Abstract
Hsp70 chaperones assist a large variety of protein folding processes within the entire lifespan of proteins. Central to these activities is the regulation of Hsp70 by DnaJ cochaperones. DnaJ stimulates Hsp70 to hydrolyze ATP, a key step that closes its substrate-binding cavity and thus allows stable binding of substrate. We show that DnaJ stimulates ATP hydrolysis by Escherichia coli Hsp70, DnaK, very efficiently to >1000-fold, but only if present at high (micromolar) concentration. In contrast, the chaperone activity of DnaK in luciferase refolding was maximal at several hundredfold lower concentration of DnaJ. However, DnaJ was capable of maximally stimulating the DnaK ATPase even at this low concentration, provided that protein substrate was present, indicating synergistic action of DnaJ and substrate. Peptide substrates were poorly effective in this synergistic action. DnaJ action required binding of protein substrates to the central hydrophobic pocket of the substrate-binding cavity of DnaK, as evidenced by the reduced ability of DnaJ to stimulate ATP hydrolysis by a DnaK mutant with defects in substrate binding. At high concentrations, DnaJ itself served as substrate for DnaK in a process considered to be unphysiological. Mutant analysis furthermore revealed that DnaJ-mediated stimulation of ATP hydrolysis requires communication between the ATPase and substrate-binding domains of DnaK. This mechanism thus allows DnaJ to tightly couple ATP hydrolysis by DnaK with substrate binding and to avoid jamming of the DnaK chaperone with peptides. It probably is conserved among Hsp70 family members and is proposed to account for their functional diversity.
Hsp70 proteins are highly versatile chaperones that assist a large variety of folding processes, ranging from folding of newly synthesized proteins to facilitation of proteolytic degradation of unstable proteins (1–4). This functional diversity requires Hsp70 proteins to associate promiscuously with misfolded proteins as well as selectively with folded substrates, including low-abundance regulatory proteins. Correct selection of substrates is postulated to be a role for DnaJ cochaperones, which control the ATPase cycle of Hsp70 proteins.
ATP regulates the affinity of Hsp70 for substrates. Hsp70 exhibits low affinity and fast exchange rates for substrates in the ATP state, and high affinity and low exchange rates in the ADP state (5, 6). Hydrolysis of ATP thus locks substrates into the substrate-binding cavity of Hsp70, and this key step is positively controlled by DnaJ (7–11). Substrate release is mediated by ADP/ATP exchange, a process that is catalyzed for some Hsp70 homologs by GrpE nucleotide exchange factors (8, 9, 12, 13).
The assumed role for DnaJ cochaperones is to target Hsp70 partner proteins to their substrates by catalyzing ATP hydrolysis by Hsp70 (2, 14). Cells encode multiple DnaJ homologs which share a conserved stretch of ≈75 residues, defined as J domain, but differ with respect to the other domains (15–17). The J domain is required for stimulation of ATP hydrolysis by Hsp70 (18, 19). The other domains appear to permit selective association of DnaJ homologs with defined subgroups of the Hsp70 substrates (16).
Little is known about the mechanism by which DnaJ exerts its targeting function in the Hsp70 chaperone cycle (2, 19). For the Escherichia coli DnaJ homolog and its Hsp70 partner, DnaK, it has been shown by NMR analysis that the J domain of DnaJ (DnaJ2–75) interacts with the ATPase domain of DnaK (20). Furthermore, the J domain is insufficient to stimulate ATP hydrolysis by DnaK. This fragment in addition required either the Gly/Phe-rich region of DnaJ, located C-terminal of the J domain, or a peptide substrate provided in trans (18, 19).
We investigated central features of the mechanism by which DnaJ cooperates with Hsp70, using E. coli DnaJ and DnaK as a model. We show that DnaJ tightly couples ATP hydrolysis with binding of protein substrate by DnaK, through a mechanism that involves communication between the ATPase and substrate-binding domains of DnaK. DnaJ also allows the DnaK system to discriminate between peptide and protein substrates and thus to prevent jamming with peptides. A mechanistic model for the action of DnaJ is proposed that is likely to be valid also for eukaryotic Hsp70 members.
MATERIALS AND METHODS
Refolding of Denatured Luciferase.
Firefly luciferase (Sigma) was denatured in 6 M guanidinium hydrochloride (Gdn⋅HCl) and refolded (80 nM luciferase, final concentration) essentially as published (21), in renaturation buffer containing DnaK (800 nM), GrpE (400 nM), ATP (5 mM), and DnaJ as indicated. Luciferase was refolded for 30 min at 30°C and activity was monitored at room temperature.
ATPase Assays.
For standard determinations, DnaK⋅ATP complexes were prepared and tested for ATP hydrolysis in single-turnover assays as described (9), with a final concentration of DnaK of ≈500 nM. For quenched flow measurements at 25°C, equal volumes of ATP (8 μM) and nucleotide-free DnaK (9.6 μM) were preincubated for 10 s before 1:1 mixing with various concentrations of DnaJ in a QUENCHED flow apparatus (KinTek). The reaction was quenched after 0.5 s with perchloric acid, and the ATP/ADP ratio was determined as described (22). Rate constants were determined with the program grafit (Erithacus Software, Staines, U.K.).
Mutant Design and Protein Purification.
Construction of dnaK mutants and purification of DnaK and GrpE proteins were as described (23, 24). The DnaJ purification protocol will be published elsewhere (H. J. Schönfeld, personal communication).
Crosslinking.
Cysteine-424 of DnaK-Q424C was reduced with DTT (25 mM) for 1 h at 30°C, and excess DTT was removed by microdialysis against 25 mM Hepes–KOH, pH 7.0/50 mM KCl/1 mM EDTA. 4-(2-Iodoacetamido)benzophenone (BPIA) in 2-fold excess was added for 1 h in the dark. Uncoupled BPIA was quenched by 4-fold excess of 2-mercaptoethanol for 10 min. The sample was dialyzed at 4°C for 3 h against 25 mM Hepes–KOH, pH 7.6/50 mM KCl/5 mM MgCl2. Crosslinking mixtures contained DnaK-Q424C-BPIA (5 μM), DnaJ (5 μM or 0.5 μM), σ32 (5 μM), peptide (25 μM), or ATP (5 mM) in a volume of 20 μl, preincubated at 30°C for 2 min and irradiated with UV light on ice for 5 min (365 nm, 100 W; Ultraviolet Products, Model B-100AP) at a distance of 5 cm. Crosslinking products were analyzed on an SDS/10% polyacrylamide gel and visualized by silver staining or immunoblotting using DnaK-, σ32- or DnaJ-specific antiserum, alkaline phosphatase-conjugated secondary antibodies, and ECF (Amersham Pharmacia Biotech). Fluorescence was detected in a fluorimager (FLA 2000, Fuji) and quantified by using MacBas software (Fuji). The signal was linear over a range of 2.5 orders of magnitude.
RESULTS AND DISCUSSION
Differing DnaJ Requirements for DnaK ATPase Stimulation and DnaK Chaperone Activity.
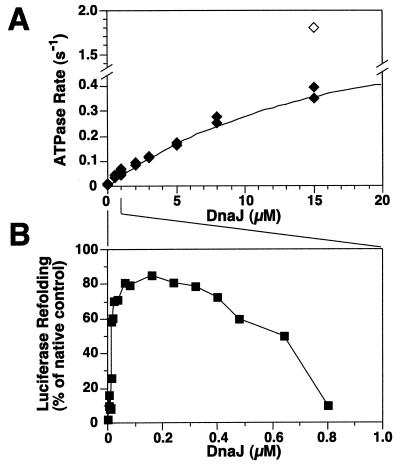
To analyze the functional interaction between DnaK and DnaJ we first determined the full potential of DnaJ to stimulate ATP hydrolysis by DnaK, using single-turnover quenched flow experiments. We prepared nucleotide-free DnaK, which was injected in the flow chamber, followed by rapid mixing with first ATP and then DnaJ, and finally by quenching with perchloric acid. At the conditions used, the ATP hydrolysis rate of 6 × 10−4 s−1 was stimulated by DnaJ to a maximal value of 0.79 ± 0.11 s−1, obtained by fitting the quadratic solution into the data by nonlinear regression (Fig. 1A). To reach efficient stimulation, high DnaJ concentrations (≥15 μM), a >6-fold molar excess of DnaJ over DnaK, were required. The high DnaJ concentration needed for maximal stimulation of ATP hydrolysis cannot be due to absence of GrpE in these assays because GrpE is known to stimulate nucleotide exchange, but not hydrolysis of γ-phosphate that is monitored here (8, 9, 12, 13).
Figure 1.
Differences in DnaJ requirements for stimulation of ATP hydrolysis by DnaK and luciferase refolding. (A) Effect of DnaJ on stimulation of ATP hydrolysis. Hydrolysis rates dependent on DnaJ (filled diamonds) were determined by quenched flow experiments and yielded a fitted maximal value of 0.79 s−1. The open diamond indicates the additional effect of σ32 (final concentration 2 μM) at the highest DnaJ concentration. (B) Efficiency of luciferase refolding by the DnaK system dependent on DnaJ. Chemically denatured luciferase was refolded in buffer containing constant concentrations of DnaK, GrpE, and ATP, and various concentrations of DnaJ. The lines between A and B indicate the differences in the DnaJ concentration used in the two experiments.
The high DnaJ concentration required for efficient stimulation of ATP hydrolysis does not reflect the cellular situation because in E. coli the DnaJ concentration is an order of magnitude lower than the DnaK concentration (25). A shift of E. coli cells to heat shock temperature does not change this stoichiometry and causes only 2- to 3-fold increase in DnaJ and DnaK concentrations, suggesting a similar situation under stress. Furthermore, the high concentrations of DnaJ necessary for efficient ATPase stimulation are unproductive for chaperone activity in vitro, as determined by DnaK-dependent refolding of unfolded luciferase. The efficiency of luciferase refolding was maximal at DnaJ concentrations that were low (around 160 nM) and substoichiometric to DnaK, and low at DnaJ concentrations that were high (≥800 nM) and optimal for ATP hydrolysis stimulation (Fig. 1B). The several hundredfold difference between the DnaJ concentrations optimal for chaperone activity and efficient hydrolysis stimulation (without substrate) is consistent with two alternative explanations. Either weak stimulation of the DnaK ATPase by DnaJ is sufficient for optimal chaperone activity, or efficient stimulation of the DnaK ATPase by DnaJ requires presence of chaperone substrates.
Efficient Stimulation of the DnaK ATPase Requires DnaJ and Protein Substrate.
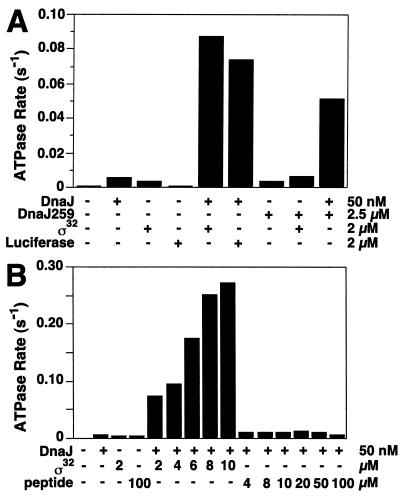
We tested whether DnaJ requires the presence of chaperone substrates to efficiently stimulate the ATPase of DnaK. We determined in single-turnover ATPase assays whether two protein substrates, heat shock transcription factor σ32 (26) and unfolded luciferase (21, 27), affect the ability of DnaJ to stimulate ATP hydrolysis. DnaJ was added at low concentration (50 nM; 0.1:1, DnaJ:DnaK) which allows efficient luciferase refolding but not stimulation of ATP hydrolysis. In the absence of DnaJ, σ32 and unfolded luciferase (at 2 μM) stimulated hydrolysis poorly (5.6- and 1.1-fold, respectively, Fig. 2A). In the presence of DnaJ, they strongly stimulated ATP hydrolysis, by ≈140 and ≈120-fold at 2 μM σ32 and luciferase, respectively (Fig. 2A). Such strong stimulation was also observed at various concentrations of DnaJ and DnaK (e.g., Fig. 1A). We also determined in single-turnover experiments the stimulation of ATP hydrolysis in a luciferase refolding reaction at optimal refolding conditions by adding DnaK⋅[γ-32P]ATP complexes at various time points. Stimulation was high at the onset of the refolding reaction, between 0.1 s−1 and 0.15 s−1, which is 160- to 250-fold above the intrinsic rate, and decreased toward the end of the reaction (data not shown). Stimulation of ATP hydrolysis by DnaJ and refolding activity of DnaK are thus correlated.
Figure 2.
Stimulation of DnaK ATPase by DnaJ requires protein substrate. (A) Effects of DnaJ, DnaJ259, σ32, and unfolded luciferase (final concentrations indicated) on ATP hydrolysis rates of DnaK as determined by single-turnover assays. Luciferase was unfolded in 3 M Gdn⋅HCl and diluted into refolding buffer. The final Gdn⋅HCl concentration (180 mM) had no effect on unstimulated and DnaJ-stimulated hydrolysis rates (data not shown). (B) Effects of increasing σ32 and peptide substrate [σ32-Q132-Q144 (28)] on ATP hydrolysis rates of DnaK in the presence of DnaJ. The ATP hydrolysis rates were determined in standard single-turnover assays.
To determine the maximally stimulated rate of ATP hydrolysis by DnaK, we titrated protein substrate to constant concentrations of DnaK and DnaJ. Such experiments were performed with σ32 because it is a DnaK substrate in the absence of denaturant (26), thus allowing titration of this substrate. With increasing σ32, the rate of ATP hydrolysis increased up to 0.27 s−1 at 10 μM σ32 (450-fold stimulation, Fig. 2B). The maximally stimulated rates were similar to those reached at a high concentration of DnaJ without substrate (Fig. 1A).
These results indicate that DnaJ and protein substrates act synergistically to provide two signals required for stimulation of ATP hydrolysis by DnaK. This finding is consistent with, but goes well beyond, the previous result (19) that a fragment of DnaJ comprising the J domain can cooperate with a peptide to stimulate the DnaK ATPase activity. The previous study suggested that the Gly/Phe-rich region of DnaJ provides a second signal required for stimulation of the DnaK ATPase. Our findings demonstrate that full-length DnaJ, at low stoichiometry with respect to DnaK as found in vivo, must act synergistically with a protein substrate to fully stimulate ATP hydrolysis by DnaK. Because ATP hydrolysis closes the substrate-binding cavity of DnaK (5, 6, 14), this synergism allows DnaJ to tightly couple hydrolysis of ATP with locking-in of substrates in complex with DnaK.
In similar set of ATPase experiments we replaced protein substrates by peptide substrates. The tested peptide (σ32-Q132-Q144) is a substrate for DnaK (Kd = 100 nM) (28) and stimulated DnaK ATP hydrolysis by 8.9-fold (Fig. 2B). However, it does not act synergistically with DnaJ in stimulation of hydrolysis, even when added at saturating concentration up to 100 μM. A similar result was obtained with another peptide substrate (σ32-M195-N207) (28) (data not shown). These results contrast with those obtained with protein substrates. They indicate that the DnaJ-dependent mechanism of stimulation of ATP hydrolysis, and thus of locking-in of substrates in the substrate-binding cavity of DnaK, is efficient with the proteins but not the peptides tested, and thus allows DnaK to discriminate in these cases between the two types of substrates. Although the physiological relevance of this discriminatory function of DnaJ is unclear at present, we propose that it avoids unproductive jamming of the DnaK chaperone system with peptides, as generated in vivo by proteolysis. The mechanism by which DnaJ allows DnaK to discriminate between protein and peptide substrates is unclear as well. The distinction between peptide and protein substrates may already be established by the DnaJ–substrate interaction. DnaJ has been found to have lower affinity for peptides (σ32-Q132-Q144; Kd = 2 μM) than for proteins [σ32; Kd = 19 nM (29)]. It may also be possible that the DnaJ-to-DnaK transfer of a protein substrate with multiple binding sites for DnaJ and DnaK is kinetically favorable. It is intriguing that peptides at very high concentrations can cooperate with the J-domain fragment to stimulate the ATPase activity of DnaK (19). It may therefore be that segments of DnaJ that are C-terminal to the J domain are involved in forming a transient complex with DnaK, which disfavors the efficient locking-in of peptide substrates into the substrate-binding cavity. Such a complex may involve a direct contact between DnaJ’s C terminus and the substrate-binding domain of DnaK.
Mutations in the Substrate-Binding Pocket of DnaK Affect Stimulation of ATPase Activity by DnaJ.
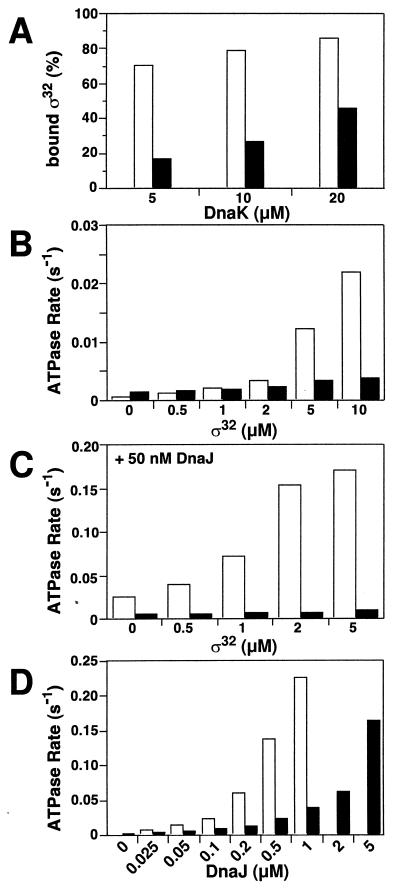
Two mechanisms may account for the synergistic action of DnaJ and protein substrate. Substrate binding by DnaJ may convert DnaJ to an active form capable of stimulating ATP hydrolysis by DnaK. Alternatively, substrate binding by DnaK may provide the second signal allowing DnaJ to stimulate ATP hydrolysis. To distinguish between these alternatives we tested the second mechanism. It predicts that a mutation in DnaK affecting substrate binding should also affect DnaJ action. We constructed a DnaK mutant protein with a Val-436 to Phe exchange (DnaK-V436F). Phe-436 is predicted by modeling into the atomic structure of the substrate-binding domain of DnaK (30) to block the central hydrophobic pocket of the substrate-binding cavity. The V436F mutation did not cause global structural changes within DnaK, as judged by circular dichroism spectroscopy and partial proteolysis (data not shown). Furthermore, the mutational alteration did not cause strong changes in substrate specificity as judged by scanning of DnaK binding to peptide libraries. However, the overall affinity for substrates was lower, 17-fold for σ32 (Fig. 3A), which was caused by a 20-fold decrease in the association rate.
Figure 3.
Defects of the DnaK-V436F mutant in substrate binding and DnaJ-dependent stimulation of ATP hydrolysis. (A) Efficiency of binding of σ32 to wild-type DnaK and DnaK-V436F as determined by gel filtration according to Gamer et al. (29). [3H]σ32 (1 μM) was incubated with the indicated amount of DnaK at 30°C for 2 h prior to separation of the σ32⋅DnaK complex and free σ32 on a Superdex 200 FPLC column (Amersham Pharmacia Biotech). The dissociation constant (Kd) of the DnaK⋅[3H]σ32 complex was calculated from the ratio of DnaK-bound and free [3H]σ32 to be 1.4 and 24 μM for wild-type DnaK and DnaK-V436F, respectively. (B and C) Single-turnover ATP hydrolysis rates of wild-type DnaK and DnaK-V436F in the presence of σ32 without (B) or with 50 nM DnaJ (C). (D) Effects of increasing DnaJ concentrations on single-turnover ATP hydrolysis rates of wild-type DnaK and DnaK-V436F. Open bars, wild-type DnaK; filled bars, DnaK-V436F.
σ32 was much less efficient in stimulating ATP hydrolysis by DnaK-V436F as compared with wild-type DnaK (Fig. 3B). Importantly, DnaJ, with and without protein substrate (σ32), only poorly stimulated ATP hydrolysis by DnaK-V436F (Fig. 3 C and D). DnaJ titration revealed that, compared with wild-type DnaK, the mutant required 5- to 10-fold more DnaJ to reach similar stimulation (Fig. 3D). This finding strongly suggests a mechanism in which stimulation of ATP hydrolysis by DnaJ requires binding of substrate in the substrate-binding cavity of DnaK.
Mutations in DnaK Altering the Interdomain Communication Affect Stimulation of ATPase Activity by DnaJ.
We considered that the mechanism by which DnaJ and protein substrate cooperate to stimulate ATP hydrolysis operates through the known interdomain communication of DnaK that couples ATP binding/hydrolysis in the ATPase domain with substrate binding in the adjacent domain (6, 9, 11, 19). This possibility cannot be stringently concluded from the data available so far.
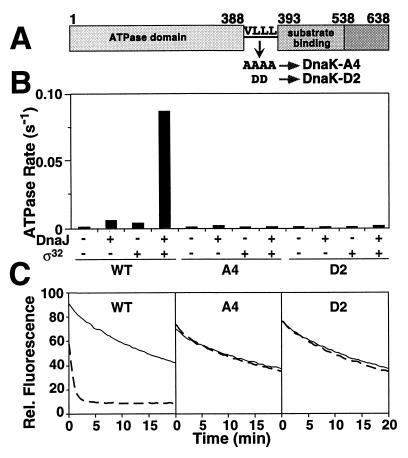
To test for participation of interdomain communication, two DnaK mutant proteins altered in the putative linker connecting these domains, DnaK-VLLL389,390,391,392AAAA (DnaK-A4) and DnaK-LL390,391DD (DnaK-D2) (Fig. 4A) were analyzed. These mutant proteins showed no overall structural and functional alterations (data not shown), but they lacked interdomain communication as judged by their inability to release substrate upon addition of ATP (Fig. 4C). Importantly, ATP hydrolysis by these mutant proteins was normal in the unstimulated state but could not be stimulated by DnaJ with or without protein substrate (σ32) (Fig. 4B). This defect was not caused by alterations in nucleotide binding and release as determined by kinetic measurements (data not shown). This indicates that the interdomain coupling mechanism of DnaK is also essential for DnaJ action.
Figure 4.
Mutations in the putative linker connecting ATPase and substrate-binding domains of DnaK abolish interdomain communication and stimulation by DnaJ. (A) Schematic representation of the DnaK linker mutants. (B) Effects of σ32 and DnaJ on ATP hydrolysis of wild type and DnaK mutants. The rates of ATP hydrolysis in the presence or absence of σ32 (2 μM) and DnaJ (50 nM) as indicated were determined by single-turnover assays. (C) Effects of ATP on dissociation rates of fluorescent labeled peptide from complex with wild-type DnaK and DnaK mutants. DnaK was prebound to fluorescent labeled peptide (σ32-Q132-Q144-C-IAANS) (28) in a 1:1 ratio. At time zero, a 100-fold excess of unlabeled peptide in buffer with or without ATP was added and the decrease in fluorescence was monitored.
DnaJ-Mediated Crosslinking of Substrates into the Substrate-Binding Cavity of DnaK.
The two-signal mechanism of DnaJ action is in apparent contradiction to earlier findings that DnaJ at high concentrations efficiently stimulates ATP hydrolysis by DnaK, even in the absence of substrate (9, 19). One explanation is that DnaJ itself, when provided at high concentration, serves as substrate for DnaK in the absence of other substrates.
To investigate this possibility we tested whether the DnaJ259 mutant protein, which fails to stimulate ATP hydrolysis by DnaK because of a point mutation in the HPD motif of the J domain (18), can still provide the second signal as substrate when present at high concentration (Fig. 2B). When added separately, DnaJ259 at high concentration and wild-type DnaJ at low concentration only poorly stimulated ATP hydrolysis. When added together they efficiently stimulated ATP hydrolysis, indicating that DnaJ and DnaJ259 act synergistically, similar to the action of DnaJ and protein substrate.
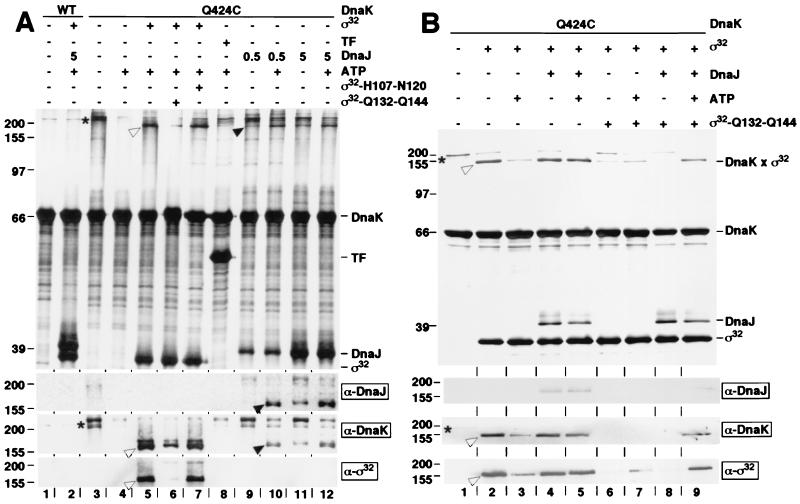
We then tested directly the ability of DnaJ to associate with the substrate-binding cavity of DnaK. Residue Gln-424 of DnaK was mutated to Cys (DnaK-Q424C) to position cysteine-specific, heterobifunctional crosslinkers for crosslinking of substrates. DnaK-Q424C had no detectable functional and structural defects in vivo and in vitro (data not shown). After coupling with crosslinker BPIA, DnaK-Q424C was incubated with substrates, DnaJ, and ATP in various combinations, followed by photocrosslinking (Fig. 5A). Nine nonsubstrate proteins tested (e.g., BSA, IgG, trigger factor) failed to crosslink to DnaK-Q424C with or without ATP (Fig. 5A, lane 8 for trigger factor with ATP; Western blot for trigger factor not shown). DnaK-Q424C dimers were crosslinked in absence of ATP and competing substrate (Fig. 5A, lane 3) and dissociated in ATP (Fig. 5A, lane 4). Dimerization of DnaK through binding of another DnaK molecule in the substrate-binding cavity was expected, given the characteristics of DnaK oligomerization, which suggest DnaK–DnaK contacts to be DnaK–substrate contacts (31). When σ32 was present, heterodimeric crosslinking products between DnaK-Q424C and σ32 formed (Fig. 5A, lane 5). The efficiency of crosslinking decreased upon addition of ATP and high-affinity peptide substrate (σ32-Q132-Q144; Kd = 100 nM) but not low-affinity peptide (σ32-H107-N120; Kd ≥ 10 μM) (28) (Fig. 5A, lanes 6 and 7).
Figure 5.
Cysteine-specific crosslinking of DnaJ and substrate into the substrate-binding cavity of DnaK. DnaK-Q424C, coupled to the heterobifunctional crosslinker BPIA by means of Cys-424, and similarly treated wild-type DnaK (WT) were incubated in the dark at 30°C with combinations of proteins [σ32, 5 μM; trigger factor (TF), 5 μM; DnaJ, 0.5, 1, or 5 μM], peptides (σ32-H107-N120; σ32-Q132-Q144, 25 μM), and ATP (5 mM) as indicated by + and − above the lanes. Crosslinking was induced by UV and products were analyzed on SDS/10% polyacrylamide gels (A and B). (Upper) Silver-stained SDS/polyacrylamide gels; (Lower) immunoblots developed with DnaK-, DnaJ-, and σ32-specific antisera (α-DnaK, etc.). Indicated are positions of molecular weight markers (left) and DnaK, trigger factor (TF), DnaJ, and σ32 (right). Asterisks and open and filled arrowheads denote DnaK-DnaK, DnaK-σ32, and DnaK-DnaJ crosslinks, respectively.
When DnaJ was added to DnaK-Q424C, crosslinking products between DnaK-Q424C and DnaJ (identified by immunoblots) formed, more populated at high DnaJ concentration and in ATP (Fig. 5A, lanes 9–12). This result suggests that DnaJ at high concentration associates with the substrate-binding cavity of DnaK. DnaJ therefore can provide the second signal, normally provided by protein substrate, that is required for stimulation of ATP hydrolysis by DnaK. The presence of competing protein substrate (e.g., σ32) decreased binding of DnaJ into the substrate-binding cavity of DnaK (compare α-DnaJ immunoblots Fig. 5A, lane 10, and Fig. 5B, lane 5; data not shown). DnaK–DnaJ heterodimers are thus unlikely to be intermediates in the DnaK functional cycle. These findings strongly suggest that the ability of DnaJ to stimulate ATP hydrolysis by DnaK in the absence of chaperone substrates results from an artificial situation in vitro, involving high DnaJ concentrations and perhaps low amounts of misfolded DnaJ that serve as substrate for DnaK.
In separate experiments we used cysteine-specific crosslinking to obtain physical evidence for a role of DnaJ to allow DnaK to discriminate between protein and peptide substrates. In the absence of DnaJ with or without ATP, a 5-fold molar excess of σ32-Q132-Q144 peptide substrate over σ32 was sufficient to prevent the association of σ32 with DnaK (Fig. 5B, lanes 6 and 7). In contrast, in the presence of DnaJ and ATP, this peptide was highly inefficient in competing with σ32 for DnaK binding (Fig. 5B, lane 9), despite the facts that the affinity of DnaK is at least 10-fold higher for this peptide than for σ32, and the peptide concentration used (25 μM) was in 5-fold molar excess over σ32. DnaJ in the absence of ATP was not effective (Fig. 5B, lane 8), suggesting that the DnaJ-mediated stimulation of ATP hydrolysis is crucial for recovery of the DnaK-σ32 crosslink. These data support the above conclusion that the DnaJ-dependent coupling mechanism allows DnaK to discriminate protein and peptide substrates.
Mechanistic Model for Functional Interaction Between DnaK and DnaJ.
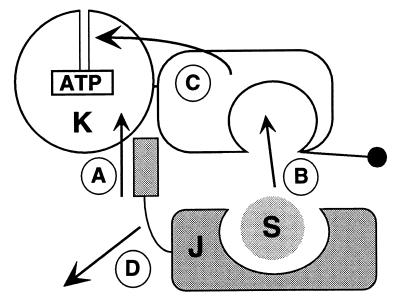
Together with earlier findings and in extension of previous models (5, 9, 19, 29, 32, 33), our findings suggest a mechanism for DnaK regulation in which DnaJ is a coupling factor for DnaK connecting ATP hydrolysis with substrate binding (Fig. 6). It relies on multiple steps, including A, interaction of DnaJ with DnaK, presumably through interaction of the J domain with the DnaK ATPase domain (20, 34, 35); B, transfer of protein substrate from DnaJ into the DnaK substrate-binding cavity; C, communication of substrate binding to the ATPase domain, which results in ATP hydrolysis and closing of the substrate-binding cavity; and D, dissociation of DnaJ from the DnaK⋅ADP⋅substrate complex. Consistent with our model, DnaJ can act catalytically to stimulate the ATPase activity of DnaK (36) and target DnaK to substrates (33).
Figure 6.
Model for the mechanism of regulation of DnaK by DnaJ. At least four steps (A–D), of unclear order with respect to steps A and B, build up the basic mechanism allowing DnaJ (J) to couple ATP hydrolysis by DnaK (K) with binding of protein substrates (S). See text for details.
This mechanism is most likely conserved within the Hsp70 family, since the involved structures of DnaJ and DnaK are conserved (2). Differences exist for the mechanism by which DnaJ homologs interact with substrates. Some homologs associate directly with substrates similar to E. coli DnaJ, others may act indirectly to position the J domain, and thus Hsp70 partner proteins, in the neighborhood of potential substrates (15, 16). Evolutionary conservation of this mechanism is supported by experiments for the uncoating of clathrin-coated vesicles, which requires Hsc70 and the DnaJ homolog auxilin (37). Stimulation of ATP hydrolysis by Hsc70 occurs only when auxilin and clathrin are present (38). Similarly, binding of yeast Hsp70, BiP (Kar2p), to immobilized substrates was greatly enhanced in an ATP-dependent manner by co-immobilization of the J domain of Sec63 (39). It is tempting to speculate that the coupling mechanism employed by DnaJ is of broad importance within the Hsp70 family and accounts for functional diversity of Hsp70 chaperone systems in cells.
Acknowledgments
We thank S. Rüdiger and Xun Zhao for DnaK mutant design; K. Paal for protein purification; H. J. Schönfeld for the DnaJ purification protocol; C. Schirra for cloning of dnaK mutants; and S. Rüdiger, C. Gässler, and A. Buchberger for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft with grants to B.B. and J.R.
ABBREVIATION
- BPIA
4-(2-iodoacetamido)benzophenone
References
- 1.Bukau B, editor. Molecular Chaperones and Folding Catalysts—Regulation, Cellular Function and Mechanisms. Amsterdam: Harwood; 1999. [Google Scholar]
- 2.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 3.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 4.Gething M-J. Nature (London) 1997;388:329–331. doi: 10.1038/40979. [DOI] [PubMed] [Google Scholar]
- 5.Schmid D, Baici A, Gehring H, Christen P. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 6.Flynn G C, Chappell T G, Rothman J E. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 7.Cheetham M E, Jackson A P, Anderton B H. Eur J Biochem. 1994;226:99–107. doi: 10.1111/j.1432-1033.1994.tb20030.x. [DOI] [PubMed] [Google Scholar]
- 8.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 10.Minami Y, Höhfeld J, Ohtsuka K, Hartl F U. J Biol Chem. 1996;271:19617–19624. doi: 10.1074/jbc.271.32.19617. [DOI] [PubMed] [Google Scholar]
- 11.Ziegelhoffer T, Lopez-Buesa P, Craig E. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]
- 12.Theyssen H, Schuster H-P, Bukau B, Reinstein J. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- 13.Packschies L, Theyssen H, Buchberger A, Bukau B, Goody R S, Reinstein J. Biochemistry. 1997;36:3417–3422. doi: 10.1021/bi962835l. [DOI] [PubMed] [Google Scholar]
- 14.Buchberger A, Reinstein J, Bukau B. In: Molecular Chaperones and Folding Catalysts—Regulation, Cellular Function and Mechanisms. Bukau B, editor. Amsterdam: Harwood; 1999. pp. 609–635. [Google Scholar]
- 15.Kelley W L. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 16.Laufen T, Zuber U, Buchberger A, Bukau B. In: Molecular Chaperones in Proteins: Structure, Function, and Mode of Action. Fink A L, Goto Y, editors. New York: Dekker; 1998. pp. 241–274. [Google Scholar]
- 17.Silver P A, Way J C. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- 18.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 19.Karzai A W, McMacken R. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- 20.Greene M K, Maskos K, Landry S J. Proc Natl Acad Sci USA. 1998;95:6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl F U. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klostermeier D, Seidel R, Reinstein J. J Mol Biol. 1998;279:841–853. doi: 10.1006/jmbi.1998.1816. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 24.Buchberger A, Schröder H, Büttner M, Valencia A, Bukau B. Nat Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- 25.Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Mol Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]
- 26.Gamer J, Bujard H, Bukau B. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-m. [DOI] [PubMed] [Google Scholar]
- 27.Schröder H, Langer T, Hartl F-U, Bukau B. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty J S, Rüdiger S, Schönfeld H-J, Schneider-Mergener J, Nakahigashi K, Yura T, Bukau B. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 29.Gamer J, Multhaup G, Tomoyasu T, McCarty J S, Rüdiger S, Schönfeld H-J, Schirra C, Bujard H, Bukau B. EMBO J. 1996;15:607–617. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Zhao X, Burkholder W F, Gragerov A, Ogata C M, Gottesman M, Hendrickson W A. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schönfeld H-J, Schmidt D, Schröder H, Bukau B. J Biol Chem. 1995;270:2183–2189. doi: 10.1074/jbc.270.5.2183. [DOI] [PubMed] [Google Scholar]
- 32.Wall D, Zylicz M, Georgopoulos C. J Biol Chem. 1995;270:2139–2144. doi: 10.1074/jbc.270.5.2139. [DOI] [PubMed] [Google Scholar]
- 33.Liberek K, Wall D, Georgopoulos C. Proc Natl Acad Sci USA. 1995;92:6224–6228. doi: 10.1073/pnas.92.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gässler C S, Buchberger A, Laufen T, Mayer M P, Schröder H, Valencia A, Bukau B. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suh W-C, Burkholder W F, Lu C Z, Zhao X, Gottesman M E, Gross C A. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierpaoli E V, Sandmeier E, Schönfeld H-J, Christen P. J Biol Chem. 1998;273:6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 37.Ungewickell E, Ungewickell H, Holstein S E, Lindner R, Prasad K, Barouch W, Martin B, Greene L E, Eisenberg E. Nature (London) 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 38.Barouch W, Prasad K, Greene L, Eisenberg E. Biochemistry. 1997;36:4303–4308. doi: 10.1021/bi962727z. [DOI] [PubMed] [Google Scholar]
- 39.Misselwitz B, Staeck O, Rapoport T A. Mol Cell. 1998;2:593–603. doi: 10.1016/s1097-2765(00)80158-6. [DOI] [PubMed] [Google Scholar]