Abstract
Although de novo protein design is an important endeavor with implications for understanding protein folding, until now, structures have been determined for only a few 25- to 30-residue designed miniproteins. Here, the NMR solution structure of a complex 73-residue three-helix bundle protein, α3D, is reported. The structure of α3D was not based on any natural protein, and yet it shows thermodynamic and spectroscopic properties typical of native proteins. A variety of features contribute to its unique structure, including electrostatics, the packing of a diverse set of hydrophobic side chains, and a loop that incorporates common capping motifs. Thus, it is now possible to design a complex protein with a well defined and predictable three-dimensional structure.
Protein folding is a complex process involving van der Waals and hydrophobic interactions, electrostatics, and hydrogen bonding networks. One approach to understanding protein folding is to design from scratch a particular protein fold, thoroughly characterize its solution properties, and determine its three-dimensional structure. The field of de novo protein design (1, 2) has experienced some recent exciting successes in the redesign of natural proteins to incorporate novel, functional metal-binding sites (3, 4). Also, the redesign of proteins patterned after the sequence or three-dimensional structural motifs such as the zinc finger (5–8), coiled coils (9), or other small protein domains (10, 11) has progressed quite significantly. Unnatural right-handed coiled coils have been successfully designed (12), and small, marginally stable models for protein secondary (13, 14) and supersecondary structures, including helix-loop-helix (15, 16) and three-stranded β-hairpin motifs (17–20), have been designed and shown to adopt the desired conformation. However, the de novo design of larger proteins with well defined hydrophobic cores and stabilities similar to natural proteins has proven to be more difficult. Often, designed proteins have adopted more dynamic structures characteristic of a molten globule conformation (1). Such structures lack the well packed apolar cores that are characteristic of the native states of proteins and are essential to their functional properties as catalysts, transducers, mechanical devices, etc. The tight and unique packing of a protein core also can be discerned in its thermodynamic and spectroscopic properties, including cooperative protein unfolding curves, well dispersed NMR spectra, and a lack of binding of hydrophobic dyes. Even more discriminating features include the change in heat capacity for unfolding, the rates and mechanism of hydrogen exchange, and—most importantly—the adoption of a unique native-like structure.
A few helical bundles (21, 22) and a coiled coil (12, 23) have passed many or all of these tests. However, the structures of only a few of these are known at high resolution. The topology of one dimeric four-helix bundle was found to differ from the design (21), and it was not possible to calculate a unique structure for a second dimeric four-helix bundle by using NMR distance restraints (24). Also, the structure of a four-helix bundle consisting of four identical α-helices (originally designed to solubilize membrane proteins) interconnected by Gly-rich loops has been determined by x-ray crystallography (22). However, the loops were not resolved in the structure, so it was impossible to determine whether the helices adopted a clockwise or counterclockwise topology. Thus, the design and structure determination of a single-chain, native-like protein of more than ≈30 residues has remained an important, unresolved problem.
The three-helix bundle occurs ubiquitously in nature as a robust scaffold for molecular recognition. First observed in the helical IgG-binding domains of Staphylococcal aureus (25), this family has grown to include DNA-binding proteins, enzymes, and structural proteins (26). Surprisingly, despite its widespread utility, there have been few attempts to design single-chain antiparallel three-helix bundles (27, 28). Therefore, α3C has recently been designed (27) by using as a starting point the crystal structure of a de novo designed antiparallel three-stranded coiled coil, “Coil-Ser” (29). In a hierarchic approach (1), the helices of Coil-Ser were shortened to a length typical of globular three-helix bundles, N-terminal capping boxes (30, 31) were included, and the electrostatic interactions between the helices were rearranged to stabilize the desired counterclockwise topology (32). Finally, the hydrophobic core was repacked with a diverse set of amino acids by using a genetic side-chain packing algorithm (33), yielding α3C. This protein was native-like as assessed from its cooperative thermal unfolding, hydrogen-deuterium exchange, and the chemical shift dispersion of its NMR spectra. Dutton and colleagues (28) recently also designed a three-helix bundle that appears native-like as judged by its NMR chemical shift dispersion.
In this report, we describe the structure determination of α3D, a derivative of α3C (Fig. 1; ref. 27). α3D contains 19 of the 20 naturally occurring amino acids (it lacks a cysteine), and it is native-like by all thermodynamic criteria. α3D differs from the α3C sequence by the following amino acid changes: M1, G2, E9Q, S16T, and S65D. Positions 9, 16, and 65 are surface-exposed and were changed to decrease the sequence homology between the α-helices.
Figure 1.
Sequences of the α3 family and of Coil-Ser. The residues are aligned with their corresponding heptad position in a coiled coil (26). α3A through α3C and Coil-Ser were chemically synthesized whereas α3D was cloned and expressed in E. coli. The residues that are different between α3D and α3C are labeled in bold.
MATERIALS AND METHODS
Expression and Characterization of α3D.
A synthetic gene for α3D was cloned into pET-16b (Novagen) and expressed in Escherichia coli strain BL21(DE3) (Novagen). After sonication and heat denaturation, α3D was purified to homogeneity on a C18 preparative reverse phase HPLC. Molecular weight was determined by electrospray mass spectroscopy (theoretical, 7977.4; determined, 7977.2). Thermal unfolding curves show that α3D is fully folded at room temperature, between pH 3.0 and pH 7.0 with melting temperature (Tm ranging from 80 to 95°C, and a heat capacity change (ΔCp) of 12 cal⋅mol−1⋅K−1 per residue. Guanidine hydrochloride denaturation in 10 mM sodium acetate (pH 5.0) and 30°C yields a free energy change for unfolding (ΔGu) of 5.1 kcal⋅mol−1 (m = 2.4 kcal⋅ mol−1⋅M−1, Cm = 2.8 M). Differential scanning caloriometry studies conducted on α3D at pH 3.0 show a two-state reversible transition (ΔHvan’t Hoff = ΔHDSC) with a ΔH of −44 kcal⋅mol−1 and a ΔCp of 10 cal⋅mol−1⋅K−1 per residue (S.T.R.W., S. F. Betz, P. Liebman, and W.F.D., unpublished results).
NMR Sample Preparation.
Isotopically enriched α3D for NMR studies was grown on M9 minimal media containing 15NH4Cl (1.0 g/liter, Isotec) with or without [U-13C]glucose (2.0 g/liter, Isotec) as the sole nitrogen and carbon sources, respectively. NMR samples included an 15N-α3D (2 mM, 92:8% H2O to 2H2O), 15N,13C-α3D (3 mM, 92:8% H2O to 2H2O), and 15N, 13C-α3D (2 mM, 99.9% 2H2O) all in 50 mM deuterated sodium acetate and 0.05% sodium azide, with a pH uncorrected for the isotope effect of 5.0.
NMR Spectroscopy.
NMR spectra were collected at 30°C on a Bruker (Bellerica, MA) DMX 600-MHz spectrometer equipped with a 5-mm x,y,z-shielded pulsed field gradient triple resonance probe. Aromatic experiments (also at 30°C) were collected on a Varian INOVA 750-MHz spectra equipped with a 5-mm z-shielded pulsed field gradient triple resonance probe. A series of three-dimensional experiments [HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, CBCA(CO)NH, C(CO)NH, H(CCO)NH, 15N-nuclear Overhauser effect (NOE) spectroscopy–heteronuclear single-quantum coherence, 15N-TOCSY–heteronuclear single-quantum coherence, HCCH-TOCSY, aromatic-13C-HMQC-TOCSY] were collected for complete resonance assignments (34). Stereospecific assignments of the methyl groups of valine and leucine residues were determined by using a 10% 13C-α3D sample as described (35).
Structure Determination.
Distance restraints were obtained from 15N- and 13C-resolved NOE spectroscopy experiments. The NOE distant restraints were divided into four classes: strong (0–2.7 Å), medium (2.7–3.3 Å), weak (3.30–4.0 Å), and very weak (4.0–6.0 Å). An HNHA experiment was used to determine 3JαN coupling constants for φ angles (36). χ1 angles were determined from NOE patterns, and the 3Jαβ coupling constants were determined from the HNHB (37) and HACAHB-COSY (38) experiments. Hydrogen exchange rates were determined by diluting a 15N-α3D sample into 99.9% 2H2O and measuring the decrease in signal intensities in serially acquired 1H,15N–heteronuclear single-quantum coherence spectra. Hydrogen exchange protection factors were calculated versus random coil exchange rates (39). α3D structure calculations were calculated by using dynamical simulated annealing protocol (40) using the program x-plor (41).
RESULTS
α3D Structure Determination.
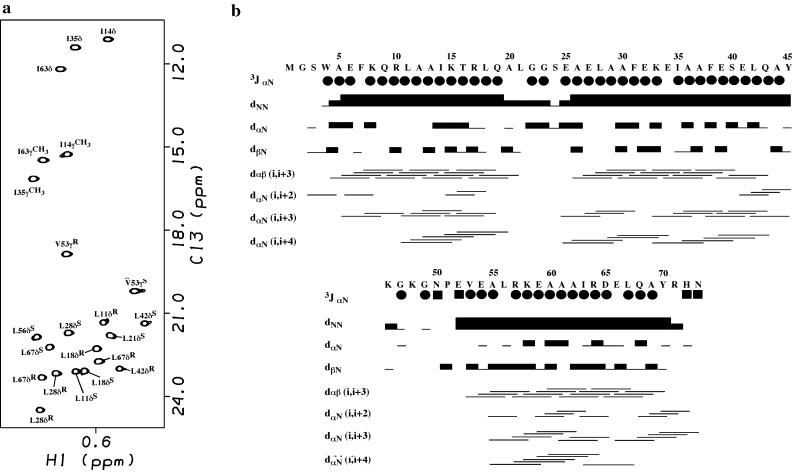
The NMR spectra of α3D showed good chemical shift dispersion typical of a well folded native proteins (Fig. 2a). The solution structure of α3D was determined by using heteronuclear multidimensional NMR methods (34). Structures were calculated on the basis of 1,143 NOE distance restraints, 48 hydrogen bonds (96 restraints), 55 φ angle restraints, and 14 χ1 angle restraints (Table 1). Sixty structures were generated by using the simulated annealing algorithm in the program x-plor (41). All of the structures showed the same overall topology, and 13 structures converged to good covalent geometry with no distance or dihedral angle violation >0.35 Å or 5°, respectively (Table 1). The structure was well defined, with ≈16 restraints per residue (very few NOEs were observed for the residues in the loops and at the termini). The sequential and short range NOEs along with the 3JαN coupling constants are represented in Fig. 2b. The stereochemistry of the structures was checked by using the program procheck-nmr (ref. 42; Table 1). The rms deviations of the well defined areas of α3D for the backbone (N, Cα, C) and backbone with side chain atoms (hydrophobic residues except W4, Y45, and Y70) were 0.75 and 1.03 Å, respectively (Table 1).
Figure 2.
(a) NMR spectrum of α3D (1H,13C-CT–heteronuclear single-quantum coherence) illustrating the assignments of the methyl groups. Prochiral assignments of the methyl groups of valine and leucine were obtained by using a 10% 13C labeled sample (35). (b) Summary of the α3D sequential NOEs. The size of the bar corresponds to the intensity of the NOE. The NOEs were taken from the 15N- and 13C-resolved nuclear Overhauser effect spectroscopy spectra. The three bond 3JαN coupling constants determined for α3D are displayed as filled circles and boxes corresponding to 3JαN coupling constants <6.0 Hz or >8.0 Hz, respectively.
Table 1.
Structural statistics for the family of 13 α3D structures
| Experimental restraints* | |
| Intraresidue (|i − j| = 0) | 459 |
| Short-to-medium range (1 < |i − j| < 5 residues) | 511 |
| Long range (|i − j| > 5 residues) | 173 |
| Hydrogen bonds | 48 |
| Φ angles | 55 |
| χ1 angles | 14 |
| Total | 1,260 |
| 〈SA〉 ± SD | |
| Mean rms deviations from experimental restraints | |
| Distance restraints, Å | 0.0209 ± 0.0005 |
| Dihedral angle, ° | 0.378 ± 0.032 |
| Mean rms deviations from ideal geometry | |
| Bonds, Å | 0.00351 ± 0.00003 |
| Angles, ° | 0.609 ± 0.015 |
| Impropers, ° | 0.721 ± 0.014 |
| Ramachandran statistics from Procheck-NMR† | |
| Residues in most favored regions | 86.2% |
| Residues in additional allowed regions | 11.4% |
| Residues in generously allowed regions | 1.4% |
| Residues in disallowed regions | 1.1% |
| Rms deviations from the mean structure, Å | |
| Backbone atoms (residues 1–73, N, Cα, C) | 1.06 |
| Backbone atoms (residues 4–21, 24–45, 51–70, N, Cα, C) | 0.75 |
| All nonhydrogen atoms (residues 1–73) | 1.61 |
| Backbone atoms (residues 4–21, 24–45, 51–70, N, Cα, C, O) and Hydrophobic core (except 4, 45, 70) | 1.03 |
None of the structures exhibit distance violations >0.35 Å or dihedral angle violation >5°.
The final values of the square well NOE and dihedral-angle potential were calculated with a force constant of 50 kcal⋅mol−1⋅Å−2 and 200 kcal⋅mol−1⋅rad−2, respectively.
The programs aqua and procheck-nmr (42) were used to check the overall quality of the structures.
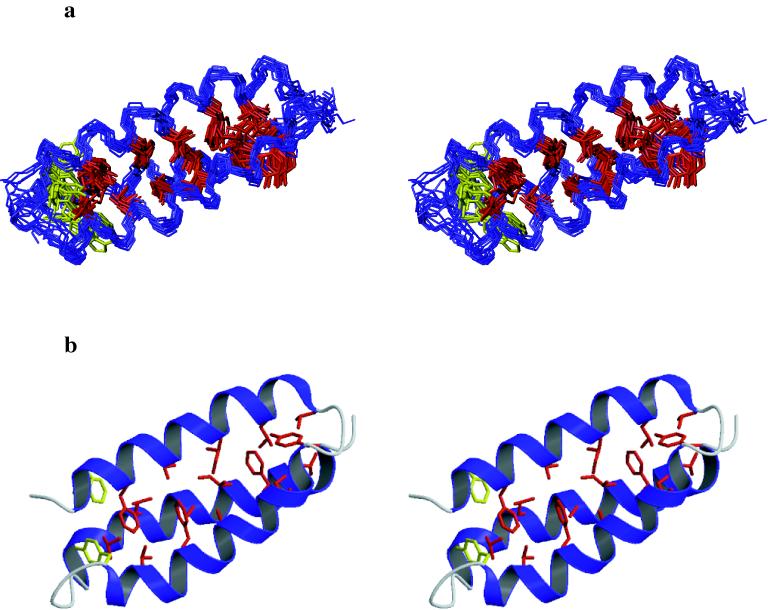
The structure of α3D reasonably agreed well with the design, showing an overall rms deviation of 1.9 Å for the backbone atoms (N, Cα, C, O) of the lowest energy structure of α3D (residues 4–21, 25–45, 51–70) versus the original model of α3C. When viewed down their axes, helical bundles may wind in a clockwise or a counterclockwise manner (32). The topology of α3D was designed to adopt a counterclockwise bundle, in contrast to the common clockwise bacterial IgG-binding proteins (25), and the counterclockwise topology indeed was observed in the structure (Fig. 3). The positions of the helices in the observed structure spanned residues 4–21, 25–45, and 51–70 and were in excellent agreement with the design (residues 4–21, 25–46, 51–71). The interhelical tilt angles were also in excellent agreement with the design: Ω1,2 = 165°, Ω1,3 = −14°, and Ω2,3 = 171°. As expected from its asymmetric sequence and nonuniform hydrophobic core, the structure deviated from a canonical coiled coil (43), showing less curvature in helices 1 and 2 and less uniformity in the interhelical crossing angles.
Figure 3.
(a) Stereo diagrams of the 13 superimposed α3D structures are shown with the hydrophobic core residues depicted in red and W4 and Y45 in yellow. The structures were aligned by using only the backbone atoms (residues 4–21, 24–45, and 51–70). The figure was generated by using the program molmol (58). (b) Stereo display of a ribbon diagram with the hydrophobic residues in red and W4 and Y45 in yellow of the lowest energy structure of α3D. The figure was created by using the programs molscript (59) and raster3d (60).
As in native proteins, the apolar side chains in the core of α3D were generally well packed. It was possible to define 14 of the 18 side chain χ1 torsional angles for the hydrophobic residues from the coupling constants (3Jαβ). These side chains, as well as two additional side chains that were defined by NOE restraints alone, adopted a single predominant rotamer in the core of the protein. The remaining two residues (W4 and Y45) showed excursions from the predominant rotameric state in half of the calculated structures. W4 was partially buried (λem = 342 nm) and appeared to adopt two distinct, well populated orientations (χ1 = −60.0° and 180°). Similar behavior has been observed for a natural three-helix bundle containing protein (44). Similarly, an IgG-binding domain shows two distinct rotamers for an interfacial phenylalanine residue (45). In summary, α3D showed a high degree of order in the packing of its core, although there were some local areas of disorder, as has been observed in native proteins of comparable structure and stability.
Hydrogen Exchange.
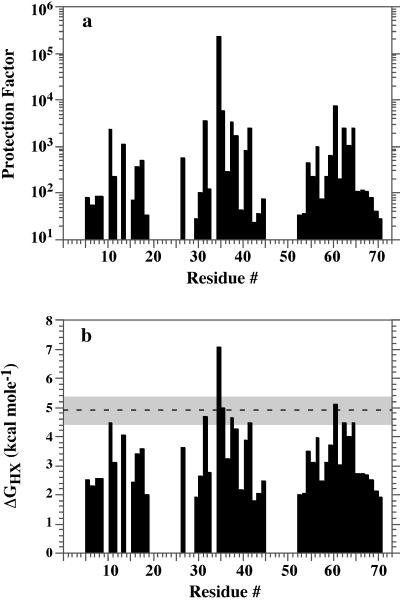
The rates at which a protein’s amide protons exchange with solvent deuterons after dissolution in 2H2O provide important structural and dynamic information. Although molten globules show relatively rapid exchange mediated by local unfolding events, exchange in native proteins is slow. Proteins often contain a subset of amides that exchange only when the protein transiently adopts a fully unfolded conformation and hence are slowed by a factor (the protection factor) approximately consistent with the equilibrium constant for folding. Indeed, nine amide protons in α3D showed protection factors within 0.5 kcal⋅mol−1 of that expected from its global stability (Fig. 4). Eight of these positions were found within helices 2 and 3, suggesting that this is the most stable cooperative unit in the protein. Also, I35 showed a degree of protection that was 2 kcal⋅mol−1 higher than expected, indicative of some residual structure in the unfolded state.
Figure 4.
(a) Protection factors for the main-chain amide protons of α3D determined from hydrogen–deuterium exchange at 15°C, pH* 5.0 (in 50 mM deuterated acetate). The protection factors were calculated from the observed exchange rates and the intrinsic exchange rates (39). Absence of a bar indicates residues with amide protons exchanging in the dead time of the experiment (18 minutes) or a lack of a well resolved amide proton cross peak. (b) Assuming an EX2 exchange mechanism, the apparent free energy, ΔGHX, was calculated from the protection factors, Pi, as RTln(Pi) (39). The dashed line represents the ΔGu at 15°C determined calorimetrically by extrapolation of the Gibbs-Helmoltz equation, ΔGT = ΔHTm (1 − T/Tm) + (T − Tm) ΔCp − TΔCpln(T/Tm). The shaded region denotes the estimated standard error in this parameter of ±0.5 kcal⋅mol−1.
DISCUSSION
These results illustrate many of the features required for the formation of a uniquely folded protein. One feature that appears to have been particularly important for stabilizing a unique topology was the incorporation of interfacial charged groups to stabilize the predicted structure, and—even more importantly—to destabilize the clockwise topology or other alternatively folded structures (46). This strategy appeared to be successful. The interfacial charged side chains align at appropriate positions for favorable interhelical electrostatic interactions, and no evidence has been obtained for alternatively folded structures. A second design feature included two interhelical connections featuring capping boxes, which serve to define the trajectory of the loops in the desired direction. The first loop (Fig. 1) is well ordered and shows the NOE patterns characteristic of an N-terminal capping box (S-X-X-E box; ref. 31). The second, longer loop is largely disordered (Fig. 3a) and provides an excellent site for evolution of function. Thus, interfacial electrostatic interactions and hydrogen bonding appear to contribute to the unique fold of α3D.
The packing of the hydrophobic side chains in the core of the protein also may contribute to the unique structure of α3D. The hydrogen exchange protection factors of an earlier version of α3D containing only Leu sidechains in the core were considerably lower than expected, and it also showed a somewhat low value of ΔCp for unfolding (27). By contrast, the diverse collection of side chains in α3D pack into a more well defined structure as assessed by its thermodynamic, spectroscopic, and structural properties. However, it is interesting to note that more than half of the rotamers observed in the structure of α3D were incorrectly predicted by the early version of the roc repacking program (33) used in its design. This finding indicates that it was possible to design a native conformation without explicitly defining the intricate details of the sidechain packing geometries. Thus, although packing is essential for stability, there is sufficient flexibility in the side chain conformations that their geometries need not be completely specified in a successfully designed protein (2, 47–49). This finding is consistent with earlier successes in de novo protein design. For example, the conformations of only three of seven interior sidechains were correctly predicted in the de novo design of a peptide based on the zinc finger motif (7).
These findings suggest that the stability was not optimized in the initial design. Since the design of α3D, there have been many major improvements in algorithms for sidechain repacking and protein design (50–56), some of which allow flexibility in the backbone conformation. Preliminary data suggest that even more stable and well ordered structures may be obtained by using similar algorithms.
CONCLUSIONS
These data clearly indicate that it is now possible to design complex, structurally defined proteins, beginning with simple, easily parameterized motifs such as an antiparallel coiled coil. Earlier successful examples of de novo-designed proteins were either multistranded coiled coils (12, 57) or had only marginal stabilities. For example, previously designed mimics of the zinc finger motif and a three-stranded β-sheet have equilibrium constants favoring the folded form by less than a factor of 10 (7, 8). Even this low level of stability was achieved only after careful optimization of the sequences. Thus, it might be difficult to evolve their sequences for the introduction of a novel function without greatly affecting their folded structures. By contrast, the stability of α3D, which is similar to natural proteins, should be much more forgiving of mutations. Hence, this protein should provide an excellent framework for the evolution of new functions within a completely novel sequence.
Acknowledgments
We thank A. Joshua Wand for time on the 750-MHz magnet (State University of New York, Buffalo, NY), Jeffrey L. Urbauer, Jack J. Skalicky, and Peter F. Flynn for NMR assistance, and R. Blake Hill for helping with structure calculations and suggestions on the manuscript. Also, thanks go out to Gregg R. Dieckmann and James D. Lear for help in the calculations of interhelical crossing angles. This work was supported by grants from the National Institutes of Health (Grant GM-54616 to W.F.D. and Grant GM-35926 to H.R.) and also was supported in part by the Materials Research Science and Engineering Centers program of the National Science Foundation (Grant DMR96-32598 to W.F.D.). The NMR facility of the Fox Chase Cancer Center was supported by grants from the National Institutes of Health (Grant CA-06927) and the Kresge Foundation.
ABBREVIATIONS
- NOE
nuclear Overhauser effect
- Tm
melting temperature
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 2a3d).
References
- 1.Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 2.Beasley J R, Hecht M H. J Biol Chem. 1997;272:2031–2034. doi: 10.1074/jbc.272.4.2031. [DOI] [PubMed] [Google Scholar]
- 3.Hellinga H. Fold Des. 1998;3:1–8. doi: 10.1016/S1359-0278(98)00001-7. [DOI] [PubMed] [Google Scholar]
- 4.Regan L. Structure (London) 1998;6:1–4. doi: 10.1016/s0969-2126(98)00001-x. [DOI] [PubMed] [Google Scholar]
- 5.Choo Y, Castellanos A, Garcia-Hernandez B, Sanchez-Garcia I, Klug A. J Mol Biol. 1997;273:525–532. doi: 10.1006/jmbi.1997.1339. [DOI] [PubMed] [Google Scholar]
- 6.Pomerantz J L, Wolfe S A, Pabo C O. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- 7.Dahiyat B I, Mayo S L. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 8.Struthers M D, Chang R P, Imperiali B. J Am Chem Soc. 1996;118:3073–3081. [Google Scholar]
- 9.Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 10.Mayo K H, Ilyina E, Park H. Protein Sci. 1996;5:1301–1315. doi: 10.1002/pro.5560050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddle D S, Santiago J V, Bray-Hall S T, Doshi N, Grantcharova V P, Yi Q, Baker D. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 12.Harbury P B, Plecs J J, Tidor B, Alber T, Kim P S. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin R L. Biophys Chem. 1995;55:127–135. doi: 10.1016/0301-4622(94)00146-b. [DOI] [PubMed] [Google Scholar]
- 14.Alba E A, Jimenez M A, Rico M, Nieto J L. Fold Des. 1996;1:133–144. doi: 10.1016/s1359-0278(96)00022-3. [DOI] [PubMed] [Google Scholar]
- 15.Fezoui Y, Connolly P J, Osterhout J J. Protein Sci. 1997;6:1869–1877. doi: 10.1002/pro.5560060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda Y, Nakai T, Ohkubo T. J Mol Biol. 1994;236:862–865. doi: 10.1006/jmbi.1994.1194. [DOI] [PubMed] [Google Scholar]
- 17.Kortemme T, Ramirez M, Serrano L. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 18.Gellman S H. Curr Opin Chem Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 19.Schenck H L, Gellman S H. J Am Chem Soc. 1998;120:4869–4870. [Google Scholar]
- 20.Das C, Raghothama S, Balaram P. J Am Chem Soc. 1998;120:5812–5813. [Google Scholar]
- 21.Hill R B, DeGrado W F. J Am Chem Soc. 1998;120:1138–1145. [Google Scholar]
- 22.Schafmeister C E, LaPorte S L, Miercke J W, Stroud R M. Nat Struct Biol. 1997;4:1039–1046. doi: 10.1038/nsb1297-1039. [DOI] [PubMed] [Google Scholar]
- 23.O’Shea E K, Lumb K J, Kim P S. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 24.Brive L, Dolphin G T, Baltzer L. J Am Chem Soc. 1997;119:8598–8607. [Google Scholar]
- 25.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 26.Schneider J P, Lombardi A, DeGrado W F. Fold Des. 1998;3:29–40. doi: 10.1016/S1359-0278(98)00011-X. [DOI] [PubMed] [Google Scholar]
- 27.Bryson J W, Dejarlais J R, Handel T M, DeGrado W F. Protein Sci. 1998;7:1404–1414. doi: 10.1002/pro.5560070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson J S, Gibney B R, Skalicky J J, Wand A J, Dutton P L. J Am Chem Soc. 1998;120:3881–3886. [Google Scholar]
- 29.Lovejoy B, Seunghyon D C, McRorie D K, DeGrado W F, Eisenberg D. Science. 1993;259:1288–1293. doi: 10.1126/science.8446897. [DOI] [PubMed] [Google Scholar]
- 30.Harper E T, Rose G D. Biochemistry. 1993;32:543–546. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H X, Lyu P, Wemmer D E, Kallenbach N R. Proteins Struct Funct Genet. 1994;18:1–7. doi: 10.1002/prot.340180103. [DOI] [PubMed] [Google Scholar]
- 32.Efimov A V. Curr Opin Struct Biol. 1993;3:379–384. [Google Scholar]
- 33.Desjarlais J R, Handel T M. Proein Sci. 1995;4:2006–2018. doi: 10.1002/pro.5560041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavanagh J, Fairbrother W J, Palmer A G, III, Skelton N J. Protein NMR Spectroscopy. San Diego: Academic; 1996. [Google Scholar]
- 35.Neri D, Otting G, Wüthrich K. Tetrahedron. 1990;46:3287–3296. [Google Scholar]
- 36.Vuister G, Bax A. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 37.Archer S J, Ikura M, Torchia D A, Bax A. J Magn Reson. 1991;95:636–641. [Google Scholar]
- 38.Grzesiek S, Kuboniwa H, Hinck A P, Bax A. J Am Chem Soc. 1995;117:5312–5315. [Google Scholar]
- 39.Bai Y, Milne J S, Mayne L, Englander S W. Proteins Struct Funct Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilges M, Clore G M, Gronenborn A M. FEBS Lett. 1988;239:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 41.Brünger A T. X-PLOR, Version 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 42.Laskowski R A, Rullmann J A C, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 43.Seo J, Cohen C. Proteins Struct Funct Genet. 1993;15:223–234. doi: 10.1002/prot.340150302. [DOI] [PubMed] [Google Scholar]
- 44.Wong K B, Daggett V. Biochemistry. 1998;37:11182–11192. doi: 10.1021/bi980552i. [DOI] [PubMed] [Google Scholar]
- 45.Starovasnik M A, Skelton N J, O’Connell M P, Kelley R F, Reilly D, Fairbrother W J. Biochemistry. 1996;35:1558–15569. doi: 10.1021/bi961409x. [DOI] [PubMed] [Google Scholar]
- 46.Lumb K J, Kim P S. Science. 1995;268:436–439. doi: 10.1126/science.7716550. [DOI] [PubMed] [Google Scholar]
- 47.Bloomberg S, Dill K A. Protein Sci. 1994;3:997–1009. doi: 10.1002/pro.5560030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behe M J, Lattman E E, Rose G D. Proc Natl Acad Sci USA. 1991;88:4195–4199. doi: 10.1073/pnas.88.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Axe D D, Foster N W, Fersht A R. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazar G A, Dejarlais J R, Handel T M. Protein Sci. 1997;6:1167–1178. doi: 10.1002/pro.5560060605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harbury P B, Tidor B, Kim P S. Proc Natl Acad Sci USA. 1995;92:8408–8412. doi: 10.1073/pnas.92.18.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dahiyat B I, Gordon D B, Mayo S L. Protein Sci. 1997;6:1333–1337. doi: 10.1002/pro.5560060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su A, Mayo S L. Protein Sci. 1997;6:1701–1707. doi: 10.1002/pro.5560060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahiyat B I, Mayo S L. Proc Natl Acad Sci USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahiyat B I, Sarisky C A, Mayo S L. J Mol Biol. 1997;273:789–796. doi: 10.1006/jmbi.1997.1341. [DOI] [PubMed] [Google Scholar]
- 56.Street A G, Mayo S L. Fold Des. 1998;3:253–258. doi: 10.1016/S1359-0278(98)00036-4. [DOI] [PubMed] [Google Scholar]
- 57.Ogihara N L, Weiss M S, DeGrado W F, Eisenberg D. Protein Sci. 1997;6:80–88. doi: 10.1002/pro.5560060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 59.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 60.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]