Abstract
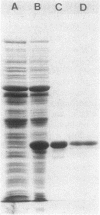
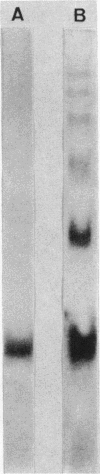
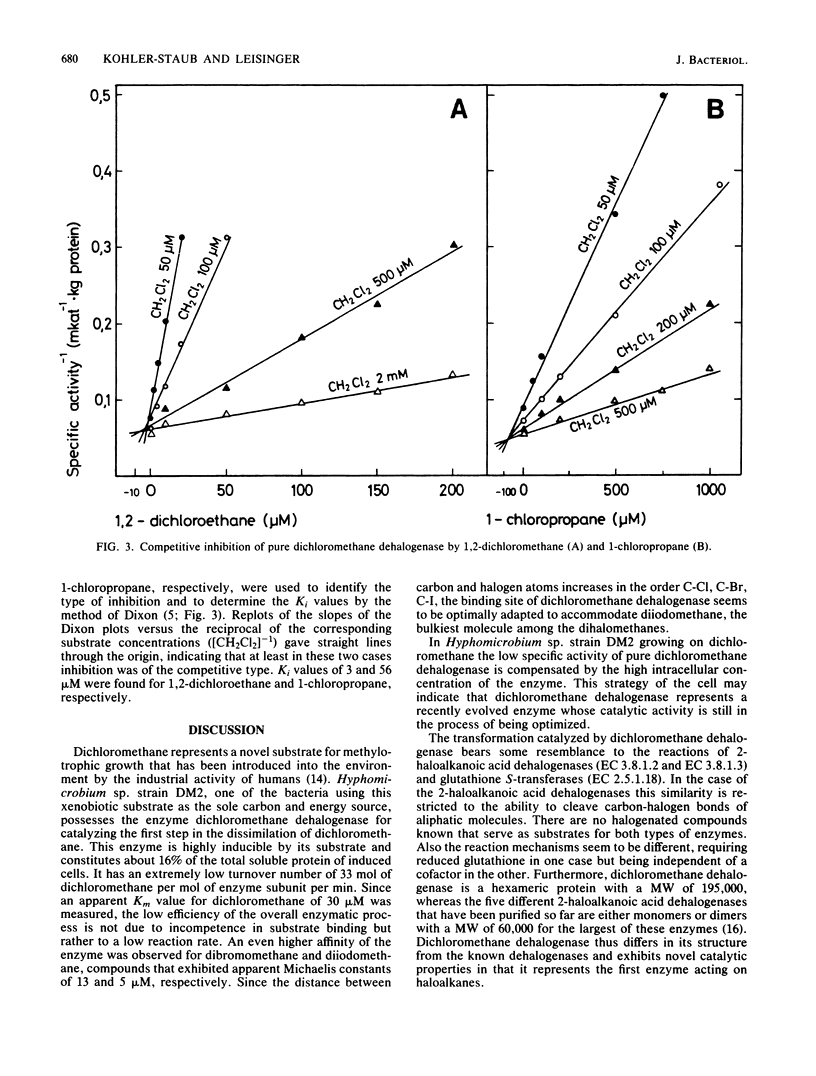
Dichloromethane dehalogenase, a highly inducible glutathione-dependent enzyme catalyzing the conversion of dichloromethane into formaldehyde and inorganic chloride, was purified fivefold with 60% yield from Hyphomicrobium sp. strain DM2. The electrophoretically homogeneous purified enzyme exhibited a specific activity of 17.3 mkat/kg of protein. Its pH optimum was 8.5. The enzyme was stable at -20 degrees C for at least 6 months. A subunit molecular weight of 33,000 was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gel filtration of native dichloromethane dehalogenase yielded a molecular weight of 195,000. Subunit cross-linking with dimethyl suberimidate confirmed the hexameric tertiary structure of the enzyme. Dichloromethane dehalogenase was highly specific for dihalomethanes. Its apparent Km values were 30 microM for CH2Cl2, 15 microM for CH2BrCl, 13 microM for CH2Br2, 5 microM for CH2I2, and 320 microM for glutathione. Several chlorinated aliphatic compounds inhibited the dichloromethane dehalogenase activity of the pure enzyme. The Ki values of the competitive inhibitors 1,2-dichloroethane and 1-chloropropane were 3 and 56 microM, respectively.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. E., Anders M. W. Metabolism of dihalomethanes to formaldehyde and inorganic halide--II. Studies on the mechanism of the reaction. Biochem Pharmacol. 1978;27(16):2021–2025. doi: 10.1016/0006-2952(78)90061-8. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brunner W., Staub D., Leisinger T. Bacterial degradation of dichloromethane. Appl Environ Microbiol. 1980 Nov;40(5):950–958. doi: 10.1128/aem.40.5.950-958.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIXON M. The determination of enzyme inhibitor constants. Biochem J. 1953 Aug;55(1):170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes C. S. Studies on plant amylases: The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem J. 1932;26(5):1406–1421. doi: 10.1042/bj0261406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecka G. M. Fate and effects of methylene chloride in activated sludge. Appl Environ Microbiol. 1982 Sep;44(3):701–707. doi: 10.1128/aem.44.3.701-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPat-Polasko L. T., McCarty P. L., Zehnder A. J. Secondary substrate utilization of methylene chloride by an isolated strain of Pseudomonas sp. Appl Environ Microbiol. 1984 Apr;47(4):825–830. doi: 10.1128/aem.47.4.825-830.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leisinger T. Microorganisms and xenobiotic compounds. Experientia. 1983 Nov 15;39(11):1183–1191. doi: 10.1007/BF01990355. [DOI] [PubMed] [Google Scholar]
- Meijer J., DePierre J. W., Rannug U. Measurement of drug-metabolizing systems in Salmonella typhimurium strains G46, TA15135, TA100, TA1538 and TA98. Chem Biol Interact. 1980 Sep;31(3):247–254. doi: 10.1016/0009-2797(80)90013-7. [DOI] [PubMed] [Google Scholar]
- Motosugi K., Soda K. Microbial degradation of synthetic organochlorine compounds. Experientia. 1983 Nov 15;39(11):1214–1220. doi: 10.1007/BF01990358. [DOI] [PubMed] [Google Scholar]
- Rittmann B. E., McCarty P. L. Utilization of dichloromethane by suspended and fixed-film bacteria. Appl Environ Microbiol. 1980 Jun;39(6):1225–1226. doi: 10.1128/aem.39.6.1225-1226.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel S. Hydrophobic chromatography. Methods Enzymol. 1974;34:126–140. doi: 10.1016/s0076-6879(74)34012-8. [DOI] [PubMed] [Google Scholar]
- Summer K. H., Göggelmann W., Greim H. Glutathione and glutathione S-transferases in the Salmonella mammalian-microsome mutagenicity test. Mutat Res. 1980 May;70(3):269–278. doi: 10.1016/0027-5107(80)90018-4. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]