Abstract
Oxygen availability affects the transcription of a number of genes in nearly all organisms. Although the molecular mechanisms for sensing oxygen are not precisely known, heme is thought to play a pivotal role. Here, we address the possibility that oxygen sensing in yeast, as in mammals, involves a redox-sensitive hemoprotein. We have found that carbon monoxide (CO) completely blocks the anoxia-induced expression of two hypoxic genes, OLE1 and CYC7, partially blocks the induction of a third gene, COX5b, and has no effect on the expression of other hypoxic or aerobic genes. In addition, transition metals (Co and Ni) induce the expression of OLE1 and CYC7 in a concentration-dependent manner under aerobic conditions. These findings suggest that the redox state of an oxygen-binding hemoprotein is involved in controlling the expression of at least two hypoxic yeast genes. By using mutants deficient in each of the two major yeast CO-binding hemoproteins (cytochrome c oxidase and flavohemoglobin), respiratory inhibitors, and cob1 and ρ0 mutants, we have found that the respiratory chain is involved in the anoxic induction of these two genes and that cytochrome c oxidase is likely the hemoprotein “sensor.” Our findings also indicate that there are at least two classes of hypoxic genes in yeast (CO sensitive and CO insensitive) and imply that multiple pathways/mechanisms are involved in modulating the expression of hypoxic yeast genes.
With the exception of strict anaerobes, all organisms depend on oxygen and have had to evolve sensory systems to gauge oxygen availability and mechanisms for enduring periods of hypoxia or anoxia. The hypoxic response for most organisms comprises a complex biochemical and genetic program that includes the differential expression of a large number of genes (1, 2). Thus, oxygen availability affects the intracellular levels, and often activities, of a large number of proteins in most organisms. Currently, the sensory mechanisms for gauging oxygen availability have not been precisely defined, nor is it known whether similar oxygen-sensing mechanisms are found among different taxonomic groups. However, one component that appears to be involved in most, if not all, organisms is heme (2).
In vertebrates, heme likely functions as a redox-sensitive component of a hemoprotein oxygen sensor. This conclusion is supported by two findings (1–5): (i) carbon monoxide (CO) blocks the induction of hypoxic genes in many cells and cell lines at low oxygen concentrations; and (ii) transition metals, which compromise the ability of hemoproteins to bind O2, induce these same genes under normoxic conditions. So far, two possible hemoprotein oxygen sensors have been implicated. The first is a multisubunit plasma-membrane-bound cytochrome b NAD(P)H oxidase that is thought to reduce oxygen to superoxide (6–10). Superoxide and subsequently generated reactive oxygen species (ROS) may act as second messengers, controlling the activity of the trans-acting factor(s) that regulate the transcription of hypoxic genes (2, 10, 11). The second is cytochrome a3 of the mitochondrial respiratory chain (12–17). Although the signaling pathway from cytochrome c oxidase is not yet clearly defined, a recent report (18), which appeared during the revision of this manuscript, suggests that ROS may be involved here too.
Heme is also involved in regulating the transcription of oxygen-responsive genes in the yeast Saccharomyces cerevisiae (2, 19, 20). The effects of heme have been shown to be mediated, in part, by several transcription factors. These include Hap1p and the Hap2/3/4/5p complex, which activate the transcription of many aerobic and some hypoxic genes, and Rox1p, which represses the transcription of most hypoxic genes (19–22). Previously, it was noted that there are at least two ways that heme might function in regulating the transcription of oxygen-responsive genes (2, 23). In the first, heme would serve as a ligand that binds to a transcription factor and thus heme concentration would be important. In the second pathway, heme would function as a redox-sensitive component of a hemoprotein, and thus heme redox state would be important, as has been proposed for vertebrate cells. Current models for oxygen sensing in yeast favor the first type of pathway (19, 22). These models propose that oxygen availability affects cellular heme levels, and that this, in turn, affects the intracellular levels and/or activities of these transcription factors (see discussions in refs. 19, 22, 24).
Recent studies have revealed that oxygen concentration affects the expression of several S. cerevisiae genes over a range of oxygen concentrations in which heme concentration is not expected to vary (20, 25). Therefore, it is likely that other mechanisms besides those that affect heme concentration are involved in oxygen sensing in this organism. In this study, we have sought to determine whether redox-sensitive hemoprotein(s), similar to those proposed for mammalian cells, function in oxygen sensing in this yeast.
MATERIALS AND METHODS
Yeast Strains and Media.
The following S. cerevisiae strains were used: JM43 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 [ρ+]) (26); JM43 ρ0 (MAT α his4–580 trp1–289 leu2–3 112 ura3–52 [ρ0] (27); JM43-GD5ab (MAT α his4–580 trp1–289 leu2–3, 112 ura3–52 cox5b∷LEU2 cox5a∷URA3 [ρ+](28); DR11 (MATα his4–580 trp1–289 leu2–3, 112 ura3–52 yhb1∷ URA3 [ρ+] (29); and aM7–40-5b (MAT a ade1 [omega+ cob1− ρ+]), a cytb− mutant (30). Strains were grown in SSG-TEA, a semisynthetic galactose medium supplemented with Tween 80, ergosterol, silicon antifoam, and containing amino acids and uracil, as needed (31).
Growth Conditions.
Precultures were grown aerobically in a shaker (200 rpm) at 28°C to midlogarithmic phase. For experiments using gas mixtures, midlogarithmic phase precultures were used to inoculate a New Brunswick Scientific BioFloIIc fermentor containing 3.5 l of SSG-TEA media. Temperature, pH, agitation, and sparge rate were maintained at 28°C, 5.0, 300 rpm, and 4 l/min, respectively. The dissolved oxygen concentration in the fermentor was monitored with an Ingold (Wilmington, MA) polarographic oxygen electrode (12 mm). Anoxic cultures were grown in the dark (cf. ref. 30), and the gas mixture was passed through an OxyClear O2 scrubber (LabClear, Oakland, CA) to prevent trace oxygen from entering the fermentor. Aerobic cultures were grown in the fermentor, by using air as the sparge gas, or in Delong flasks containing a volume of medium of ≤1/4 the flask volume on a shaker (200 rpm) at 28°C. Cell growth was followed by measuring turbidity with a Klett–Summerson colorimeter fitted with a no. 54 green filter. Cultures were harvested during midlogarithmic phase at a density of 60 to 100 Klett units (1 to 2.5 × 107 cells/ml). For some studies, cells were harvested in stationary phase (≈300 Klett units), as indicated. To harvest cells, cultures were quick-chilled to −4°C, pelleted by centrifugation (4°C), washed once with ice-cold diethylpyrocarbonate-treated distilled water, and flash-frozen in liquid N2.
RNA Isolation and Hybridization.
Total RNA was isolated as described previously (32). RNA samples (−30 μg) were denatured and separated on 1.5% agarose gels containing Mops–formaldehyde buffer (33). The RNA was transferred to Nytran Plus membranes (Schleicher & Schuell) and hybridized as described previously (34). DNA probes were prepared by random-primer labeling of double-stranded DNA fragments by using α-32P-dCTP (DuPont NEN; cf. ref. 35). The following probes were used: a 520-bp StyI fragment of ACT1, the gene encoding actin; a 370-bp AccI/BglII fragment of COX5b, the gene encoding subunit Vb of cytochrome c oxidase; a 400-bp KpuI/XhoI fragment of CYC7, the gene encoding iso-2-cytochrome c; and a 400- and 420-bp EcoRI fragment of OLE1, the gene encoding Δ-9 fatty acid desaturase. Hybridization and stringency washes were performed as described previously (36). Signal intensity was quantitated with an AMBIS Radioanalytical Imaging System or a Molecular Dynamics STORM 860 Phosphorimager. To quantify transcript levels, signal intensity was normalized to that for ACT1 mRNA. For most experiments, two or more independent RNA blot analyses were performed for each growth condition and transcript examined. The variance in transcript level was typically within 15%.
Other.
All gases (except air) were Matheson-certified standards obtained from U.S. Welding (Denver, CO). The following gas mixtures were used: 2.5% CO2 in O2-free N2, 12.5% CO2 in O2-free N2, and 100% CO. For incubations with 80% CO, 100% CO was mixed with 12.5% CO2 in O2-free N2 by using a solenoid valve running a duty cycle of 80/20, respectively.
RESULTS
Effect of CO on the Expression of Oxygen-Regulated Genes After a Shift from Normoxia to Anoxia.
CO has remarkable specificity in biological systems, binding noncovalently to ferrous heme groups in a small number of hemoproteins. CO markedly reduces or blocks the induction of hypoxic genes in mammalian cells, a finding that is consistent with the involvement of the redox or spin state of a hemoprotein in the oxygen-sensing pathway for these genes (2, 3, 37, 38). To address whether similar hemoprotein-dependent oxygen-sensing mechanisms are found in yeast, we examined the effects of CO on the transcription of ten hypoxic genes and four aerobic genes when cells are shifted from aerobic to anoxic conditions (Table 1). CO affected the expression of only three of these genes. All three (COX5b, CYC7, and OLE1) are hypoxic genes. It should be noted that the expression of OLE1 was examined under fatty acid-repressing conditions (39) because Tween 80, which is required for anaerobic growth, was present in the growth medium. CO had no effect on the induction of the other seven hypoxic genes examined (HEM13, HMG1, HMG2, ERG11, CPR1 [NCP1], ANB1, and AAC3) or on the anoxia-induced decline in transcript levels (ref. 25; K.E.K., P.V.B., and R.O.P., unpublished observations) from the four aerobic genes (COX5a, TIF51A, AAC2, or ROX1) (data not shown).
Table 1.
Effect of CO on expression of oxygen-regulated yeast genes after a shift from normoxic to anoxic conditions
| Gene | Protein | Activator* | Repressor* | O2 effect* | Co effect† |
|---|---|---|---|---|---|
| Oxidative phosphorylation | |||||
| COX5a | Cytochrome c oxidase subunit Va | Hap2/3/4/5 | Induced | None | |
| COX5b | Cytochrome c oxidase subunit Vb | Rox1 | Repressed | Partial inhibition | |
| CYC7 | iso-2-cytochrome c | Hap1 | Rox1 | None/repressed | Total inhibition |
| Heme, sterol, and fatty acid synthesis | |||||
| HEM13 | Coproporphyrinogen III oxidase | Hap1 | Rox1 | Repressed | None |
| HMG1 | 3-hydroxy-3-methylglutaryl CoA reductase | Hap1 | Rox1 | Induced | None |
| HMG2 | 3-hydroxy-3-methylglutaryl CoA reductase | Rox1 | Repressed | None | |
| ERG11 | Cytochrome P450 lanosterol 14α-demethylase | Hap1 | Rox1 | Repressed | None |
| CPR1 (NCP1) | NADPH cytochrome P450 reductase | Rox1 | Repressed | None | |
| OLE1 | Δ-9 fatty acid desaturase | Rox1 | Repressed | Total inhibition | |
| Translation factors | |||||
| TIF51a | elF-5a | Hap1 | Induced | None | |
| ANB1 | elF-5b | Rox1 | Repressed | None | |
| Mitochondrial adenine translocases | |||||
| AAC2 | ADP/ATP translocator | Hap2/3/4/5 | Induced | None | |
| AAC3 | ADP/ATP translocator | Rox1 | Repressed | None | |
| Transcriptional regulators | |||||
| ROX1 (REO1) | DNA-binding transcriptional repressor | Hap1 | Induced | None | |
Hypoxic genes are repressed by Rox 1 p in air; aerobic genes and some hypoxic genes are activated (induced) by either Hap1 or Hap2/3/4/5p (20).
Determined by quantitating the effect of CO on transcript levels every 2 or 4 h over a 24 h period.
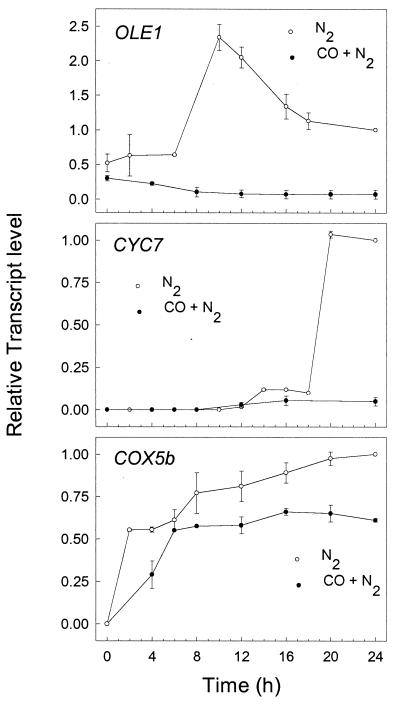
Fig. 1 shows the change in mRNA levels of OLE1, CYC7, and COX5b after shifting cells from air to anoxic conditions in the presence or absence of CO. Each gene is induced with different kinetics in the absence of CO. The level of OLE1 transcript increases gradually to a level that is >2-fold higher than its anoxic level at 10 h and then declines to it anoxic level by 16 h. The level of CYC7 transcript remains low for 12 h and then increases rapidly to its anoxic level at about 20 h, and COX5b mRNA increases continuously from the time of the shift, reaching its anoxic level at about 24 h. In the presence of CO, the anoxic induction of both OLE1 and CYC7 is completely blocked, while the anoxic induction of COX5b is partially blocked. No further increase in the levels of these transcripts was observed at 30 h, and incubations with CO did not affect anoxic growth rates (data not shown). Together, these findings clearly show that CO inhibits the anoxic induction of a subset of yeast hypoxic genes and suggest that an O2- and CO-binding hemoprotein is involved in regulating their expression.
Figure 1.
Effect of CO on the anoxic induction of OLE1, CYC7, and COX5b. S. cerevisiae strain JM43 was grown aerobically and then inoculated into a fermentor that was preequilibrated with 2.5% CO2 in O2-free N2 (N2, open symbols) or 80% CO with 2.5% CO2 in O2-free N2 (CO + N2, filled symbols). At the indicated times, cells were harvested and total RNA was isolated, fractionated, and hybridized as described in Materials and Methods. The level of each transcript was normalized to ACT1 mRNA, and this ratio is expressed relative to the same ratio in steady state anoxic cells. Data are expressed as means ± error ranges (n = 2).
Effects of Transition Metals on the Expression of Hypoxic Genes.
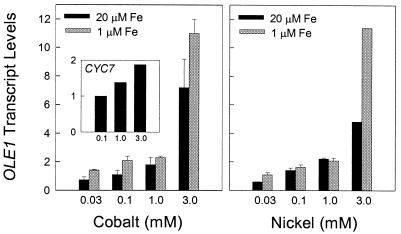
Another way to assess the involvement of hemoproteins in the transcription of these genes is to examine the effects of cobalt and nickel on their expression under normoxic conditions. In mammalian cells, these transition metals induce a number of hypoxic genes under normoxic conditions (2, 3, 40, 41). They have been shown to serve as substrates for ferrochelatease and to be incorporated into protoporphyrin IX in place of Fe, both in vivo and in cultured cells (42, 43). Nickel-substituted hemoproteins do not bind O2 (44), while cobalt-substituted hemoproteins have an exceedingly low affinity for O2 (45). To determine whether these metals have a similar effect in yeast, we examined the two hypoxic genes (OLE1 and CYC7) whose induction was completely inhibited by CO. Fig. 2 shows that both CoCl3 and NiCl3 give a concentration-dependent increase in OLE1 transcript levels. In contrast to its anoxic induction, OLE1 was maximally induced by these transition metals within 2 h. Over this short incubation period, neither cobalt nor nickel affects growth rate, and no further increase in transcripts was observed by increasing the concentration of either metal. The level of OLE1 induction achieved with NiCl3 was, in general, similar to that achieved with the same concentration of CoCl3. In nearly all cases, the level of OLE1 induction was higher in media containing 1 μM Fe than in media containing 20 μM Fe. This finding is similar to results obtained with the erythropoietin gene in cultured Hep3B cells (41); it has been interpreted to mean that Fe suppresses the normoxic induction of hypoxic genes by competing with Co or Ni as substrates for ferrochelatase. However, in yeast cells, higher concentrations of these metals are required for maximal induction of hypoxic genes than in mammalian cells (4, 41). It is not known whether this difference between yeast and mammalian cells is caused by differences in rates of cellular uptake, intracellular chelation, or incorporation into protoporphyrin IX.
Figure 2.
Effect of transition metals on the expression of OLE1 and CYC7 under normoxic conditions. Cells were grown aerobically to midlogarithmic phase for OLE1 or stationary phase for CYC7 in SSG-TEA media on a shaker at 28°C. They were incubated for 2 h with CoCl3 or NiCl3, in either low- (1 μM, shaded bars) or high- (20 μM, filled bars) iron media. Total RNA was isolated, fractionated, and hybridized as described in Materials and Methods. Transcript levels were normalized to the level of ACT1 mRNA and are presented as a decimal percent of their steady-state aerobic levels in the absence of transition metals. Data are presented as means ± error ranges for two independent experiments, except for CYC7 (n = 1).
Cobalt chloride also induced the expression of CYC7 under aerobic conditions (Fig. 2 Left Inset), but the level of induction was modest compared with that observed for OLE1. Interestingly, this induction was observed only with cultures grown to high cell density (i.e., stationary phase) and not with cells grown to midlogarithmic phase (data not shown) as was the case with OLE1. At present, it is unclear why growth phase would affect the ability of cobalt to induce CYC7 but not OLE1. Nickel had little, if any, effect on CYC7 expression.
Expression of OLE1 and CYC7 in Cytochrome c Oxidase- or Flavohemoglobin-Deficient Strains.
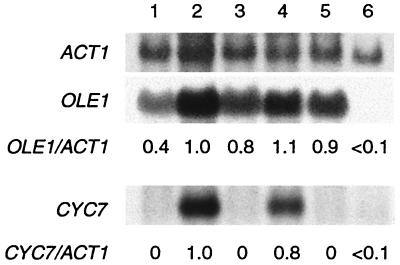
The above findings support the hypothesis that a CO-binding hemoprotein is involved in mediating the oxygen-sensitive expression of OLE1, CYC7, and possibly COX5b. As a first step toward identifying this hemoprotein, we examined the expression of OLE1 and CYC7 in strains that lack cytochrome c oxidase or flavohemoglobin, proteins that have been implicated in O2-sensing pathways in other organisms (2), and the most abundant CO-binding hemoproteins expressed in aerobic yeast cells (27, 29, 46, 47). From the results shown in Fig. 3, it is clear that transcripts of both OLE1 and CYC7 are induced under anoxic conditions in DR11, a strain that carries a null mutation in YHB1 and that lacks any detectable flavohemoglobin (29), just as they are in the wild-type strain JM43. In contrast, neither gene is induced under anoxic conditions in JM43-GD5ab, a strain with null mutations in both COX5a and COX5b and that lacks assembled cytochrome c oxidase (36). These findings indicate that cytochrome c oxidase, but not flavohemoglobin, is required for the anoxic induction of OLE1 and CYC7. Interestingly, aerobic transcript levels from OLE1 were elevated 2-fold in JM43-GD5ab, suggesting that the expression of this gene may be partially repressed by cytochrome c oxidase in aerobic cells.
Figure 3.
Effect of null mutations in cytochrome c oxidase and flavohemoglobin on the expression of OLE1 and CYC7 under aerobic and anoxic conditions. Cells were grown for at least six generations under aerobic or anoxic conditions and harvested in midlogarithmic phase. Total RNA was isolated, fractionated, and hybridized as described in Materials and Methods. All transcript levels were normalized to ACT1 mRNA and are presented as a decimal percent relative to that found in strain JM43 under steady-state anoxic conditions (lane 2), as indicated below each lane. Lane 1, JM43 (wild type) in air; lane 2, JM43 in N;, lane 3, DR11 (Δyhb1 ρ+) in air; lane 4, DR11 in N2; lane 5, JM43-GD5ab (Δcox5aΔcox5b ρ+) in air; and lane 6, JM43-GD5ab in N2.
A Functional Respiratory Chain Is Required for the Anoxic Induction of OLE1 and CYC7.
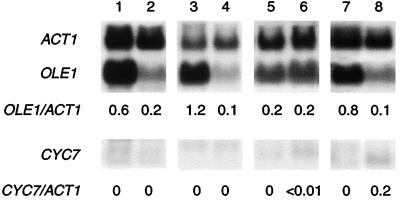
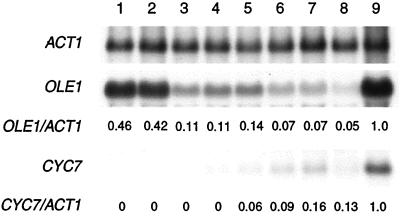
To further explore how cytochrome c oxidase might function in the anoxic induction of OLE1 and CYC7, we asked whether mitochondrial respiration per se is also involved. Fig. 4 shows that the respiratory inhibitors antimycin A and cyanide block the anoxic induction of CYC7. Although there was a slight increase in CYC7 transcript level under anoxia, with cyanide the level of induction was several-fold lower (>100-fold) than that observed in the wild-type strain (Fig. 3, lanes 1 and 2). These inhibitors also block the anoxic induction of OLE1. Compared with untreated JM43 cells (Fig. 3, lane 2) the anoxic expression of OLE1 was reduced 5-fold by both antimycin A and cyanide. The anoxic induction of these genes is also dramatically reduced in a cytochrome b-deficient mutant, aM7–40-5B, and in JM43ρ0, which lacks a mitochondrial genome and a functional respiratory chain. Neither these inhibitors nor mutations blocked the anoxic induction of those hypoxic genes that are unaffected by CO (i.e., HEM13, HMG1, HMG2, ERG11, CPR1 [NCP1], ANB1, and AAC3). Although aerobic transcript levels of CYC7 were similar to those in the wild-type strain with all treatments, aerobic transcript levels of OLE1 were elevated slightly in JM43 cells treated with antimycin A, JM43ρ0, and in aM7–40-5B, compared with untreated JM43 (Fig. 3, lane 1). In contrast, aerobic transcripts of OLE1 were reduced in the presence of cyanide. These data, together with those shown in Fig. 3, indicate that the mitochondrial respiratory chain is involved in the anoxic induction of both OLE1 and CYC7 and support the hypothesis that the CO-binding hemoprotein involved is cytochrome c oxidase.
Figure 4.
Effect of respiratory inhibitors and mutations in the respiratory chain on the expression of OLE1 and CYC7 under aerobic and anoxic conditions. Total RNA was isolated from cells grown for six generations, under aerobic or anoxic conditions, and quantitated as in the legend to Fig. 3. All transcript levels were normalized to ACT1 mRNA and are presented as a decimal percent relative to that found in strain JM43 grown under anoxic conditions, as indicated below each lane. Lane 1, JM43 (wild type) grown in air with 1 μM antimycin A; lane 2, JM43 grown in N2 with 1 μM antimycin A; lane 3, aM7–40-5B (cob1−) in air; lane 4, aM7–40-5B in N2; lane 5, JM43 grown in air with 1 mM KCN; lane 6, JM43 grown in N2 with 1 mM KCN; lane 7, JM43ρ0 in air; and lane 8, JM43ρ0 in N2.
Effects of Transition Metals on OLE1 Expression in Respiration-Deficient Yeast Strains.
To determine whether the effect of transition metals on the expression of OLE1 (Fig. 2) are mediated by the respiratory chain, we examined the ability of cobalt to induce OLE1 transcripts in respiration-deficient strains. From Table 2 it is clear that although cobalt induces the normoxic expression of OLE1 in both JM43–GD5ab and JM43ρ0, the level of induction is substantially lower than that observed in the respiration-proficient parent JM43. These results suggest that a substantial fraction of OLE1 induction by cobalt is mediated by the mitochondrial respiratory chain, but that nonmitochondrial mechanisms also contribute. These results are in contrast to results from a recent study with Hep3b cells (18), in which investigators found no attenuation in the level of induction of EPO or HIF-1 transcripts by cobalt in ρ0 cells. These findings point to uncertainty as to the precise mechanism(s) by which transition metals induce the expression of hypoxic genes in different cell types but suggest that the effect of these metals is mediated largely by the mitochondrion in yeast cells.
Table 2.
Induction of OLE1 transcripts by cobalt in respiratory-deficient strains
| Yeast strain | OLE1 induction |
|---|---|
| JM43 | 7.3 ± 1.7* |
| JM43-GD5ab | 2.3 ± 0.2 |
| JM43ρ0 | 2.1 ± 0.2 |
After a 2-h aerobic incubation with 3 mM CoC13 OLE1, transcripts were measured and normalized to ACT1 mRNA. Data are presented as a decimal percent relative to no added cobalt (mean ± error range, n = 2).
Does the Respiratory Chain Produce a Signal During the Transition to Anoxia?
The requirement of a functional respiratory chain and cytochrome c oxidase for the anoxic induction of OLE1 and CYC7 is puzzling, because yeast cells grown under anaerobic conditions lack both cytochrome n oxidase and a functional mitochondrial respiratory chain (48). Thus, this requirement may be transient and may occur during the shift from aerobic to anaerobic conditions. During this shift, oxygen concentration in the fermentor drops to zero within 1 h (25, 40). To ask whether O2 is required for the generation of the signal involved in the induction of OLE1 and CYC7, we shifted cells from aerobic to anoxic conditions, added an anoxic solution of cyanide after anaerobiosis was achieved, and followed their expression over a 24-h period. From Fig. 5 it is clear that OLE1 transcript levels decrease after the addition of cyanide to anoxic cells, just as they did in the presence of CO (Fig. 1). Cyanide also severely inhibited the induction of CYC7 in anoxic cells, although there was a slight increase in its transcript levels toward the end of the 24-h incubation. These findings suggest that the “signal” produced by the respiratory chain can be generated in the absence of oxygen.
Figure 5.
Effect of cyanide on the induction of OLE1 and CYC7 after a shift to anoxic conditions. Strain JM43 was grown aerobically for at least six generations to early log-phase; the sparge gas was then switched from air to 2.5% CO2 in O2-free N2. After achieving anaerobiosis (1 h), an anoxic solution of KCN (1 mM final concentration) was added. Cells were harvested at the indicated times, and total RNA was isolated and subjected to Northern blot analyses as described in Materials and Methods. Transcript levels were normalized to the level of ACT1 mRNA and are presented as a decimal percent of their steady-state levels under anoxic conditions (lane 9). Lane 1, aerobic cells before the shift; lane 2, 1 h after the shift, just before KCN addition; Lanes 3–8, 5 h, 9 h, 13 h, 17 h, 21 h, and 25 h, respectively, after the shift.
DISCUSSION
The results presented here show that CO blocks the anoxic induction of at least two hypoxic genes (OLE1, CYC7) and partially inhibits the induction of a third (COX5b) in S. cerevisiae. Given the specificity of CO for ferrous hemoproteins that reversibly bind oxygen, these findings support the hypothesis that an O2-binding hemoprotein is involved in the induction of these hypoxic genes. Further support for this conclusion comes from the finding that transition metals induce the expression of OLE1 and CYC7 under normoxic conditions. Finally, our studies with respiratory inhibitors and mutants indicate that the respiratory chain is involved in the signaling pathway, and that cytochrome c oxidase may act as an oxygen sensor for the induction of these hypoxic genes.
Two Classes of Hypoxic Genes in Yeast.
The finding that CO inhibits the induction of some, but not all, hypoxic genes indicates that there are at least two classes of hypoxic genes in yeast: CO sensitive and CO insensitive. This finding implies that there are multiple mechanisms/pathways involved in modulating the expression of hypoxic genes in yeast, as in mammals (2, 9), and that one of these involves cytochrome c oxidase and the respiratory chain. This latter pathway appears to be independent of the hypoxic transcription factor Rox1p because it affects three genes (OLE1, CYC7, and COX5b) that are under its control but has no effect on the expression of seven other hypoxic genes (HEM13, HMG1, HMG2, ERG11, CPR1 [NCP1], ANB1, and AAC3) that are also regulated by this factor.
Oxygen Sensing by Mitochondria and Cytochrome c Oxidase.
Three findings support the conclusion that cytochrome c oxidase is the CO-binding hemoprotein involved in the anoxic induction of OLE1 and CYC7. First, their induction is absent in strains (e.g., JM43-GD5ab and JM43ρ0) that lack holocytochrome c oxidase. Second, cyanide, which forms a complex with cytochrome a33+, blocks the anoxic induction of these genes. Third, respiratory inhibitors and mutations that block the respiratory chain upstream of cytochrome c oxidase also inhibit this induction. What all of these treatments have in common is blockage of electron transport through cytochrome c oxidase. Cytochrome c oxidase also has been implicated as a probable oxygen sensor and in the regulation of hypoxic genes in a number of mammalian cells, including carotid body cells (12–14, 49), hepatocytes (17, 18, 50), and cardiomyocytes (51).
The mitochondrial respiratory chain and cytochrome c oxidase are well suited for a role in oxygen sensing, because they are consumers of most of the oxygen that is used by eucaryotic cells and because the flux of electrons through both the respiratory chain and cytochrome c oxidase is affected by oxygen concentration in several cell types, including yeast (31) and mammalian cells (50, 52–54). Moreover, both the apparent Km for oxygen binding to cytochrome c oxidase and the rate of electron transfer from heme a to a3 are affected by oxygen concentration (53). Currently, it is not clear how cytochrome c oxidase may “sense” oxygen as cells are shifted into hypoxic or anoxic conditions, what signal is released from the mitochondrion during this process, and what pathway is involved in transducing this signal into an effect on nuclear gene expression. There are several possibilities. First, as noted by Wilson (51), mitochondrial oxidative phosphorylation depends on oxygen concentration; hence, mitochondria can convert information concerning cytosolic oxygen concentration into a metabolic signal [e.g., [ATP]/[ADP] [Pi], NAD(P)H/NAD(P)]. This type of signal, in turn, can affect virtually every aspect of cell function, including the phosphorylation of transcription factors or other members of a signal transduction pathway. This model is intriguing because the threshold for expression of hypoxic yeast genes is low, 0.25 μM O2 (refs. 25 and 31; K.E.K., P.V.B., and R.O.P., unpublished observations), and approximates the apparent Km for oxygen binding to yeast cytochrome c oxidase (R.O.P., C. E. Trueblood, and D. F. Wilson, unpublished observations). However, respiratory inhibitors and mutations affect expression of OLE1 and CYC7 under strictly anoxic conditions in which the respiratory chain is already inhibited by oxygen deprivation. These findings argue against a signaling pathway that involves global changes in cellular energy charge or redox but do not preclude a pathway linked to the respiratory chain itself. A second possibility, proposed for mammalian cells (18, 55), is that there is a decrease in the Vmax of cytochrome c oxidase in response to hypoxia (56), and that this decrease in Vmax increases the reduction state of mitochondrial electron carriers that are upstream of cytochrome c oxidase and the generation of ROS (55). These ROS, which have been implicated in the crosstalk between the mitochondrion and nucleus (57), would then function to activate members of signal transduction pathways, (e.g., stress kinases, nuclear transcriptions factors, etc.). Although we cannot rule out a role for ROS in oxygen sensing in yeast, it seems unlikely that this type of signaling is involved in the anoxic induction of OLE1 and CYC7, because cyanide blocks their induction when added to anaerobic cells, in which ROS should be absent (58). Moreover, the hypoxic isoform, Vb, of yeast cytochrome c oxidase subunit V functions to increase, not decrease, the Vmax of yeast holocytochrome c oxidase (59, 60). A third possibility is that a protein involved in a signal-transduction pathway serves as an electron acceptor or donor for cytochrome c oxidase, and that this protein is released under hypoxic or anoxic conditions directly into the cytosol, where it activates or inactivates transcription factors that regulate the expression of hypoxic genes. Precedent for this sort of pathway comes from the fact that mitochondria export proteins and peptides (61), and that cytochrome c, which is immediately upstream of cytochrome c oxidase in the respiratory chain, is easily released from mitochondria under certain conditions (e.g., apoptosis) (62, 63). A fourth possibility is that the functional integrity of cytochrome c oxidase or other components of the respiratory chain is compromised as cells are shifted from aerobic into anaerobic conditions, and that respiratory chain subunits or subcomplexes are released from the mitochondrion and serve directly as “signals” of hypoxia. It is well known that there is a coordinate loss of cytochromes after cells are shifted from aerobic to anoxic conditions (64), but it is not known how fast this loss occurs, and whether it is fast enough to explain the kinetics of induction observed here. Clearly, more experimental work is required to decide among these possibilities.
Acknowledgments
We thank Drs. Charles Martin (YEP352/OLE4.8) and Alexander Tzagoloff (aM7-40-5B) for providing plasmids and yeast strains. This work was supported by grant GM30228 from the National Institutes of Health to R.O.P., a grant from the Council for Tobacco Research, and a postdoctoral fellowship from the American Heart Association of Colorado to K.E.K.
ABBREVIATION
- ROS
reactive oxygen species
References
- 1.Hochachka P W, Buck L T, Doll C J, Land S C. Proc Natl Acad Sci USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunn H F, Poyton R O. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg M A, Dunning S P, Bunn H F. Science. 1988;242:1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 4.Maxwell P H, Pugh C W, Ratcliffe P J. Proc Natl Acad Sci USA. 1993;90:2423–2427. doi: 10.1073/pnas.90.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G L, Semenza G L. Proc Natl Acad Sci USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross A R, Henderson L, Jones O T, Delpiano M A, Hentschel J, Acker H. Biochem J. 1990;272:743–747. doi: 10.1042/bj2720743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorlach A, Holtermann G, Jelkmann W, Hancock J T, Jones S A, Jones O T, Acker H. Biochem J. 1993;290 (Pt 3):771–776. doi: 10.1042/bj2900771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acker H. Ann N Y Acad Sci. 1994;718:3–10. doi: 10.1111/j.1749-6632.1994.tb55698.x. [DOI] [PubMed] [Google Scholar]
- 9.Acker H, Xue D. News Physiol Sci. 1995;10:211–215. [Google Scholar]
- 10.Bunn H, Gu J, Huang L, Park J W. J Exp Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratcliffe P, Rourke J, Maxwell P. J Exp Biol. 1998;201:1153–1162. doi: 10.1242/jeb.201.8.1153. [DOI] [PubMed] [Google Scholar]
- 12.Mulligan E, Lahiri S, Storey B T. J Appl Physiol. 1981;51:438–446. doi: 10.1152/jappl.1981.51.2.438. [DOI] [PubMed] [Google Scholar]
- 13.Obeso A, Almaraz L, Gonzalez C. Brain Res. 1985;348:64–68. doi: 10.1016/0006-8993(85)90360-9. [DOI] [PubMed] [Google Scholar]
- 14.Wilson D F, Mokashi A, Chugh D, Vinogradov S, Osanai S, Lahiri S. FEBS Lett. 1994;351:370–374. doi: 10.1016/0014-5793(94)00887-6. [DOI] [PubMed] [Google Scholar]
- 15.Lahiri S, Buerk D G, Chugh D, Osanai S, Mokashi A. Brain Res. 1995;684:194–200. doi: 10.1016/0006-8993(95)00420-u. [DOI] [PubMed] [Google Scholar]
- 16.Lahiri S, Chugh D K, Mokashi A, Vinogradov S, Osanai S, Wilson D F. Adv Exp Med Biol. 1996;388:213–217. doi: 10.1007/978-1-4613-0333-6_26. [DOI] [PubMed] [Google Scholar]
- 17.Chandel N S, Budinger G R, Choe S H, Schumacker P T. J Biol Chem. 1997;272:18808–18816. doi: 10.1074/jbc.272.30.18808. [DOI] [PubMed] [Google Scholar]
- 18.Chandel N S, Maltepe E, Goldwasser E, Mathieu C E, Simon M C, Schumaker P T. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zitomer R S, Lowry C V. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwast K E, Burke P V, Poyton R O. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 21.de Winde J H, Grivell L A. Prog Nucleic Acid Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- 22.Pinkham J L, Keng T. In: Heme-Mediated Gene Regulation in Saccharomyces cerevisiae. Winkelmann G, Winge D R, editors. New York: Dekker; 1994. pp. 455–501. [Google Scholar]
- 23.Poyton R O, Burke P V. Biochim Biophys Acta. 1992;1101:252–256. [PubMed] [Google Scholar]
- 24.Zhang L, Guarente L. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burke P V, Raitt D C, Allen L A, Kellogg E A, Poyton R O. J Biol Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 26.Cumsky M G, Ko C, Trueblood C E, Poyton R O. Proc Natl Acad Sci USA. 1985;82:2235–2239. doi: 10.1073/pnas.82.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterland R A, Basu A, Chance B, Poyton R O. J Biol Chem. 1991;266:4180–4186. [PubMed] [Google Scholar]
- 28.Trueblood C E, Poyton R O. Genetics. 1988;120:671–680. doi: 10.1093/genetics/120.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X-J, Raitt D, Burke P V, Clewell A S, Kwast K E, Poyton R O. J Biol Chem. 1996;271:25131–25138. doi: 10.1074/jbc.271.41.25131. [DOI] [PubMed] [Google Scholar]
- 30.Tzagoloff A, Akai A, Needleman R B, Zulch G. J Biol Chem. 1975;250:8236–8242. [PubMed] [Google Scholar]
- 31.Burke P V, Kwast K E, Everts F, Poyton R O. J Appl Environ Microbiol. 1998;64:1040–1044. doi: 10.1128/aem.64.3.1040-1044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elder R T, Loh E Y, Davis R W. Proc Natl Acad Sci USA. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang S S, Yin X, Guzzzo-Arkuran C, Jones V S, Davison A J. BioTechniques. 1993;14:380–381. [PubMed] [Google Scholar]
- 34.Kwast K E, Burke P V, Brown K, Poyton R O. Curr Genet. 1997;32:377–383. doi: 10.1007/s002940050291. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 10.14–10.15. [Google Scholar]
- 36.Trueblood C E, Poyton R O. Mol Cell Biol. 1987;7:3520–3526. doi: 10.1128/mcb.7.10.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kietzmann T, Schmidt H, Unthan-Fechner K, Probst I, Jungermann K. Biochem Biophys Res Commun. 1993;195:792–798. doi: 10.1006/bbrc.1993.2115. [DOI] [PubMed] [Google Scholar]
- 38.Goldberg M A, Schneider T J. J Biol Chem. 1994;269:4355–4359. [PubMed] [Google Scholar]
- 39.Choi J-Y, Stukey J, Hwang S-Y, Martin C E. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- 40.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho V T, Bunn H F. Biochem Biophys Res Comm. 1996;223:175–180. doi: 10.1006/bbrc.1996.0865. [DOI] [PubMed] [Google Scholar]
- 42.Sinclair P, Gibbs A H, Sinclair J F, de Matteis F. Biochem J. 1979;178:529–538. doi: 10.1042/bj1780529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinclair P R, Sinclair J F, Bonkowsky H L, Gibbs A H, Matteis F D. Biochem Pharmacol. 1982;31:993–999. doi: 10.1016/0006-2952(82)90333-1. [DOI] [PubMed] [Google Scholar]
- 44.Shibayama N, Morimoto H, Kitagawa T. J Mol Biol. 1986;192:331–336. doi: 10.1016/0022-2836(86)90368-2. [DOI] [PubMed] [Google Scholar]
- 45.Yonetoni T, Yamamoto H, Woodrow G V. J Biol Chem. 1974;249:682–690. [PubMed] [Google Scholar]
- 46.Zhu H, Riggs A F. Proc Natl Acad Sci USA. 1992;89:5015–5019. doi: 10.1073/pnas.89.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford M J, Sherman D R, Goldberg D E. J Biol Chem. 1995;270:6991–6996. doi: 10.1074/jbc.270.12.6991. [DOI] [PubMed] [Google Scholar]
- 48.Linnane A W, Haslam J M. Curr Top Cell Regul. 1970;2:101–172. [Google Scholar]
- 49.Lahiri S. News Physiol Sci. 1994;9:161–165. [Google Scholar]
- 50.Wilson D F, Rumsey W L, Green T J, Vanderkooi J M. J Biol Chem. 1988;263:2712–2718. [PubMed] [Google Scholar]
- 51.Budinger G R, Duranteau, Chantel N S, Schumaker P T. J Biol Chem. 1998;273:3320–3326. doi: 10.1074/jbc.273.6.3320. [DOI] [PubMed] [Google Scholar]
- 52.Rumsey W L, Schlosser C, Nuutinen E M, Robiolio M, Wilson D F. J Biol Chem. 1990;265:15392–15399. [PubMed] [Google Scholar]
- 53.Verkhovsky M I, Morgan J E, Puustinen A, Wikstrom M. Nature (London) 1996;380:268–270. doi: 10.1038/380268a0. [DOI] [PubMed] [Google Scholar]
- 54.Gnaiger E, Steinlechner-Maran R, Mendez G, Eberl T, Margreiter R. J Bioenerg Biomembr. 1995;27:583–596. doi: 10.1007/BF02111656. [DOI] [PubMed] [Google Scholar]
- 55.Duranteau J, Chandel N S, Kulisz A, Shao Z, Schumacker P T. J Biol Chem. 1998;273:11619–11624. doi: 10.1074/jbc.273.19.11619. [DOI] [PubMed] [Google Scholar]
- 56.Chandel N S, Budinger G S, Schumacker P T. J Biol Chem. 1996;271:18672–18677. doi: 10.1074/jbc.271.31.18672. [DOI] [PubMed] [Google Scholar]
- 57.Poyton R O, McEwen J E. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 58.Boveris A. In: Tissue Hypoxia and Ischemia. Reivich M, Coburn R, Lahiri S, Chance B, editors. New York: Plenum; 1977. pp. 67–82. [Google Scholar]
- 59.Allen L A, Zhao X J, Caughey W, Poyton R O. J Biol Chem. 1995;270:110–118. doi: 10.1074/jbc.270.1.110. [DOI] [PubMed] [Google Scholar]
- 60.Burke P V, Poyton R O. J Exp Biol. 1998;201:1163–1175. doi: 10.1242/jeb.201.8.1163. [DOI] [PubMed] [Google Scholar]
- 61.Poyton R O, Sevarino K A, McKee E E, Duhl D J M, Cameron V, Goehring B. Adv Mol Cell Biol. 1996;17:247–280. [Google Scholar]
- 62.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 63.Kluck R M, Bossy-Wetzel, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 64.Rep M, Grivell L A. Curr Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]