Abstract
We have isolated a protein, mature RRFHCP, from chloroplasts of spinach (Spinacia oleracea L.) that shows 46% sequence identity and 66% sequence homology with ribosome recycling factor (RRF) of Escherichia coli. RRF recycles ribosomes through disassembly of the posttermination complex. From the cDNA analysis and from the amino-terminal sequencing of the isolated protein, the mature RRFHCP was deduced to have a Mr of 21,838 with 193 aa. It lacks the 78-aa chloroplast targeting sequence encoded by the RRFHCP cDNA sequence. The RRFHCP synthesized in vitro was imported into isolated chloroplasts with simultaneous conversion to the mature RRFHCP. Transcription of the gene coding for RRFHCP was not dependent on light, yet it was limited mostly to photosynthetic tissues in which only one transcript size was detected. Mature RRFHCP exerted a bactericidal effect on E. coli carrying temperature-sensitive RRF at the permissive temperature whereas wild-type E. coli was not affected.
The plant chloroplast translational system (reviewed in refs. 1 and 2) is similar to the prokaryotic one, in that chloroplasts and bacterial ribosomes have common features (3, 4), and are sensitive to the same antibiotics. Chloroplast mRNAs are similar to that of prokaryotes in that they are not m7G-capped and can be polycistronic (5, 6). Only a few chloroplast protein synthesis factors, like the elongation factor G (EF-G) homologue (7, 8), have been characterized. No information is available about disassembly of the post-termination complex of chloroplast ribosomes (1).
In bacteria, despite numerous studies on translation, the importance of disassembly of the post-termination complex of ribosomes has been overlooked (9). Two factors are necessary for the disassembly of the post-termination complex: ribosome recycling factor (RRF) (10) and either EF-G (11) or release factor 3 (RF3) (12). RRF, which is essential for bacteria (13), was purified (14) and cloned (15), and its characteristics have been reviewed (9, 10, 16). RRF increased translation 4- to 8-fold by recycling ribosomes from one round of translation to another (17–19). Inactivation of RRF in vivo is bactericidal or bacteriostatic depending on the growth phase (20). Every prokaryote genome sequenced so far has the RRF gene (frr) homologue except those belonging to Archae (see reviews in refs. 10, 16, and 21). Mycoplasma genitalium, the smallest free-living organism, retains RRF, suggesting a key role of this factor for bacterial life (discussed in ref. 16).
We report here the characterization of a nuclear-encoded RRF homologue in a plant. The spinach RRF homologue (RRFHCP) is processed to mature RRFHCP, which is a chloroplast protein. Transcripts of frrhcp (RRFHCP gene) were found in photosynthetic tissues, but transcription was not light-dependent. The expression of frrhcp in an Escherichia coli mutant with temperature-sensitive RRF was bactericidal even at the permissive temperature whereas it had no effect on the wild-type E. coli. This description of an “inhibitor” of prokaryotic RRF suggests that mature RRFHCP may function in the recycling of ribosomes in chloroplasts in a fashion similar to prokaryotic RRF.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
The strains and plasmids prepared for this study are shown in Table 1. The other bacterial strains and plasmids used in this study are MC1061[frr+ Smr, Δlac(IPOZYA)X74 (22), B834(DE3)pLysS (Novagen), LJ3 [MC1061 frr1 (frame shift), Δ(srl-recA)306::Tn10] (20), LJ14 [MC1061 frr14 (Ts)] (20), XL1-Blue MRF′ (Stratagene), pKK233–2 (Pharmacia), pET15b (Novagen), pCRII (Invitrogen), pBR322 (New England Biolabs), pIB279 (23), pUC19 (New England Biolabs), pRR2 (24), pMR2 (pSC101 replicon, wild-type frr and its promoter, Cmr) (13), and pRR1 (15).
Table 1.
Strains and plasmids
| Name | Characteristics |
|---|---|
| Strains | |
| LJ2708 | LJ3 transformed with pPEN1054sacBneo plasmid |
| LJ2211 | LJ3 transformed with pPEN1054sacBneo and pMR2 plasmids |
| LJ2846 | MC1061 harboring the F′ from XL1-Blue MRF′ |
| LJ2221 | LJ14 harboring the F′ from XL1-Blue MRF′ |
| Plasmids | |
| pKK233-2RRFM | pKK233–2 derivative carrying a fragment of the plant frr cDNA as a 0.58-kbp NcoI fragment encoding the mature part of the plant RRF |
| pET15b-RRFM | pET15b derivative carrying a fragment of the plant frr cDNA as a 0.55-kbp BamHI fragment encoding the mature part of the plant RRF fused to an N-terminal His-tag sequence |
| pCRII-RRF | pCRII derivative carrying a fragment of the plant frr cDNA as a 0.9-kbp EcoRI fragment (nucleotide residues 1–898 of the plant cDNA) |
| pPEN1054 | pBR322 carries frr (Wt: wild type) as a 0.9-kb EcoRI-SmaI fragment from pRR1 between its EcoRI-ScaI sites; tetracyclin resistance; ColE1 replicon |
| pPEN1054 sacBneo | pPEN1054 carries the sacB-Neor cassette from pIB279 as a 3.1-kb BamHI fragment inserted into its single BamHI site; Kmr; ColE1 replicon |
Immunological Screening, cDNA Isolation, and Sequencing.
Immunoscreening of the spinach cDNA library in λgt11 (25) (total of 1.5 × 105) was performed with antibodies against a subfraction of purified envelope proteins from spinach chloroplast (26). The nucleotide sequence on both strands of this isolated subclone (KpnI/SacI fragment) was determined and analyzed by using the pc/gene release 6.8 program (IntelliGenetics).
Southern and Northern Blot Analysis.
DNA or RNA was extracted from spinach plants grown as described (27). Northern and Southern blot analyses were performed by using Hybond-N+ membranes (Amersham). Hybridization was allowed to proceed overnight at 65°C. Membranes were washed in 2× SSC (0.3 M NaCl/0.03 M Na3-citrate)/0.1% SDS at room temperature and 1× SSC/0.1% SDS for 15 min at 65°C.
Expression of Recombinant Mature RRFHCP in E. coli and Antibody Production.
Histidine-tagged partial RRFHCP was expressed in B834(DE3)pLysS E. coli harboring pET15b-RRFM; pET15b (Novagen) carrying the PCR-amplified and BamHI-digested fragment (primers AGTGGATCCTGAAAAGTCGTTGATAG and CTGGGATCCTTAGACTTTCATTAACTC) coding for the 185 C-terminal residues of RRFHCP (the region homologous to the entire E. coli RRF) (15). The construction allows expression in E. coli of a fusion protein of 210 residues (23,726 Da) including 25 residues encoded by the pET15b plasmid. The highly expressed RRFHCP derivative was purified to homogeneity by metal affinity chromatography (NTA; Novagen), PD10 column (Pharmacia), and CM-acryl IBF column (IBF, Villeneuve-la-Garenne, France). The pure recombinant protein thus prepared (named His6-partial RRFHCP, 2 mg) was used to obtain a rabbit polyclonal antibody (Eurogentec, Brussels) against mature RRFHCP.
Purification of Antibodies Against Mature RRFHCP and Western Blot Analysis.
IgGs were purified from the rabbit antiserum (28) and subsequently subjected to affinity chromatography on His6-partial RRFHCP bound to a CNBr-activated Sepharose 4B (Pharmacia) (29). Western blot analysis was performed by using the goat anti-rabbit alkaline phosphatase-conjugated antibody (dilution, 1:8,000; Sigma).
Subcellular Fractionation of Spinach Leaves.
Spinach chloroplasts and mitochondria were purified starting with spinach leaves purchased from markets as described (30, 31). Envelope, stroma, and thylakoid subfractions from the chloroplasts were purified and checked for purity (30).
Import of RRFHCP into Intact Chloroplasts with Simultaneous Maturation.
The cDNA fragment (from nucleotides l–898; see Fig. 1B) was inserted into the EcoRI site of the plasmid pCRII (Invitrogen) downstream from the sp6 promoter. This plasmid (PCRII-RRFHCP) was linearized and transcribed with sp6 RNA polymerase (Boehringer Mannheim). The transcripts were translated in wheat germ extracts (Boehringer Mannheim) with [35S]methionine (37 Tbq/mmol; Amersham). The labeled RRFHCP that was made (in 2.5 μl of the translation assay mixture) was incubated with pea chloroplasts corresponding to 18 μg chlorophyll at 25°C for 15 min in 100 μl of a reaction mixture (32). The mixture was incubated further with or without thermolysin (0.1 mg/mg of chlorophyll) in the presence of 0.5 mM CaCl2. Intact chloroplasts then were purified from the reaction mixture as described (32) and were analyzed by SDS/PAGE (33) and fluorography.
Figure 1.
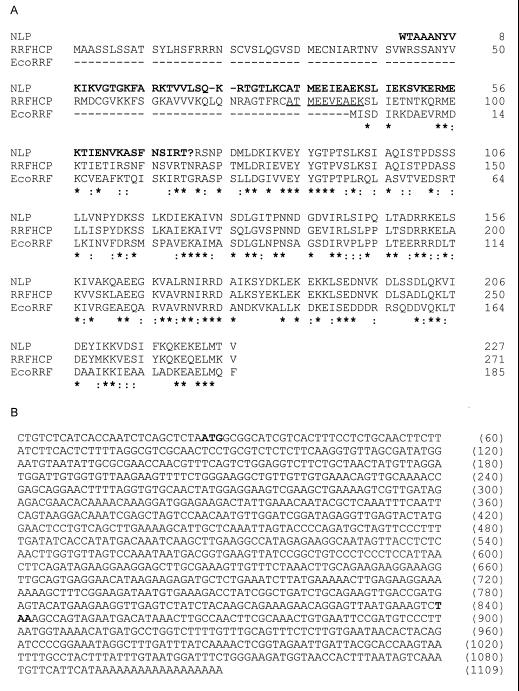
(A) Amino acid sequence of RRFHCP and NLP (from carrot) in comparison with E. coli RRF sequence (EcoRRF). The NLP amino acid sequence is not deduced from the original sequence (GenBank accession no. X72384) but from a corrected sequence in which 1 nt (G216) has been removed. The modified residues are represented in bold and as a “?”. Two gaps (–) have been inserted in the NH2-terminal part of the NLP sequence. The NH2-terminal sequence of mature RRFHCP (10 residues, underlined letters) was determined by sequencing of mature RRFHCP from spinach chloroplasts. Identical (∗) and similar (two dots) residues are indicated. Conservative amino acid substitutions were grouped as follows: I-L-M-V, K-R-H, A-S-P-T-G, Y-F-W, N-Q, and D-E. (B) DNA sequence of RRFHCP cDNA. The initiation (ATG) and the termination (TAA) codons are in bold.
Purification of Mature RRFHCP from Spinach Chloroplast.
Starting from 3-kg leaves, we obtained 600 mg of stromal proteins (30). RRFHCP from this fraction was isolated on a CM-trisacryl (IBF) column, preequilibrated with 50 mM ammonium acetate buffer (pH 6.9). An ammonium acetate (pH 8.0) gradient, 50–250 mM, was used to elute mature RRFHCP, which was detected at the beginning of the gradient with the antibody against His6-partial RRFHCP. The protein was measured by the Bradford method (34).
Assay of E. coli RRF and Complementation Assay of E. coli RRF Mutants with the Plant RRF Homologue.
The RRF activity was measured as described (35, 36) at 32°C. E. coli polysomes were isolated and treated with puromycin to remove the nascent peptide chains. Conversion of these polysomes to monosomes was catalyzed by EF-G and RRF and used for determining RRF activity. Plasmid pKK233–2RRFM was constructed by linearization of pKK233–2 (Pharmacia) with NcoI and ligation to a PCR-amplified fragment (primers TTTACCATGGCAACTAT GGAGGAAGTC and ATTCCCATGGCTTTAGACTTTCATTAAC) corresponding to amino acids 79–271 of RRFHCP. E. coli LJ2708 [RecA− lacI− with frameshifted frr in the chromosome and harboring a pPEN1054sacBneo, carrying kanamycin-resistance gene (Kmr) and wild-type E. coli frr] was transformed with pKK233–2RRFM by using Apr as a selection marker. Similarly, LJ2211 [LJ2708 harboring additional plasmid pMR2 carrying wild-type frr and chloramphenicol-resistance gene (Cmr)] was transformed with pKK233–2RRFM. For the complementation assay, the presence of the resident plasmid, pPEN1054sacBneo, was examined by kanamycin resistance.
Bactericidal Action of Mature RRFHCP on LJ14.
MC1061 [lacI− with wild-type frr (22)] and its temperature-sensitive RRF derivative, LJ14 (20), were transformed with pKK233–2RRFM carrying frrhcp, the expression of which is controlled by the lac promoter. LJ14 carries frr14, which codes for temperature-sensitive RRF because of the amino acid change at Val(117)Asp. Where indicated, the F′[proAB lacIqΔ(lacZ)M15 Tn10] was transferred to the strain by conjugation (37) from E. coli XL1-Blue MRF′ (Stratagene) to obtain strains LJ2846 or LJ2221 by using streptomycin (50 μg/ml, for recipient) and tetracycline [10 μg/ml, for selection of bacteria containing F′, which carries the tetracycline-resistance gene (Tn10)] for double-selection of the transconjugants.
RESULTS
Molecular Cloning of a Plant RRF Homologue.
A spinach cDNA expression library in λgtll was screened with antibodies against a protein fraction from chloroplast envelope membranes for clones expressing envelope proteins. Three independent, identical positive clones containing a 1.1-kbp cDNA insert were found. Northern blot analysis on total spinach leaf RNA by using this cDNA as a probe revealed a single band of approximately the same size (1.1 kbp). The λgt11 DNA insert was subcloned and sequenced. A protein of 271 aa residues (30,431 Da) (Fig. 1A) is coded for by the 1,109-bp nucleotide sequence (Fig. 1B). The putative initiation methionine at nucleotides 27–29 is followed by an alanine residue that is common in most plant proteins (38). This protein has a significant homology to E. coli RRF (15): the C-terminal sequence (residues 87–271) of the spinach protein possesses 46% identity (66% homology) with the sequence of E. coli RRF (Fig. 1A). This protein was named RRFHCP. The N terminus of the deduced amino acid sequence of RRFHCP is 86 aa residues longer than that of the bacterial RRF (Fig. 1A). This sequence is compatible with mitochondria- and chloroplast-targeting sequences (39).
Spinach Contains a Single Gene for RRFHCP That Is Highly Expressed in Photosynthetic Tissues.
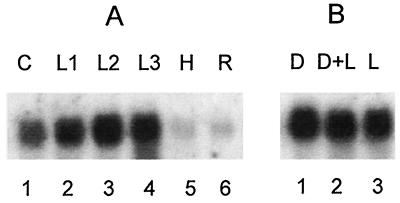
A Southern blot of genomic digests was probed with an internal fragment of the RRFCP cDNA. One band only was detected in the EcoRI, HindIII, and EcoRV digests (data not shown), suggesting the presence of one gene per haploid genome. Northern blot analysis performed on total RNA from various spinach tissues revealed a constitutive expression of the gene during plant development in both cotyledons and leaves (Fig. 2A), whereas the expression in hypocotyles and roots was very low. Thus, expression of the RRFHCP gene appears to be almost restricted to photosynthetic tissues. Because high expression of RRFHCP also was detected in spinach leaves grown in the dark (Fig. 2B), the expression appears to be light-independent but tissue-specific.
Figure 2.
Northern blot analysis of plant RRF homologue. (A) Tissue-specific expression of RRFHCP. Northern blots were performed on total RNA (10 μg each) from cotyledons (C), leaves (L), hypocotyls (H), and roots (R) isolated from 20-day-old spinach plants. L1, 2, and 3 represent different ages of leaves, with L1 as the oldest (approximately 10 days). (B) Expression of RRFHCP is not light-dependent. Lanes: 1, 18 days in the dark (D); 2, 14 days in the dark followed by 4 days in the light (D+L); 3, 18 days in the light (L). Methylene blue staining of total RNA and serial hybridizations with different probes were used as loading controls (not shown). The 32P-labeled DNA probe corresponds to nucleotides 269–477 of RRFHCP cDNA.
The Plant RRF Homologue Is Localized Within the Chloroplast.
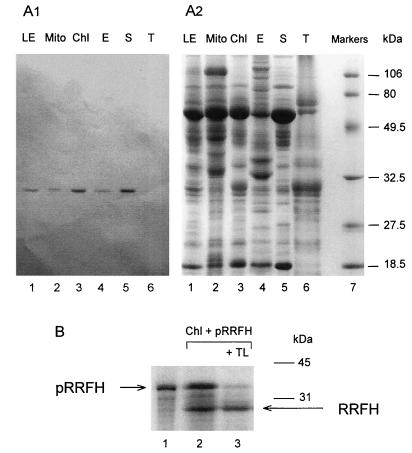
We expressed histidine-tagged spinach RRF (His6-partial RRFHCP) in E. coli cells and raised antibodies against this protein. Western blot analysis performed on leaf extracts (Fig. 3) showed that a single protein reacted with antibodies raised against His6-partial RRFHCP. Its apparent molecular mass was smaller (26,500 Da; Fig. 3A1 and A2) than the size deduced from the nucleotide sequence (30,300 Da). This was expected because of the presence of an additional N-terminal sequence in RRFHCP, as compared with bacterial RRF (Fig. 1). We call the 26,500-Da protein “mature RRFHCP.” This means that RRFHCP is a “precursor ” to the “mature” RRFHCP.
Figure 3.
Subcellular localization of RRFHCP. (A) Analysis of polypeptides from different spinach leaf subfractions. (A1) Western blot analysis of polypeptides (50 μg protein) from spinach leaf subfractions separated by SDS/PAGE with antibody (dilution of 1:1,500) against His6-partial RRFHCP. (A2) Coomassie blue staining of the corresponding gel. Lanes: 1, total leaves (LE); 2, mitochondria (Mito); 3, chloroplast (Chl); 4, chloroplast envelope membranes (E); 5, stroma (S); 6, thylakoid membranes (T); 7, molecular mass markers (low range; Bio-Rad). (B) Import of the [35S]methionine-labeled RRFHCP into isolated pea chloroplasts. The mixtures for the in vitro import reaction were analyzed by SDS/PAGE and fluorography. Lanes: 1, in vitro translation products of RRFHCP cDNA inserted into pCRII; 2, 35S-labeled RRFHCP (10-fold more than lane 1) was incubated with pea chloroplasts and chloroplasts were isolated after the import reaction; 3, the same as lane 2 except that the chloroplasts were treated further with thermolysin (TL) to remove proteins bound to the cytosolic face of the chloroplast outer envelope. The mature and precursor proteins are indicated as RRFH and pRRFH, respectively.
Western blot analysis allows the detection of mature RRFHCP in both chloroplasts and mitochondria (Fig. 3), but mature RRFHCP was enriched only in chloroplasts. In chloroplasts, the major proportion of mature RRFHCP was found in the stroma, some RRFHCP was associated with the envelope subfraction, but no RRFHCP was detected in the thylakoids (Fig. 3A1). The significance of RRFHCP association with envelope membranes is unclear and presently under investigation. All these observations were confirmed further by immunolabeling of RRFHCP within isolated intact spinach chloroplasts followed by electron microscopy (results not shown).
After its import into isolated chloroplasts, RRFHCP (apparent Mr of 35,000) was processed into a protein with an apparent Mr of 26,500 (Fig. 3B), a value identical to that of the mature RRFHCP (Fig. 3A). The processed protein was not digested by thermolysin (Fig. 3B), indicating that it was imported into and processed within the chloroplast. Thermolysin does not cross the envelope membranes and is used to digest the proteins that are accessible on the cytosolic face of the outer envelope membrane (40). It therefore appears that the N-terminal sequence of RRFHCP functions as a chloroplast-targeting sequence.
Characterization of Mature RRFHCP from Spinach Chloroplast.
To characterize further mature RRFHCP, we purified the protein from spinach chloroplast stroma. A single polypeptide reacting with the antibody raised against His6-partial RRFHCP was eluted from a CM-acryl column. Starting from 600 mg stromal proteins (i.e., 3 kg spinach leaves), we obtained 0.3 mg pure, mature RRFHCP protein. This indicates that RRFHCP is a minor chloroplast protein. The N-terminal sequence of the purified protein (NH2-ATMEEVEAEK-COOH) corresponds to residues 79–88 of RRFHCP deduced from the cDNA sequence (Fig. 1). Therefore, we concluded that the mature RRFHCP is composed of 193 aa residues (21,838 Da).
The Mature RRFHCP Cannot Function for E. coli RRF.
We prepared a plasmid (pKK233–2RRFM) carrying truncated (t) frrhcp and Apr. t-frrhcp codes for the mature RRFHCP. We examined whether this could functionally replace the resident plasmid A (pPEN1054sacBneo) carrying E. coli frr and Kmr. For this, the resident plasmid was placed in a strain of E. coli (LJ2708) whose chromosomal RRF gene (frr) is nonfunctional because of a frameshift mutation in frr (13). This strain was transformed with pKK233–2RRFM (incoming plasmid). As can be seen from Table 2 (line 1, column 4), 100% of the ampicillin-resistant transformants carrying the plant frr homologue were kanamycin-resistant because of the presence of plasmid A, which carried wild-type frr, despite its incompatibility with the incoming plasmid (pKK233–2RRFM). This indicates the absolute requirement for the wild-type E. coli frr, even in the presence of the plant frr homologue: t-RRFHCPfrr does not function in E. coli. The situation was identical when an empty vector (pUC19) was the incoming plasmid (Table 2, column 4, line 5). Line 3 in Table 2 indicates what can be expected if the incoming plasmid functions successfully, leading to the loss (87.5%) of the resident plasmid A (only 12.5% of the population was kanamycin-resistant). Line 2 shows that plasmid A and pKK233–2RRFM are incompatible because plasmid A was eliminated (only 9.4% remaining) if the cell had another resident plasmid B (pMR2, carrying functional frr), which is compatible with pKK233–2RRFM.
Table 2.
The spinach RRF gene, frrhcp, cannot functionally replace the E. coli RRF gene, frr
| E. coli strains | Incoming plasmid | Resident plasmid | Percentage of E. coli with frrhcp retaining resident plasmid A |
|---|---|---|---|
| a | pKK233-2RRFM | A | 100 |
| b | pKK233-2RRFM | A + B | 9.4 |
| a | pRR2 | A | 12.5 |
| b | pRR2 | A + B | 0 |
| a | pUC19 | A | 100 |
| b | pUC19 | A + B | 0 |
Plasmid pKK233–2RRFM carries Apr and t-frrhcp, the spinach RRF gene. Plasmid pRR2 carries E. coli frr in pUC19 (13). Two strains were transformed by these plasmids and pUC19; a, LJ2708, frr−, lac, [E. coli harboring plasmid A (pPEN1054sacBneo carrying Kmr and frr)]. b, LJ2211, the same as strain a harboring plasmids A and B (pMR2 carrying frr and Cmr). Plasmid A, pKK233-2RRFM and pRR2 have the ColE1 replicon, making them incompatible with each other; plasmid B has a different replicon (pSC101 origin), making it compatible with pKK233–2RRFM. Transformants were selected by ampicillin with six single-colony isolations and tested for Kmr to examine the presence of the resident plasmid A.
Bactericidal Action of Mature RRFHCP on E. coli with Temperature-Sensitive RRF.
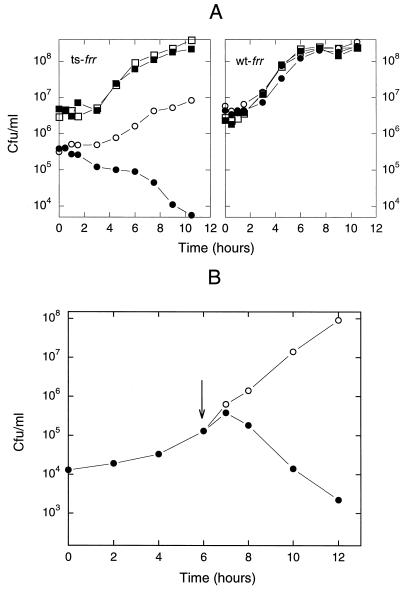
In Fig. 4, E. coli LJ2221 (with temperature-sensitive RRF), harboring pKK233–2RRFM carrying t-frrhcp, was exposed to isopropyl β-d-thiogalactoside, which induces the production of mature RRFHCP in both lag (Fig. 4A) and logarithmic (Fig. 4B) growth phases. The colony-forming unit decreased upon expression of the plant gene, indicating that RRFHCP is bactericidal in E. coli carrying temperature-sensitive RRF. The RRFHCP effect cannot be a simple toxic effect of the foreign protein because RRFHCP did not exert any effect on the wild-type E. coli (Fig. 4A Right). In a separate experiment, it was found that, because of the toxic effect of the plant RRF, bacteria tend to eliminate the plasmid carrying the frrcp (data not shown).
Figure 4.
The expression of the plant frr gene in E. coli carrying temperature-sensitive RRF is bactericidal even at the permissive temperature, 32°C. (A) At the lag phase. Circles, temperature-sensitive E. coli (LJ2221)/lacIq F′ with temperature-sensitive RRF (Left) or wild-type E. coli RRF (LJ2846)/lacIq F′ (Right), both harboring pKK233–2RRFM (carries t-frrhcp). Squares, the same as above except that bacteria harbored empty vector, pKK233–2. Bacteria (0.05 OD at 540 nm in LB) were divided into two aliquots, incubated with (solid symbols) or without (open symbols) 5 mM isopropyl β-d-thiogalactoside (IPTG) added at 0 time. (B) At the log phase; as in A except that the temperature-sensitive bacteria (0.05 OD at 540 nm in LB) were grown for 6 hr. At the point of the arrow, the culture was divided into two parts and IPTG was added to one of them (solid symbols). Samples were plated onto LA containing 100 μg/ml of ampicillin for viable counts.
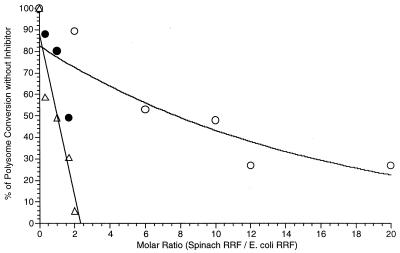
The selective inhibitory effect of RRFHCP on E. coli temperature-sensitive RRF was confirmed further by in vitro experiments (Fig. 5). The activity of temperature-sensitive RRF was almost completely inhibited by a 2-fold excess of mature RRFHCP whereas 30-fold excess inhibited wild-type RRF by only 70%.
Figure 5.
The E. coli RRF reaction is inhibited by mature RRFHCP. The activity of wild-type RRF (10 μg, ○; 60 μg, ●) and temperature-sensitive RRF (60 μg, ▵) is expressed as a percentage of that which occurs in the absence of the inhibitor. This is plotted against the molar ratio of mature RRFHCP to E. coli RRF. Because of the low activity of the temperature-sensitive RRF, it was not technically possible to use 10 μg of it.
DISCUSSION
The data presented in this paper support the hypothesis that eukaryotic RRF is an organelle protein and probably involved in protein synthesis within the organelles in the same way that bacterial RRF functions.
First, mature RRFHCP is localized in the chloroplast. This is supported by several lines of evidence. (i) Mature RRFHCP has been purified starting from Percoll-purified chloroplast, where it is enriched. (ii) The molecular mass of mature RRFHCP isolated from chloroplasts is 26,500 Da. This is similar to that predicted from the cDNA sequence of RRFHCP, assuming that RRFHCP undergoes a posttranslational maturation in chloroplasts. (iii) This assumption was proven to be correct by N-terminal sequencing of the mature protein purified from isolated chloroplasts and by in vitro import of RRFHCP into isolated intact chloroplasts with conversion to mature RRFHCP. (iv) The N-terminal sequence of RRFHCP deduced from the cDNA sequence is identified as a plant organelle-targeting signal by computer analysis (39).
Second, the only other known plant cDNA encoding an RRF homologue is NLP (for nuclear located protein) from carrot cells (GenBank accession no. X72384). Examination of the NLP sequence revealed that introduction of a single nucleotide gap in the carrot sequence (guanosine 216) allows alignment of the NH2-terminal sequences of NLP and RRFHCP, including 50 residues of RRFHCP-targeting sequence (Fig. 1A). The carrot and spinach proteins share 76% identity and 89% homology in their mature part, suggesting a function and localization of NLP in carrots that is similar to that of RRFHCP in spinach.
Third, the RRFHCP gene was constitutively expressed in photosynthetic tissues, which abound in chloroplasts. It is poorly expressed in roots and hypocotyles (Fig. 2). The specific presence of RRFHCP in chloroplasts of green leaves correlates with the huge increase of protein synthesis in photosynthesizing tissues during leaf greening (41). Interestingly, the chloroplast ribosomal protein CS1 is expressed in the same tissue-specific, light-independent manner as RRFHCP (42).
Fourth, translation in chloroplasts is similar to that of prokaryotes (1, 43). Recent evidence that yeast RRF may be a mitochondrial protein (44) is consistent with the notion that the eukaryotic RRF homologue is an organelle protein. Indeed, our preliminary studies on yeast RRF homologue suggest that it is not essential for the cytoplasmic protein synthesis but is essential for the maintenance of mitochondria (to be published elsewhere).
Fifth, mature RRFHCP has a specific inhibitory action on E. coli temperature-sensitive RRF but not on wild-type RRF. This suggests that mature RRFHCP interacts with other components of the translation machinery in a fashion similar to that of E. coli RRF so that it can exert a competitive inhibitory action on the bacterial temperature-sensitive RRF but not on wild-type RRF. One explanation would be that temperature-sensitive RRF, even at the permissive temperature, has a weaker affinity for its substrate than wild-type RRF. It is conceivable that the amino acid substitution that gave thermolability has influenced the temperature-sensitive RRF structure even at the permissive temperature.
That eukaryotic RRF is an organelle protein and therefore is not involved in the cytoplasmic protein synthesis of eukaryotes is consistent with the recent finding that Archea, despite being prokaryotes, have no frr homologue (21). These organisms have diverged very early in evolution from Eubacteria, their translation machinery is most similar to that of eukaryotes (45) and, being bacteria, they do not have organelles such as chloroplasts and mitochondria.
The bactericidal effect of RRFHCP has an important implication for the development of antimicrobial agents targeted against bacterial RRF. The irreversible inhibition of bacterial RRF resulted in a bactericidal effect regardless of the growth phase of the bacteria. This is in contrast to the bacteriostatic effect of reversible inactivation of temperature-sensitive RRF during the growth phase but correlates with the bactericidal effect during the lag phase (20). Therefore, inhibition of RRF is fatal to prokaryotes. That eukaryotic RRF is an organelle protein suggests that it is not essential for the major eukaryotic cytoplasmic protein synthesis. Antimicrobial agents targeted against bacterial RRF therefore may be harmful only to mitochondria. However, widely used antibiotics such as erythromycin and tetracycline, which influence mitochondrial protein synthesis (46, 47), are with little side effects. On the other hand, organelle and prokaryotic RRF probably have distinct functional and structural features that would make it possible to develop new bactericidal agents that do not inhibit organelle RRF of eukaryotes.
Acknowledgments
We thank Dr. J. Garin and M. Vinçon (Commissariat à l’Energie Atomique, Grenoble, France) for NH2-terminal sequencing, Dr. E. Maréchal (Commissariat à l’Energie Atomique, Grenoble, France) for preparation of the antibody, and Dr. R. Dumas (Unité Mixte de Recherche 41, Centre National de la Recherche Scientifique/Rhône Poulenc Agrochimie, Lyon, France) for providing spinach cDNA library.
ABBREVIATIONS
- RRF
ribosome recycling factor
- RRFHCP
chloroplast RRF homologue
- frr
E. coli RRF gene
- frrhcp, RRFHCP gene
Apr, Cmr, Kmr, and Smr, ampicillin, chloramphenicol, kanamycin, and streptomycin resistances, respectively
Note
The Ad Hoc Nomenclature Subcommittee gave the name RF4 (48) for ribosome recycling factor [originally called ribosome releasing factor (35)], but the committee recently agreed not to use RF4 because RRF should not be confused with the peptide release factors involved in the termination of the chain elongation. A short note will be published on this matter elsewhere.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the EMBL database (accession no. AJ133751).
References
- 1.Harris E H, Boynton J E, Gillham N W. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern D B, Higgs D C, Jianjun Y. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 3.Bohnert H J, Crouse E J, Schmitt J M. Organization and Expression of Plastic Genomes: Encyclopedia of Plant Physiology. 14B. New York: Springer; 1982. pp. 475–530. [Google Scholar]
- 4.Dorne A-M, Eneas-Fiho J, Heizmann R, Mache R. Mol Gen Genet. 1984;193:129–134. doi: 10.1007/BF00327425. [DOI] [PubMed] [Google Scholar]
- 5.Sugita M, Sugiura M. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 6.Rochaix J D. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 7.Akkaya M S, Welsch P L, Wolfe M A, Duerr B K, Becktel W J, Breinberger C A. Arch Biochem Biophys. 1994;308:109–117. doi: 10.1006/abbi.1994.1016. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Torres J, Breitenberger C A, Spielmann A, Stutz E. Biochim Biophys Acta. 1993;1174:191–194. doi: 10.1016/0167-4781(93)90114-s. [DOI] [PubMed] [Google Scholar]
- 9.Janosi L, Hara H, Zhang S, Kaji A. Adv Biophys. 1996;32:121–201. doi: 10.1016/0065-227x(96)84743-5. [DOI] [PubMed] [Google Scholar]
- 10.Kaji A, Teyssier E, Hirokawa G. Biochem Biophys Res Commun. 1998;250:1–4. doi: 10.1006/bbrc.1998.9168. [DOI] [PubMed] [Google Scholar]
- 11.Hirashima A, Kaji A. J Biol Chem. 1973;248:7580–7587. [PubMed] [Google Scholar]
- 12.Grentzmann G, Kelly P J, Laalami S, Shuda M, Firpo M A, Cenatiempo Y, Kaji A. RNA. 1998;4:973–983. doi: 10.1017/s1355838298971576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janosi L, Shimizu I, Kaji A. Proc Natl Acad Sci USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirashima A, Kaji A. Biochemistry. 1972;11:4037–4044. doi: 10.1021/bi00772a005. [DOI] [PubMed] [Google Scholar]
- 15.Ichikawa S, Kaji A. J Biol Chem. 1989;264:20054–20059. [PubMed] [Google Scholar]
- 16.Janosi L, Ricker R, Kaji A. Biochimie. 1996;78:959–969. doi: 10.1016/s0300-9084(97)86718-1. [DOI] [PubMed] [Google Scholar]
- 17.Ryoji M, Karpen J W, Kaji A. J Biol Chem. 1981;256:5798–5801. [PubMed] [Google Scholar]
- 18.Pavlov M Y, Freistroffer D V, Heurgue-Hamard V, Buckingham R H, Ehrenberg M. J Mol Biol. 1997;273:389–401. doi: 10.1006/jmbi.1997.1324. [DOI] [PubMed] [Google Scholar]
- 19.Kung H-F, Treadwell B V, Spears C, Tai P-C, Weissbach H. Proc Natl Acad Sci USA. 1977;74:3217–3221. doi: 10.1073/pnas.74.8.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janosi L, Mottagui-Tabar S, Isaksson L A, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 22.Casadaban M J, Martinez-Arias A, Shapira S K, Chou J. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 23.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu I, Kaji A. J Bacteriol. 1991;173:5181–5187. doi: 10.1128/jb.173.16.5181-5187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyunh T V, Young R A, Davis R W. In: cDNA Cloning: A Practical Approach. Glover D M, editor. Oxford: IRL; 1985. pp. 49–78. [Google Scholar]
- 26.Maréchal E, Block M A, Joyard J, Douce R. C R Acad Sci Paris. 1991;313:521–528. [Google Scholar]
- 27.Teyssier E, Block M A, Douce R, Joyard J. Plant J. 1996;10:903–912. doi: 10.1046/j.1365-313x.1996.10050903.x. [DOI] [PubMed] [Google Scholar]
- 28.Saint-Blancard J, Foucard J, Limmone F, Girot P, Boschetti E. Ann Pharm Fr. 1980;39:403–409. [PubMed] [Google Scholar]
- 29.Rolland N, Droux M, Lebrun M, Douce R. Arch Biochem Biophys. 1993;300:213–222. doi: 10.1006/abbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 30.Douce R, Joyard J. In: Methods in Chloroplast Molecular Biology. Edelman M, Hallick R, Chua N-H, editors. Amsterdam: Elsevier Science; 1982. pp. 239–256. [Google Scholar]
- 31.Douce R, Bourguignon J, Brouquisse R, Neuburger M. Methods Enzymol. 1985;148:403–415. [Google Scholar]
- 32.Waegemann K, Soll J. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Hirashima A, Kaji A. J Mol Biol. 1972;65:43–58. doi: 10.1016/0022-2836(72)90490-1. [DOI] [PubMed] [Google Scholar]
- 36.Hirashima A, Kaji A. Biochem Biophys Res Commun. 1970;41:877–883. doi: 10.1016/0006-291x(70)90165-8. [DOI] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1972. pp. 351–376. [Google Scholar]
- 38.Lutcke H A, Chew K C, Mickel F S, Moss K A, Kern H F, Scheele G A. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joyard J, Billecocq A, Bartlett S G, Block M A, Chua N-H, Douce R. J Biol Chem. 1983;258:10000–10006. [PubMed] [Google Scholar]
- 41.Mayfield S P, Yohn C B, Cohen A, Danon A. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- 42.Franzetti B, Zhou D-X, Mache R. Nucleic Acids Res. 1992;20:4153–4157. doi: 10.1093/nar/20.16.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danon A. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanai T, Takeshita S, Atomi H, Umemura K, Ueda M, Tanaka A. Eur J Biochem. 1998;256:212–220. doi: 10.1046/j.1432-1327.1998.2560212.x. [DOI] [PubMed] [Google Scholar]
- 45.Bell S D, Jackson S P. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 46.Pious D A, Hawley P. Pediatr Res. 1972;6:687–692. doi: 10.1203/00006450-197208000-00007. [DOI] [PubMed] [Google Scholar]
- 47.Doersen C J, Stanbridge E J. Biochim Biophys Acta. 1982;698:62–69. doi: 10.1016/0167-4781(82)90185-3. [DOI] [PubMed] [Google Scholar]
- 48.Clark B F C, Grunberg-Manago M, Gupta N K, Hershey J W B, Hinnebusch A G, Jackson R J, Maitra U, Mathews M B, Merrick W C, Rhoads R E, et al. Biochimie. 1996;78:1119–1122. [Google Scholar]