, 1,N6-ethenoadenine adducts or abasic sites significantly enhance accumulation of Top2 cleavage complexes at the damaged site as a result of an increased cleavage rate but a nearly normal religation rate (13). The present BaP DE dA adducts, in contrast, are strong inhibitors of religation at several sites, as shown by the failure of the cleavage complexes to religate in the presence of EDTA. Thus, the present results also demonstrate Top2 poisoning by the bulky adducts that are formed after reaction of DNA with a carcinogenic DE metabolite of a polycyclic aromatic hydrocarbon. Such adducts are known to be mutagenic, and the present observations suggest the possibility of an additional mechanism for cell transformation mediated by these adducts, namely the potential for chromosomal damage induced by Top2 poisoning.
Supplementary Material
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Top, human topoisomerase; VP-16, etoposide; BaP, benzo[a]pyrene; DE, 7,8-diol 9,10-epoxide; dA, deoxyadenosine.
References
- 1.Wang, J. C. (1996) Annu. Rev. Biochem. 65, 635-692. [DOI] [PubMed] [Google Scholar]
- 2.Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430-440. [DOI] [PubMed] [Google Scholar]
- 3.Wilstermann, A. M. & Osheroff, N. (2003) Curr. Top. Med. Chem. 3, 321-338. [DOI] [PubMed] [Google Scholar]
- 4.Champoux, J. J. (2001) Annu. Rev. Biochem. 70, 369-413. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. (1996) Nature 379, 225-232. [DOI] [PubMed] [Google Scholar]
- 6.Burden, D. A. & Osheroff, N. (1998) Biochim. Biophys. Acta 1400, 139-154. [DOI] [PubMed] [Google Scholar]
- 7.Nitiss, J. L. (1998) Biochim. Biophys. Acta 1400, 63-81. [DOI] [PubMed] [Google Scholar]
- 8.Zechiedrich, E. L., Christiansen, K., Andersen, A. H., Westergaard, O. & Osheroff, N. (1989) Biochemistry 28, 6229-6236. [DOI] [PubMed] [Google Scholar]
- 9.Berger, J. M. & Wang, J. C. (1996) Curr. Opin. Struct. Biol. 6, 84-90. [DOI] [PubMed] [Google Scholar]
- 10.Fass, D., Bogden, C. E. & Berger, J. M. (1999) Nat. Struct. Biol. 6, 322-326. [DOI] [PubMed] [Google Scholar]
- 11.Pommier, Y., Goldwasser, F. & Strumberg, D. (2001) in Cancer Chemotherapy and Biotherapy: Principle and Practice, ed. Longo, D. L. (Lippincott Williams & Wilkins, Philadelphia), pp. 538-578.
- 12.Chen, A. Y. & Liu, L. F. (1994) Annu. Rev. Pharmacol. Toxicol. 34, 191-218. [DOI] [PubMed] [Google Scholar]
- 13.Sabourin, M. & Osheroff, N. (2000) Nucleic Acids Res. 28, 1947-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denissenko, M. F., Pao, A., Tang, M. & Pfeifer, G. P. (1996) Science 274, 430-432. [DOI] [PubMed] [Google Scholar]
- 15.Thakker, D. R., Yagi, H., Akagi, H., Koreeda, M., Lu, A. H., Levin, W., Wood, A. W., Conney, A. H. & Jerina, D. M. (1977) Chem. Biol. Interact. 16, 281-300. [DOI] [PubMed] [Google Scholar]
- 16.Sayer, J. M., Kroth, H., Yagi, H., Kalena, G., Jerina, D. M., Ramos, L. A., Dipple, A., Ponten, I. & Goodman, M. F. (2002) Polycyclic Aromat. Compd. 22, 871-879. [Google Scholar]
- 17.Jerina, D. M., Chadha, A., Cheh, A. M., Schurdak, M. E., Wood, A. W. & Sayer, J. M. (1991) in Biological Reactive Intermediates IV: Molecular and Cellular Effects and Their Impact on Human Health, eds. Witmer, C. M., Snyder, R., Jollow, D. J., Kalf, G. F., Kocsis, J. J. & Sipes, L. G. (Plenum, New York), pp. 533-553.
- 18.Lovett, B. D., Strumberg, D., Blair, I. A., Pang, S., Burden, D. A., Megonigal, M. D., Rappaport, E. F., Rebbeck, T. R., Osheroff, N., Pommier, Y. G. & Felix, C. A. (2001) Biochemistry 40, 1159-1170. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, Y., Strissel, P., Strick, R., Chen, J., Nucifora, G., Le Beau, M. M., Larson, R. A. & Rowley, J. D. (2002) Proc. Natl. Acad. Sci. USA 99, 3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pommier, Y., Laco, G. S., Kohlhagen, G., Sayer, J. M., Kroth, H. & Jerina, D. M. (2000) Proc. Natl. Acad. Sci. USA 97, 10739-10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buening, M. K., Wislocki, P. G., Levin, W., Yagi, H., Thakker, D. R., Akagi, H., Koreeda, M., Jerina, D. M. & Conney, A. H. (1978) Proc. Natl. Acad. Sci. USA 75, 5358-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zegar, I. S., Chary, P., Jabil, R. J., Tamura, P. J., Johansen, T. N., Lloyd, R. S., Harris, C. M., Harris, T. M. & Stone, M. P. (1998) Biochemistry 37, 16516-16528. [DOI] [PubMed] [Google Scholar]
- 23.Volk, D. E., Rice, J. S., Luxon, B. A., Yeh, H. J., Liang, C., Xie, G., Sayer, J. M., Jerina, D. M. & Gorenstein, D. G. (2000) Biochemistry 39, 14040-14053. [DOI] [PubMed] [Google Scholar]
- 24.Yeh, H. J., Sayer, J. M., Liu, X., Altieri, A. S., Byrd, R. A., Lakshman, M. K., Yagi, H., Schurter, E. J., Gorenstein, D. G. & Jerina, D. M. (1995) Biochemistry 34, 13570-13581. [DOI] [PubMed] [Google Scholar]
- 25.Pradhan, P., Tirumala, S., Liu, X., Sayer, J. M., Jerina, D. M. & Yeh, H. J. (2001) Biochemistry 40, 5870-5881. [DOI] [PubMed] [Google Scholar]
- 26.Pommier, Y., Cockerill, P. N., Kohn, K. W. & Garrard, W. T. (1990) J. Virol. 64, 419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier, Y., Capranico, G., Orr, A. & Kohn, K. W. (1991) J. Mol. Biol. 222, 909-924. [DOI] [PubMed] [Google Scholar]
- 28.Capranico, G., Tinelli, S., Zunino, F., Kohn, K. W. & Pommier, Y. (1993) Biochemistry 32, 145-152. [DOI] [PubMed] [Google Scholar]
- 29.Wasserman, R. A., Austin, C. A., Fisher, L. M. & Wang, J. C. (1993) Cancer Res. 53, 3591-3596. [PubMed] [Google Scholar]
- 30.Kroth, H., Yagi, H., Sayer, J. M., Kumar, S. & Jerina, D. M. (2001) Chem. Res. Toxicol. 14, 708-719. [DOI] [PubMed] [Google Scholar]
- 31.Osheroff, N. (1987) Biochemistry 26, 6402-6406. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, M. J. & Osheroff, N. (1990) Biochemistry 29, 2511-2515. [DOI] [PubMed] [Google Scholar]
- 33.Capranico, G., Kohn, K. W. & Pommier, Y. (1990) Nucleic Acids Res. 18, 6611-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pommier, Y., Capranico, G., Orr, A. & Kohn, K. W. (1991) Nucleic Acids Res. 19, 5973-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capranico, G. & Binaschi, M. (1998) Biochim. Biophys. Acta 1400, 185-194. [DOI] [PubMed] [Google Scholar]
- 36.Freudenreich, C. H. & Kreuzer, K. N. (1993) EMBO J. 12, 2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freudenreich, C. H. & Kreuzer, K. N. (1994) Proc. Natl. Acad. Sci. USA 91, 11007-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaxel, C., Capranico, G., Kerrigan, D., Kohn, K. W. & Pommier, Y. (1991) J. Biol. Chem. 266, 20418-20423. [PubMed] [Google Scholar]
- 39.Staker, B. L., Hjerrild, K., Feese, M. D., Behnke, C. A., Burgin, A. B., Jr., & Stewart, L. (2002) Proc. Natl. Acad. Sci. USA 99, 15387-15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redinbo, M. R., Stewart, L., Kuhn, P., Champoux, J. J. & Hol, W. G. J. (1998) Science 279, 1504-1513. [DOI] [PubMed] [Google Scholar]
- 41.Pommier, Y., Kohlhagen, G., Laco, G. S., Kroth, H., Sayer, J. M. & Jerina, D. M. (2002) J. Biol. Chem. 277, 13666-13672. [DOI] [PubMed] [Google Scholar]
- 42.Tian, L., Sayer, J. M., Kroth, H., Kalena, G., Jerina, D. M. & Shuman, S. (2003) J. Biol. Chem. 278, 9905-9911. [DOI] [PubMed] [Google Scholar]
- 43.Tewey, K. M., Chen, G. L., Nelson, E. M. & Liu, L. F. (1984) J. Biol. Chem. 259, 9182-9187. [PubMed] [Google Scholar]
- 44.Pommier, Y., Schwartz, R. E., Zwelling, L. A. & Kohn, K. W. (1985) Biochemistry 24, 6406-6410. [DOI] [PubMed] [Google Scholar]
- 45.Bromberg, K. D., Burgin, A. B. & Osheroff, N. (2003) J. Biol. Chem. 278, 7406-7412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.