Abstract
The von Hippel–Lindau tumor suppressor protein pVHL assembles with cullin-2 (hCUL-2) and elongin B/C forming a protein complex, CBCVHL, that resembles SKP1–CDC53–F-box protein ubiquitin ligases. Here, we show that hCUL-2 is modified by the conserved ubiquitin-like protein NEDD8 and that NEDD8–hCUL-2 conjugates are part of CBCVHL complexes in vivo. Remarkably, the formation of these conjugates is stimulated by the pVHL tumor suppressor. A tumorigenic pVHL variant, however, is essentially deficient in this activity. Thus, ligation of NEDD8 to hCUL-2 is linked to pVHL activity and may be important for pVHL tumor suppressor function.
Modification of proteins by covalent attachment of the highly conserved polypeptide ubiquitin occurs universally in eukaryotic cells (see refs. 1 and 2 for review). Proteins modified by ubiquitin tags usually are targeted for degradation by the 26S proteasome or, in the case of some plasma-membrane proteins, to the lysosome via the endocytic route. The conjugation reaction is mediated by a series of well defined enzymatic reactions and leads to the formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and the ɛ-amino group of a lysine residue of an acceptor protein. Ubiquitin is first activated by an ubiquitin-activating enzyme, E1, and subsequently transferred to an ubiquitin-conjugating enzyme, E2, also known as UBC. Ubiquitin can be transferred to a substrate either directly from the E2 or via substrate-selective ubiquitin ligases, E3s (1, 2). Recently, a class of E3 ligases that are characterized by a striking modular organization have been identified in yeast. These E3s, termed SCF (SKP1–CDC53–F-box protein), collaborate with the E2 enzyme UBC3CDC34 in substrate ubiquitination. SCF complexes contain CDC53, SKP1, and one of several so-called F-box proteins that function as substrate-recruiting factors of the ubiquitin ligase (see refs. 3 and 4 for review). Depending on the F-box protein used, SCF is known to ubiquitinate distinct proteins, and ubiquitination targets them for destruction by the 26S proteasome. SCF substrates include G1 cyclins, cyclin-dependent-kinase inhibitors, the cyclin-dependent-kinase inhibitory kinase SWE1, and other regulators (4–11).
Humans possess several CDC53-related proteins termed cullins. Of these proteins, two, hCUL-1 and hCUL-2, are known to be part of protein complexes that resemble yeast SCF. hCUL-1 associates with the human counterpart of SKP1, hSKP1, and an F-box protein termed SKP2. The formed complex seems to be the functional equivalent of yeast SCF (12, 13). hCUL-2, however, is part of a different complex that contains elongin B, a distant relative of ubiquitin, and the SKP1-related protein elongin C (14–16). The hCUL-2–elongin B/C complex, which we propose to call CBC, can assemble with the von Hippel–Lindau tumor suppressor protein, pVHL (17–19), forming CBCVHL (15, 16, 20, 21). Because of the structural resemblance to SCF, it seems likely that CBC complexes also may function as E3-type ubiquitin ligases, with pVHL representing one of several possible alternative substrate-recognition subunits (another candidate is the F-box protein elongin A; refs. 6 and 14). The gene encoding the pVHL subunit of the complex is linked to a hereditary cancer syndrome, von Hippel–Lindau disease (17). Individuals who carry germ-line mutations in the VHL gene are predisposed to a variety of benign and malignant tumors, which include clear-cell carcinomas of the kidney (RCC), pheochromocytomas, and vascular tumors of the central nervous system and retina (17, 22–24). Inactivation or loss of both VHL alleles has been reported for the majority of sporadic RCC and cerebellar hemangioblastomas, which is in keeping with Knudson’s two-hit hypothesis of tumor suppressor-mediated carcinogenesis (23, 24). It has been proposed that these pathological defects might be consequences of an impaired CBCVHL ubiquitination activity (15, 16).
Recently, the CDC53 subunit of yeast SCF was shown to be modified by conjugation to the ubiquitin-like protein RUB1 (25, 26). In contrast to ubiquitination, this modification does not seem to target the CDC53 protein for degradation (25, 26). Genetic studies have indicated that it seems to affect the cell-cycle function of the SCF ligase (25). Previous work has shown that modification by RUB1 proceeds analogously to ubiquitination and involves an E1-like ULA1/UBA3 enzyme pair for activation and a specific E2, UBC12, for conjugation of the modifier (25, 26). Intriguingly, the RUB1 conjugation pathway seems to be conserved across species. NEDD8 (27) is presumed to be the human homolog of RUB1, displaying 59% identical amino acids with the yeast protein. Furthermore, conjugation of NEDD8 to other proteins recently was shown to involve enzymes related to the yeast ULA1/UBA3 and UBC12 enzymes (28).
Here, we show that mammalian NEDD8 and yeast RUB1 are indeed functionally equivalent and that the human protein can substitute for RUB1 in yeast and modifies CDC53. Moreover, we also found that the human cullins hCUL-1 and hCUL-2 are modified by NEDD8 in vivo. Intriguingly, modification of the hCUL-2 subunit of the CBC complex by NEDD8 requires pVHL to be part of the same complex, indicating that the CBCVHL complex itself is involved in its own modification. Most importantly, we discovered that this modification is stimulated only by wild-type but not tumorigenic-mutant forms of pVHL. Thus, our data provide a clear correlation between the tumorigenic potential of pVHL mutant forms and their deficiency in supporting CBC modification. Ligation of NEDD8 to hCUL-2 therefore is linked to pVHL activity and may be important for pVHL tumor suppressor function.
MATERIALS AND METHODS
Strains and Plasmids.
Yeast strains rub1-1, ula1, and ubc12, as well as rub1-1 cdc34 mutants and plasmids expressing tagged RUB1 or CDC53, have been described (26). For expression of N-terminally hemagglutinin epitope (HA)-tagged NEDD8 (HANEDD8), mouse NEDD8 cDNA (a gift from S. Kumar, Kyoto University, Kyoto), encoding the maturated form of the protein, was triple-HA epitope-tagged by PCR and cloned downstream of the GAL1-10 promoter (YIFN8 and pSFN8). YIFN8 was integrated into the yeast URA3 locus of the various strains; for the NEDD8/CDC53 coimmunoprecipitation experiments, pSFN8 was used. To rescue the synthetic lethality of the rub1-1 cdc34 double mutant, the pGAL1-10–NEDD8 expression cassette was cloned into the yeast vector YIplac128 (YIN8), which was integrated into the yeast LEU2 locus.
A plasmid expressing HA-tagged hCUL-2 (pcDNA3–Hs-cul-2) has been described (15). NEDD8 tagged with a triple-myc epitope was constructed by PCR and subcloned into pcDNA3 (Invitrogen; pcDNA3–3mycNEDD8). Glutathione S-transferase (GST)–NEDD8 was constructed by PCR and subcloned into a pGEX–2TK vector (Amersham Pharmacia). Recombinant GST or GST–NEDD8 proteins were produced in BL21 cells and purified according to the supplier’s instructions (Amersham Pharmacia). pVHL cDNAs (pVHL or pVHLΔ157) contain a FLAG tag at the N terminus (15) and were subcloned into pcDNA3 vectors.
Cells and Cell Culture.
Human 293 cells (American Type Culture Collection) were transfected by using the calcium phosphate procedure with 2.5–5 μg of pcDNA3–3mycNEDD8 plasmid per 6-well dish at 50% confluence for 16 h; 48 h after transfection, cells were harvested and lysed in TBST (10 mM Tris·HCl, pH 7.5/150 mM NaCl/0.1% Tween 20; ref. 15), and 60 μg of total protein was analyzed by SDS/10% PAGE and transferred onto a nitrocellulose membrane. The membrane was probed with anti-myc antibody 9E10 (Babco, Richmond, CA; 1:1,000), anti-hCUL-1 antibodies (a gift from W. Krek, Friedrich-Miescher-Institute, Basel), or anti-hCUL-2 antibodies against a peptide representing amino acids 111–126 of hCUL-2. The human sporadic RCC 786-0 cell line lacks one VHL allele and expresses a truncated VHL protein (amino acids 1–104) from the second allele (16, 22). These cells express a polylinker sequence from the vector, a VHL gene bearing a naturally occurring cancer-predisposing nonsense mutation, which truncates the protein after amino acid 155 (HApVHLΔ155), or the gene for wild-type human pVHL (HApVHL; ref. 22). pVHLΔ155 and VHLΔ157 express virtually identical proteins. pVHL was HA-tagged at the N terminus. Cells were harvested, and pVHL was immunoprecipitated with anti-HA antibodies (HA11, Babco) as described (15, 16) and transferred to a nitrocellulose membrane, which was probed with anti-hCUL-2 antibodies. Blots were incubated with horseradish peroxidase-conjugated secondary antibodies, and signals were detected by using an enhanced chemiluminescence detection system (Amersham Pharmacia). For protein sequencing, the bands were sliced out from the poly(vinylidene difluoride) filter membrane and subjected to Edman degradation.
In Vitro Translation Analysis.
pcDNA3–Hs-cul-2 (1 μg) was translated simultaneously with various amounts of pVHL cDNAs (0.1–2 μg) in the presence of [35S]methionine in vitro by using a coupled transcription/translation system (50 μl; Promega). GST or GST–NEDD8 proteins (1 μg) were added at the beginning of the reaction. The reactions were then analyzed by SDS/10% PAGE and processed for autoradiography.
RESULTS
Yeast RUB1 and Human NEDD8 Are Functional Orthologs.
Because of the sequence similarity of NEDD8 and RUB1, we asked whether the two proteins are functional orthologs. To this end, we expressed HANEDD8 in yeast. As shown in Fig. 1, an immunoreactive conjugation pattern virtually identical to that of RUB1 (HARUB1) was observed. RUB1-protein conjugate formation requires the E1 heterodimer ULA1/UBA3 for RUB1 activation and the E2 enzyme UBC12 for conjugation (26). We found that NEDD8 conjugation was virtually absent in ula1, uba3, and ubc12 null mutants (Fig. 1), indicating that the yeast enzymes can collaborate with the human ubiquitin-like protein.
Figure 1.
Conjugation of mammalian NEDD8 in yeast. The total conjugation pattern of HANEDD8 (compared with HARUB1) in yeast was determined by Western blotting with HA-specific antibodies. The lower band corresponds to free RUB1 (first lane) or NEDD8 (other lanes). NEDD8 conjugates are virtually absent in ula1, uba3, and ubc12 mutants defective in RUB1 conjugation. The immunoreactive 30-kDa protein is a crossreacting yeast protein.
To confirm directly that RUB1 and NEDD8 are functional orthologs, we performed complementation assays. Yeast null mutants in the RUB1-conjugation pathway are viable and grow at wild-type rates (25, 26). However, these mutations (e.g., rub1-1) exhibit synthetic lethality with cdc34-1, a temperature-sensitive mutation of the gene encoding the UBC3CDC34 E2 enzyme, which cooperates with SCF ubiquitin ligases (25, 26). When grown at 33°C, rub1-1 cdc34-1 double mutants, but not cdc34-1 single mutants, are inviable. This growth defect could indeed be rescued by expression of NEDD8 in these cells (Fig. 2), indicating that NEDD8 can substitute for RUB1 in yeast.
Figure 2.
NEDD8 can substitute for RUB1 in yeast. Yeast mutants were streaked out on agar plates and incubated at 23°, 33°, or 37°C. Conditional cdc34-1 mutants are inviable at 37°C. Combined with the rub1-1 null mutation the double mutant (cdc34-1 rub1-1) is inviable even at 33°C. Viability could be restored by NEDD8 expression from a glucose-repressible GAL1-10 promoter (pGAL–NEDD8). The promoter is “off” on glucose (Upper) and “on” on galactose plates (Lower).
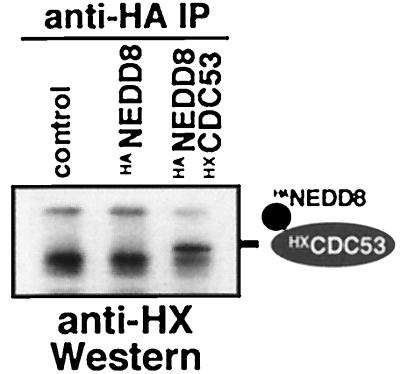
The ability to complement the yeast mutant suggests that NEDD8 may modify the same substrates as RUB1 in yeast. One of the proteins known to be modified by RUB1 is the cullin CDC53, a 94-kDa subunit of the yeast SCF ubiquitin ligase (25, 26). To test whether CDC53 can serve as a substrate for NEDD8 modification in yeast, we coexpressed in rub1-1 mutant cells epitope-tagged forms of both CDC53 and NEDD8. As shown in Fig. 3, CDC53 was indeed found to be modified by NEDD8, indicating that the striking substrate selectivity of the modification pathway was not affected by the substitution.
Figure 3.
CDC53 can be modified by NEDD8 in yeast. Western blot developed with Xpress-tag-specific antibodies of immunoprecipitates with HA-specific antibodies from yeast cells carrying vector (control), plasmids expressing HANEDD8, or HANEDD8 plus Xpress-tagged CDC53 (HXCDC53; Invitrogen). Immunoprecipitation of HANEDD8 results in a coprecipitation of HANEDD8-modified HXCDC53.
hCUL1 and hCUL2 Are Modified by NEDD8 in Vivo.
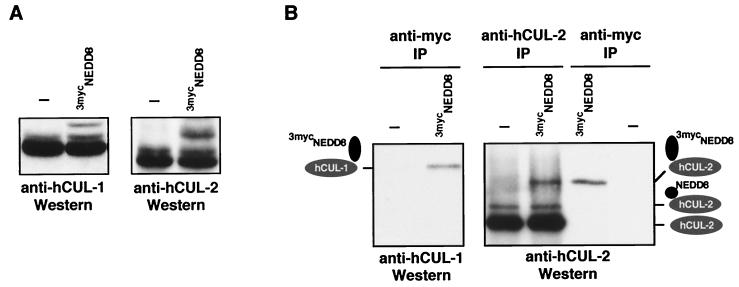
Given the conservation of RUB1 and NEDD8 pathways, an obvious question was whether human cullins (29, 30), the structural homologs of yeast CDC53, are modified by NEDD8 in mammalian cells in vivo. In fact, the size of one of the NEDD8 conjugates in human cells (≈80 kDa) is consistent with the predicted sizes of NEDD8-modified cullins (26, 31). Indeed, a Western analysis of human embryonic kidney 293 cells with hCUL-1- or hCUL-2-specific antibodies identified two closely spaced ≈80-kDa immunoreactive bands in SDS gels, suggesting that they may correspond to the unmodified and NEDD8-modified forms of both cullins, respectively (Fig. 4A, Left). When we expressed a cDNA encoding a larger 3mycNEDD8 in these cells, hCUL-1- and hCUL-2-specific antibodies identified additional, even slower-migrating bands (Fig. 4A; Right), suggesting that they may represent conjugates of 3mycNEDD8 with the human cullins hCUL-1 and hCUL-2, respectively. To test this idea, we immunoprecipitated proteins from 293 cells transfected with 3mycNEDD8 cDNA (or empty vector as control) with myc-specific antibodies and identified hCUL-1 and hCUL-2 reactive proteins by Western blotting with hCUL-1- and hCUL-2-specific antibodies. As shown in Fig. 4B, cullin-specific immunoreactive proteins were found exclusively in 3mycNEDD8-transfected cells, confirming that both human cullins, hCUL-1 and hCUL-2, are modified by 3mycNEDD8. Thus, we conclude that, in a manner strikingly analogous to the modification of CDC53 by RUB1 in yeast, the human cullins hCUL-1 and hCUL-2 are subject to modification by the mammalian RUB1 ortholog NEDD8 in vivo. Moreover, recent work has shown that another human cullin, hCUL-4A, is modified by NEDD8 (28), but the function of this cullin has remained enigmatic.
Figure 4.
hCUL-1 and hCUL-2 are modified by NEDD8 in vivo. (A) Human 293 cells transiently transfected either with empty vector (−) or with pcDNA3–3mycNEDD8 were lysed, and proteins were analyzed by immunoblotting with anti-hCUL-1 antibodies (Left) or anti-hCUL-2 antibodies (Right). (B) As in A, 293 cells transiently transfected either with empty vector (−) or with pcDNA3–3mycNEDD8 were lysed and proteins were immunoprecipitated with anti-myc (Left), anti-hCUL-2 (Right, two left lanes), or anti-myc antibodies (Right, two right lanes), followed by Western analysis with anti-hCUL-1 (Left) or anti-hCUL-2 antibodies (Right).
NEDD8 Modifies CBC Complexes.
As discussed above, hCUL-1 and hCUL-2 are subunits of different protein complexes. hCUL-1 is part of the human SCF (12, 13), whereas hCUL-2 is a component of the CBC complex that also contains elongin B/C (15, 16). CBC can assemble with pVHL, forming CBCVHL (15, 16, 20, 21).
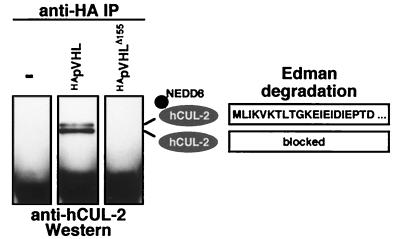
To determine whether NEDD8-modified hCUL-2 is present in CBCVHL, we used VHL-deficient (VHL −/−) RCC cell lines stably transfected with HApVHL, with a tumorigenic mutant form, or with a vector (16). Most tumorigenic VHL mutations lead to a loss in elongin B/C and hCUL-2 interaction (15, 16, 20, 21). One example is the naturally occurring mutant that we used, pVHLΔ155, which expresses a truncated protein lacking the elongin B/C interaction domain (15, 16). Immunoprecipitation with HA-specific antibodies coprecipitated a protein doublet of ≈80 kDa only from extracts of cells expressing the wild-type form HApVHL but not of cells expressing HApVHLΔ155 or vector. As shown in Fig. 5, both bands reacted with hCUL-2-specific antibodies. In a related experiment, the two bands were isolated from a preparative gel and subjected to Edman degradation. The N terminus of hCUL-2 seemed to be blocked, but we obtained a sequence from the upper band, which corresponded to the first 21 N-terminal residues of NEDD8 (Fig. 5). This result directly confirms that the slower-migrating species of hCUL-2, which are detectable by either immunoprecipitation (Figs. 4B and 5) or Western blotting (Fig. 4A), correspond to the NEDD8-modified form of the cullin. Moreover, these data indicate that NEDD8-modified hCUL-2 is a part of CBC complexes that also contain wild-type pVHL.
Figure 5.
NEDD8-modified hCUL-2 is part of the CBCVHL complex. Human RCC cells stably expressing vector (control), wild-type HApVHL, or a naturally occurring mutant HApVHLΔ155 were used for coimmunoprecipitation analysis. pVHL was immunoprecipitated by using anti-HA antibodies, and proteins were analyzed with immunoblots probed with anti-hCUL-2 antibodies. Both hCUL-2-specific bands were excised from a preparative gel blot and subjected to Edman degradation. The obtained sequence of the first 21 cycles derived from the material of the upper band is shown on the Right. The N terminus of hCUL-2 seemed to be blocked, and only the N terminus of the NEDD8 modification was accessible to sequencing. Proteolytic digestion of the hCUL-2 bands, however, yielded peptide sequences corresponding to hCUL-2 (15).
Wild-Type but Not Tumorigenic Forms of pVHL Stimulate NEDD8–hCUL2 Conjugation.
Next, we coexpressed combinations of cDNAs for hCUL-1, hCUL-2, and pVHL in the presence of [35S]methionine in a reticulocyte lysate (which endogenously contains NEDD8, elongin B, elongin C, and undetectable levels of pVHL). As shown in Fig. 6A, both hCUL-1 and hCUL-2 ran in an SDS gel as a doublet. Additional, even slower-migrating forms of both cullins were observed when GST–NEDD8 was added to the extract, resulting in a reduction of the original NEDD8–hCUL-2 conjugate. This result confirms that both cullins are modified by NEDD8 or GST–NEDD8. Remarkably, expression of pVHL stimulated conjugate formation of NEDD8 (or GST–NEDD8) with hCUL-2 about 3-fold (Fig. 6A, left lanes). In contrast, pVHL had no stimulatory effect on NEDD8–hCUL-1 conjugate formation (Fig. 6A, right lanes). Thus, pVHL levels control the conjugation of NEDD8 to the CBC subunit hCUL-2, but they do not influence conjugation of NEDD8 to the SCF subunit hCUL-1. This observation is in line with the known specificity of pVHL to associate with hCUL-2 but not with hCUL-1 protein complexes (15).
Figure 6.
Cullins are modified by NEDD8 in vitro and the hCUL-2 modification is stimulated by wild-type pVHL. (A) hCUL-1 and hCUL-2 cDNAs (1 μg each) were translated in a reticulocyte lysate system simultaneously with a pVHL cDNA (1 μg) in the presence of recombinant GST or GST–NEDD8 protein (1 μg). Reaction mixtures were processed by SDS/PAGE and autoradiography. (B) hCUL-2 cDNA was translated in a reticulocyte lysate system simultaneously with increasing amounts of pVHL or pVHLΔ157 cDNAs (0.1, 0.5, 1, 2 μg), and reaction mixtures were processed as in A. (C) The NEDD8 conjugated hCUL-2 bands were quantified by densitometry of different exposures of autoradiographs. The values were plotted as fold induction vs. the value obtained without addition of pVHL (filled bars, wild-type pVHL; open bars, pVHLΔ157).
Using a similar experimental setup, we expressed increasing levels of wild-type pVHL or its tumorigenic mutant form (pVHLΔ157) in the lysate. Intriguingly, only the wild-type pVHL protein, but not the mutant form of pVHL, stimulated conjugation of NEDD8 to hCUL-2 in a dose-dependent manner (Fig. 6B). A quantification of the data indicated that the presence of wild-type pVHL can stimulate NEDD8–hCUL-2 conjugation about 5-fold, whereas the mutant form is essentially inactive in this assay (Fig. 6C). We therefore conclude that increasing levels of wild-type pVHL in the lysate results in a built up of fully assembled CBCVHL complexes and that the hCUL-2 subunit of these complexes is a substrate for NEDD8 modification. In contrast, the tumorigenic variant, pVHLΔ157, is unable to bind to CBC and is consequently inactive in stimulating NEDD8–hCUL-2 conjugate formation.
DISCUSSION
Posttranslational modifications of proteins by the covalent attachment of another protein is more common than previously assumed. In addition to the well characterized ubiquitin system, eukaryotes possess several pathways that mediate the conjugation of ubiquitin-like modifiers to specific cellular proteins. In contrast to ubiquitin, these proteins do not seem to promote protein degradation (32, 33). The human interferon-inducible ubiquitin-like protein UCRP, for instance, seems to mediate protein targeting to the cytoskeleton (34–36). Similarly, conjugation of the mammalian ubiquitin-like protein SUMO-1 can affect the subcellular localization of the substrate (37–41). Conjugation of SUMO-1 to IκBα, however, seems to inhibit ubiquitin-dependent degradation of the substrate (42). This observation may point to a general role of ubiquitin-like proteins as modulators of the ubiquitin pathway.
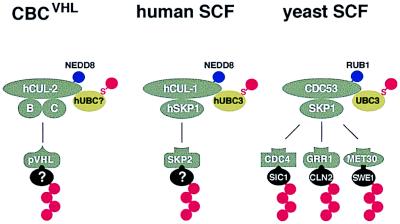
In this work we have shown that the conserved NEDD8/RUB1 modification pathway is functionally linked to the ubiquitin/proteasome system. Similar to the situation in yeast, substrates of this pathway in humans are members of the conserved family of cullin proteins. We discovered that both hCUL-1 and hCUL-2 are modified by NEDD8 in vivo. hCUL-1 is part of the human SCF ubiquitin ligase complex, whereas hCUL-2 is part of CBC. Because of the striking parallels between SCF and CBCVHL complexes with respect to protein composition and modification (Fig. 7), it seems very likely that CBC complexes also function as E3-type ubiquitin ligases in selective protein degradation pathways.
Figure 7.
Model of SCF-like protein complexes and their modification by NEDD8/RUB1. CBC and SCF complexes (gray) are structurally related. Yeast SCF ubiquitin ligase contains the cullin CDC53, SKP1, and one of several F-box proteins, which recruit specific substrates (black) to the complex leading to their ubiquitination (red chain). Ubiquitin conjugation via SCF is catalyzed by the UBC3CDC34 ubiquitin-conjugating enzyme (green), which forms a thioester-linked (S) intermediate with ubiquitin (red). Human cullin hCUL-1 is part of a similar SCF complex, whereas hCUL-2 associates with elongin B (B), the SKP1-related protein elongin C (C), and the pVHL tumor suppressor protein, which may function as a substrate-recruiting factor. Whether CBC can interact with alternative factors (other than pVHL) is not known. The cullins of all three complexes are modified by NEDD8 or RUB1 (blue).
Modification of CBC Is Linked to pVHL Tumor Suppressor Activity.
Most importantly, our data have identified a clear biochemical distinction between the wild-type and a tumorigenic mutant form of pVHL. Apparently, formation of an active CBCVHL complex containing the tumor suppressor protein pVHL is needed for efficient conjugation of the ubiquitin-like protein NEDD8 to one of CBCVHL’s own subunits, hCUL-2. Although the biological consequence of the modification of CBCVHL in not known currently, it shows striking parallels to the modification of SCF in yeast. SKP1 is required for RUB1–CDC53 conjugation, and overexpression of either the F-box protein CDC4 or the interacting E2 enzyme UBC3CDC34 stimulates this reaction (25). Apparently, modification by RUB1 influences vital functions of SCF ubiquitin ligases (see Fig. 2), possibly by affecting SCF’s subunit composition, stability, localization, or activities toward specific proteolytic substrates (25, 26). The detection of NEDD8–hCUL-1 conjugates further suggests that mammalian SCFs, and probably other cullin-containing E3s, also might be regulated in a similar fashion. Interestingly, in Arabidopsis, conjugation of a structural homolog of NEDD8/RUB1 is crucial for auxin response (43), emphasizing the importance of this conserved protein-modification pathway in growth control.
Function of CBCVHL.
The pVHL tumor suppressor protein has been proposed to play a “gatekeeper” function in certain tissues such as the kidney, presumably by controlling the number or behavior of respective tissue cells (15). Recent work has shown that CBCVHL interacts with the extracellular matrix protein fibronectin at an endoplasmic reticulum-related compartment, leading to the speculation that misfolded fibronectin molecules might be eliminated by CBCVHL-mediated endoplasmic reticulum-associated degradation (44). However, pVHL seems to have important nuclear functions as well. It is required for cell-cycle exit on serum withdrawal (45) and plays a role in the down-regulation of the expression of several proteins including vascular endothelial growth factor (VEGF), platelet derived growth factor-β (PDGF-β), and glucose transporter 1 (Glut-1; refs. 16 and 46). This function is consistent with the prominent neovascularization and high VEGF levels of VHL-associated tumors (23, 46). Cells lacking functional pVHL overexpress these hypoxia-inducible genes, which are regulated at the level of transcription and mRNA stability (46–50). HIF-1α is known to play a key role in hypoxia-induced gene expression. Thus, it seems very attractive to speculate that CBCVHL controls the half-life of this short-lived transcription factor and proteins that control mRNA stability (16, 47). Because NEDD8 is predominantly located in the nucleus (31), the influence of this modification pathway on the CBCVHL complex might be spatially restricted for nuclear functions of this putative E3 ubiquitin ligase.
Acknowledgments
We thank M. Scheffner for materials and discussions, W. Krek for hCUL-1 antibodies, S. Kumar for NEDD8 cDNA clones, and H. Ulrich and G. Pyrowolakis for comments on the manuscript. S.J. was supported by grants from Deutsche Forschungsgemeinschaft, Training and Mobility of Researchers European ubiquitin network, and Fonds der Chemischen Industrie. A.P. was supported by the Max Planck Society.
ABBREVIATIONS
- SCF
SKP1–CDC53–F-box protein
- En
enzyme n
- RCC
renal cell carcinoma
- HA
hemagglutinin epitope
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 3.Krek W. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 4.Patton E E, Willems A R, Tyers M. Trends Genet. 1998;14:236–242. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y, Jentsch S, Mann C. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 6.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S W. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 7.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Kobayashi R, Wittenberg C, Tyers M. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 8.Feldman R M R, Corell C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser P, Sia R A L, Bardes E G S, Lew D J, Reed S I. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisztwan J, Marti A, Sutterlüty H, Gstaiger M, Wirbelbauer C, Krek W. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyapina S A, Correll C C, Kipreos E T, Deshaies R J. Proc Natl Acad Sci USA. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aso T, Lane W, Conaway J W, Conaway R C. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 15.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonergan K M, Iliopoulos O, Ohh M, Kamura T, Conaway R C, Conaway J W, Kaehlin W G., Jr Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 18.Iliopoulos O, Kaelin W G., Jr Mol Med. 1997;3:289–293. [PMC free article] [PubMed] [Google Scholar]
- 19.Conaway J W, Kamura T, Conaway R C. Biochim Biophys Acta. 1998;1377:M49–M54. doi: 10.1016/s0304-419x(97)00035-8. [DOI] [PubMed] [Google Scholar]
- 20.Duan R D, Pause A, Burgess W H, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 21.Kibel A, Iliopoulos O, deCaprio J A, Kaelin W G., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 22.Iliopoulos O, Kibel A, Gray S, Kaelin W G., Jr Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 23.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y, Lee S, Pause A, Dudley C F, Latif F, Kuzmin I, et al. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 24.Maher E R, Kaelin W G., Jr Medicine (Baltimore) 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Tomooka Y, Noda M. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 28.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. Genes Dev. 1998;12:2263–2267. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathias N, Johnson S, Winey M, Adams A, Goetch L, Pringle J, Byres B, Goebl M G. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 31.Kamitani T, Kito K, Nguyen H P, Yeh E T H. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 32.Hochstrasser M. Genes Dev. 1998;2:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 33.Jentsch S, Ulrich H D. Nature (London) 1998;394:321–323. doi: 10.1038/26335. [DOI] [PubMed] [Google Scholar]
- 34.Loeb K R, Haas A L. J Biol Chem. 1992;267l:7806–7813. [PubMed] [Google Scholar]
- 35.Loeb K R, Haas A L. Mol Cell Biol. 1994;14:8408–8419. doi: 10.1128/mcb.14.12.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas A L, Ahrens P, Bright P M, Ankel H. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 37.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson P R, Hochstrasser M. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- 39.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh H, Pu R T, Dasso M. Trends Biol Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 41.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 42.Desterro J M P, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 43.del Pozo J C, Timpte C, Tan S, Callis J, Estelle M. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- 44.Ohh M, Yauch R L, Lonergan K M, Whaley J M, Stemmer-Rachmamimov A O, Louis D N, Gavin B J, Kley N, Kaehlin W G, Jr, Iliopoulos O. Mol Cell. 1998;1:959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 45.Pause A, Lee S, Lonergan K M, Klausner R D. Proc Natl Acad Sci USA. 1998;95:993–998. doi: 10.1073/pnas.95.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm J A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siemeister G, Weindel K K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 49.Mukhopadhyay D, Knebelmann B, Cohen H T, Ananth S, Sukhatme V P. Mol Cell Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L E, Gu J, Schau M, Bunn F. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]