Abstract
β-Catenin plays a dual role in the cell: one in linking the cytoplasmic side of cadherin-mediated cell–cell contacts to the actin cytoskeleton and an additional role in signaling that involves transactivation in complex with transcription factors of the lymphoid enhancing factor (LEF-1) family. Elevated β-catenin levels in colorectal cancer caused by mutations in β-catenin or by the adenomatous polyposis coli molecule, which regulates β-catenin degradation, result in the binding of β-catenin to LEF-1 and increased transcriptional activation of mostly unknown target genes. Here, we show that the cyclin D1 gene is a direct target for transactivation by the β-catenin/LEF-1 pathway through a LEF-1 binding site in the cyclin D1 promoter. Inhibitors of β-catenin activation, wild-type adenomatous polyposis coli, axin, and the cytoplasmic tail of cadherin suppressed cyclin D1 promoter activity in colon cancer cells. Cyclin D1 protein levels were induced by β-catenin overexpression and reduced in cells overexpressing the cadherin cytoplasmic domain. Increased β-catenin levels may thus promote neoplastic conversion by triggering cyclin D1 gene expression and, consequently, uncontrolled progression into the cell cycle.
β-Catenin is a major component of adherens junctions linking the actin cytoskeleton to members of the cadherin family of transmembrane cell–cell adhesion receptors (1, 2). In addition, β-catenin can translocate into the nucleus (3–7), where it can complex with transcription factors of the LEF-1 family and regulate the expression of specific genes (8, 9). By playing such a dual role—a structural role in cell–cell junctions and a regulatory role in the nucleus—β-catenin can transduce changes in cell adhesion and junction formation to control transmembrane signaling and gene expression (1, 10–12). β-Catenin-mediated signaling depends on its accumulation and subsequent translocation into the nucleus. The level of β-catenin in the cell is regulated by its association with the tumor suppressor molecule adenomatous polyposis coli (APC; refs. 13 and 14), axin (15, 16), and glycogen synthase kinase 3β (GSK-3β; ref. 17). Phosphorylation of β-catenin by the APC–axin–GSK-3β complex (18, 19) leads to its degradation by the ubiquitin–proteasome system (6, 20). The failure of this degradation in cells expressing mutant APC or β-catenin leads to the accumulation of β-catenin and is common in human colon cancer and melanoma (21–23). Elevated β-catenin levels in such tumors are suggested to confer uncontrolled activation of gene transcription by the β-catenin/LEF-1 complex that may contribute to tumor progression (1, 24, 25). The nature of the target genes of the β-catenin/LEF complex is, however, largely unknown, except for the recently discovered c-MYC that was shown to contain LEF-1-binding sequences in its promoter (26).
Cyclin D1 is a major regulator of the progression of cells into the proliferative stage of the cell cycle (27). Although the cyclin D1 gene is not amplified in human colon cancer, the expression of cyclin D1 is elevated in about 30% of human adenocarcinomas and in adenomatous polyps of the colon (28, 29), and expression of anti-sense cyclin D1 cDNA abolished the growth of SW480 colon cancer cells in nude mice, indicating a critical role for cyclin D1 in tumorigenesis (30).
In this study, we investigated the possibility that the cyclin D1 gene is a target for the β-catenin/LEF-1 complex and show that the cyclin D1 promoter contains a LEF-1 binding sequence that is activated in human colon cancer cells. We further show that transcriptional activation of the cyclin D1 gene in such cells can be inhibited by enhancing the degradation of β-catenin with wild-type APC and axin or by its binding to the cadherin cytoplasmic tail. Unscheduled activation of cyclin D1 transcription by the β-catenin/LEF-1 complex in colon cancer cells may result in uncontrolled cell proliferation and thus contribute to tumor progression in these cells.
MATERIALS AND METHODS
Plasmid Constructions.
Plasmids containing the luciferase reporter under the cyclin D1 promoter and the deletion constructions derived from it have been described (31). The mutated LEF-1 binding site at nucleotides −75 and −74 from AT to GC (−163mtLefCD1LUC) and the deletion of the LEF-1 binding site from −81 to −73 (−163ΔLefCD1LUC) were constructed by PCR-based site directed mutagenesis by using the −163CD1LUC plasmid. Reporter plasmids containing luciferase under a multimeric consensus- or mutant inactive-LEF-1 binding site (TOPFLASH and FOPFLASH; ref. 9) and vectors expressing various forms of β-catenin (7), LEF-1 (5), axin, APC (15), and the N-cadherin tail (32) have been described.
Cell Lines and Transfections.
SW480, 293T, and Neuro 2A cell lines were maintained in DMEM with 10% (vol/vol) calf serum. Transient transfections were performed by the calcium phosphate precipitation method with 293T and Neuro 2A cell lines and by Lipofectamine with SW480 cells. A β-galactosidase-expressing plasmid (0.5 μg) was included in each transfection to monitor the transfection efficiency. We found that β-galactosidase expression was not affected significantly by either LEF-1 or β-catenin cotransfection. After 48 h, the cells were lysed, and luciferase and β-galactosidase activities were determined by enzyme assay kits from Promega. Luciferase activity was normalized to β-galactosidase activity as an internal transfection control. N-cadherin-tail-expressing cell lines were generated by stably transfecting the pECE-N-cadherin tail and pSV-hygro expression vectors into SW480 cells (7, 32). Individual clones were selected for resistance to 100 μg/ml hygromycin. The green fluorescent protein (GFP)–ΔN-β-catenin and the mutant β-catenin constructs (β-cat Y33) were HA-tagged (7).
Immunoblotting.
Protein levels were determined by immunoblotting by using a monoclonal antibody against cyclin D1 (DCS-6; Neomarkers, Freemont, CA), a polyclonal antiserum against β-catenin, and monoclonal antibodies against pan cadherin (CH-19), and vinculin (h-VIN 1) as described (ref. 7; all from Sigma). Anti-LEF-1 rabbit antiserum was a gift from R. Grosschedl (University of California, San Francisco).
In Vitro Translation and DNA Binding Analysis.
In vitro translated proteins were prepared by using a coupled transcription and translation kit (Promega). β-Catenin and LEF-1 were expressed from pCIneo constructs, and the efficiency of translation was determined by a reaction containing [35S]methionine, followed by SDS/PAGE and Western blotting. For protein–DNA interaction, in vitro translated LEF-1 and β-catenin were incubated with the following 32P-labeled duplex oligonucleotide probes: CD1 (5′-CTCTGCCGGGCTTTGATCTTTGCTTAACAACA-3′), CD1TOP (5′-CTCTGCCGGCCTTTGATCTTTGCTTAACAACA-3′), and CD1FOP (5′-CTCTGCCGGGCTTTGGCCTTTGCTTAACAACA-3′). The wild-type and mutant LEF-1 binding sequences are underlined. The binding reaction contained ≈40,000 cpm of 32P-labeled DNA that was incubated for 30 min with the in vitro translated proteins in 20 mM Hepes, pH 7.9/75 mM NaCl/1 mM dithiotreitol/2 mM MgCl2/10% (vol/vol) glycerol/0.1 mg of BSA/10 μg/ml salmon sperm DNA. DNA–protein complexes were electrophoresed in 4% native acrylamide gels and visualized by autoradiography. Binding reactions also were carried out with nuclear extracts from SW480 cells (3 μg protein per reaction mixture) that were prepared as described (33).
RESULTS
Activation of the Cyclin D1 Promoter and Elevation of Cyclin D1 in Cells Transfected with β-Catenin.
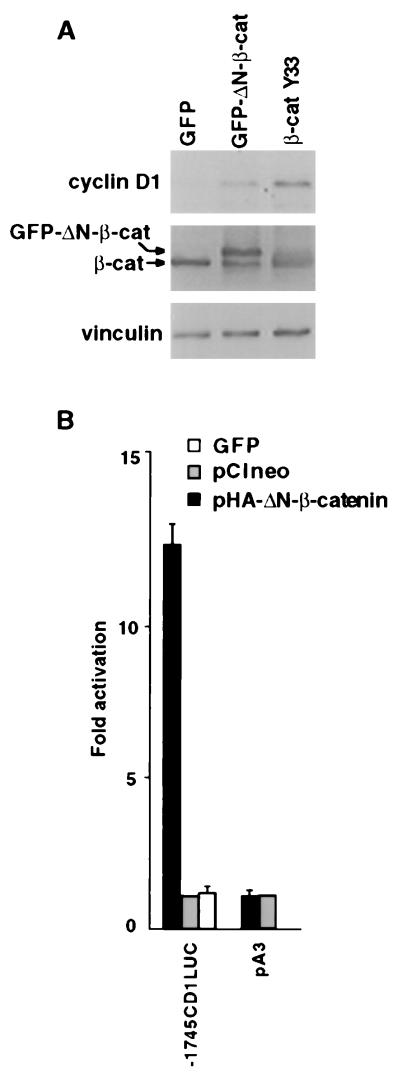
We assessed the effect of β-catenin overexpression on cyclin D1 abundance in human 293T cells, previously shown to respond by activation of β-catenin/LEF-1-driven transcription (7, 21, 22). Cells were transfected with expression vectors encoding mutant N-terminal β-catenin constructs (GFP–ΔN-β-catenin and β-cat Y33) that render it stable against degradation (7, 22, 23). Cyclin D1 protein levels were induced 2-to 3-fold after transfection with these β-catenin constructs, as compared with transfection with a construct encoding the GFP that served as control (Fig. 1A). To examine whether the cyclin D1 promoter was a direct transcriptional target for activation by the β-catenin/LEF-1 complex, an HA-tagged ΔN-β-catenin expression plasmid (pHA-ΔN-β-catenin) was cotransfected with the human cyclin D1 promoter linked upstream to a luciferase reporter (Fig. 2A). The cyclin D1 promoter was induced 12-fold by this β-catenin (Fig. 1B) and was not affected by either the empty expression plasmid (Fig. 1B; pA3) or the construct encoding GFP. The cyclin D1 promoter was also induced 10- to 15-fold by β-catenin overexpression in the human Neuro 2A neuroblastoma cell line (see below; Fig. 3).
Figure 1.
Induction of cyclin D1 and β-catenin-responsive transactivation of the cyclin D1 promoter. (A) Cells from the 293T line were transfected with 4 μg of GFP, GFP-linked ΔN-β-catenin (GFP-ΔN-β-cat), or mutant β-cat Y33. Cell lysates were analyzed by Western blotting for levels of cyclin D1, β-catenin (β-cat), and vinculin. (B) Cells from the 293T line were transfected with 0.8 μg of a reporter plasmid containing 1,745 bp of the cyclin D1 promoter (−1745CD1Luc) or with the empty pA3 plasmid, together with 4 μg of an HA-tagged ΔN-β-catenin-encoding plasmid (pHA-ΔN-β-cat), a GFP expression construct, or pCIneo. The bars represent luciferase activity in cells transfected with pHA-ΔN-β-catenin divided by the activity in cells transfected with control plasmid (pCIneo). Each transfection was carried out in duplicate plates. The means ± SD from three separate transfections are shown.
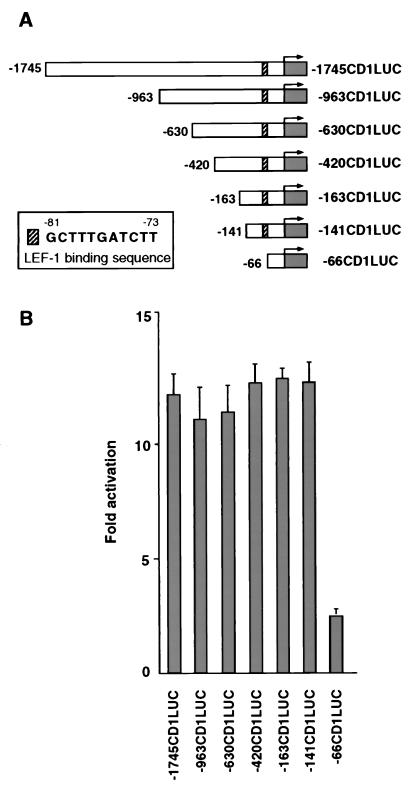
Figure 2.
Identification of the LEF-1 binding sequence in the cyclin D1 promoter. (A) Schematic representation of reporter constructs from the cyclin D1 promoter, deletion constructs, and the LEF-1-binding sequence between nucleotides −81 and −73 of the promoter (Insert). (B) The promoter deletion constructs of the cyclin D1 promoter (0.8 μg) shown in A were transfected into 293T cells as described in Fig. 1B.
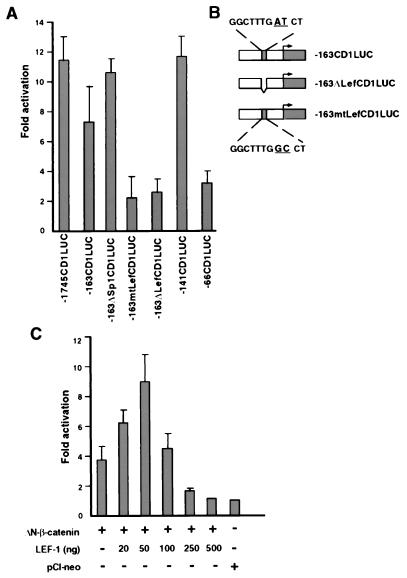
Figure 3.
The LEF-1-binding sequence in the cyclin D1 promoter is required for β-catenin-mediated transactivation. (A) Cells from the 293T line were transfected with the indicated reporter plasmid (0.8 μg), together with 4 μg of ΔN-β-catenin or the control plasmid (pCIneo). The promoter activity is presented as in Fig. 1B. (B) Schematic representation of mutations in the −163 cyclin D1 promoter construct, including an AT to GC change at nucleotides −75 and −74 (−163mtLefCD1LUC) and deletion of the LEF-1 binding site (−163ΔLefCD1LUC) between nucleotides −81 to −73. (C) Neuro 2A cells were transfected with 0.8 μg of −1745CD1Luc, 2 μg of ΔN-β-catenin, or a control plasmid (pCIneo), along with increasing amounts of a LEF-1 expression plasmid. DNA concentrations were kept constant with empty vector DNA. Transfections were carried out in triplicate and the means ± SD are presented.
Identification of a LEF-1 Binding Sequence in the Cyclin D1 Promoter.
To identify the sequences in the cyclin D1 promoter that can confer transactivation by β-catenin, a series of 5′ promoter deletion constructions were used (Fig. 2A). Such mapping identified a region between nucleotides −141 and −66 that was sufficient for transcriptional activation by β-catenin (Fig. 2B). Analysis of the sequence between nucleotides −141 and −66 indicated that nucleotides −81 to −73 represent a consensus LEF binding site (Fig. 2A, Insert). To examine the functional significance of this candidate LEF binding site, we introduced (in the context of the −163 bp reporter plasmid, −163CD1LUC) a deletion (−163ΔLefCD1LUC) and point mutations (163mtLefCD1LUC) that should render the LEF binding site in the cyclin D1 promoter inactive (Fig. 3B). β-Catenin-driven transactivation of the cyclin D1 constructs was inhibited when this putative LEF-1-binding domain was either deleted or mutated (Fig. 3A). Mutations introduced outside of this region and in the Sp1 binding site of this construct (−163ΔSp1CD1LUC; ref. 34) had no effect on β-catenin-driven activation of the promoter (Fig. 3A). Transactivation of the cyclin D1 promoter by β-catenin also depended on the level of cotransfected LEF-1. At low concentrations of the expression plasmid, LEF-1 elevated β-catenin-mediated activation of cyclin D1 a further 2.5-fold (Fig. 3C) but inhibited transcription at high plasmid concentrations, consistent with its transcription repressor role when expressed in excess over β-catenin (35, 36).
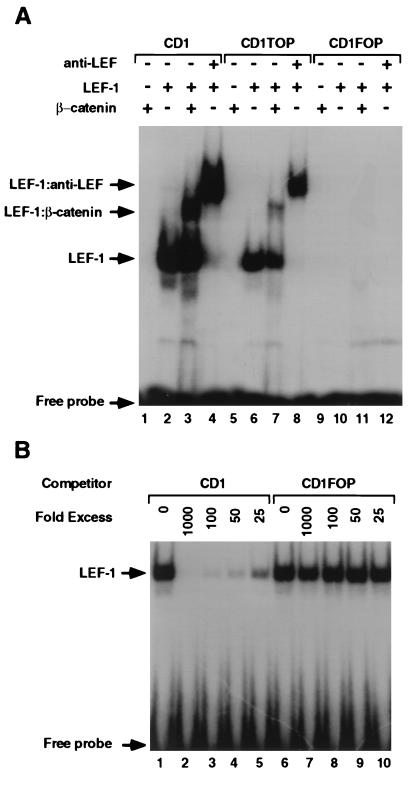
The β-Catenin/LEF-1 Complex Binds in Vitro to the LEF-1 Consensus Sequence of the Cyclin D1 Promoter.
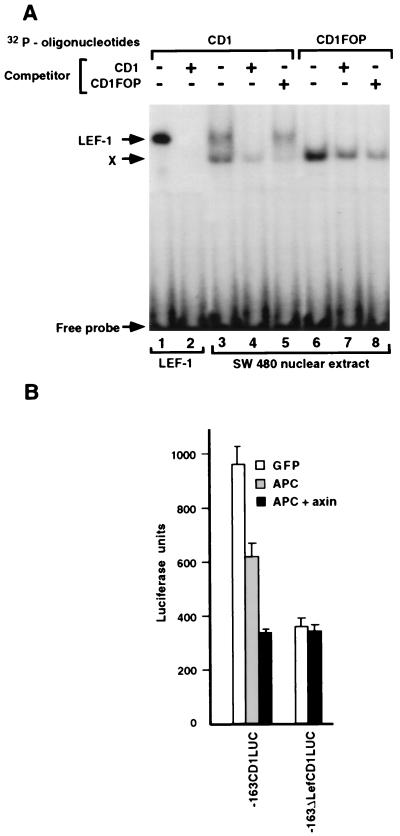
To determine whether the LEF-1 binding sequence in the cyclin D1 promoter can bind a LEF-1/β-catenin heterodimer, we conducted electrophoretic mobility-shift assays comparing the cyclin D1 LEF-1 site with a consensus LEF-1-binding site (CD1TOP; ref. 9) and a mutant LEF-1 binding site (CD1FOP). In vitro, synthesized LEF-1 bound to the LEF-1 sequence in the cyclin D1 promoter (Fig. 4A, CD1, lane 2), and the complex was supershifted with an LEF-1 antibody (Fig. 4A, lanes 4 and 8). The mutant sequence derived from the −163mtLefCD1LUC (CD1FOP), in contrast, failed to bind LEF-1 (Fig. 4A, CD1FOP, lane 10). β-Catenin transcribed and translated in vitro did not bind on its own, as expected, to this sequence in the cyclin D1 promoter (Fig. 4A, lanes 1, 5, and 9). A supershift was introduced in the electrophoretic mobility of this DNA fragment when β-catenin and LEF-1 were added together (Fig. 4A, lanes 3 and 7), implying the formation of a ternary complex consisting of the DNA, LEF-1, and β-catenin. This binding of LEF-1 to the cyclin D1 promoter was blocked by the addition of excess unlabeled CD1 but not by the mutant CD1FOP (Fig. 4B).
Figure 4.
Electrophoretic mobility-shift assays of the cyclin D1 promoter. (A) Duplex oligonucleotides containing the LEF-1 binding sequences of the cyclin D1 promoter (CD1), a consensus LEF-1 binding sequences (CD1TOP) and a substitution of nucleotides −75 and −74 from AT to GC (CD1FOP) were “end labeled” with [32P]dATP and incubated with in vitro translated LEF-1 and/or β-catenin or with anti-LEF-1 antibody. The protein–DNA complexes were separated by electrophoresis and visualized by autoradiography. (B) In vitro translated LEF-1 was incubated with the 32P-labeled CD1 oligonucleotide, and increasing amounts of unlabeled CD1 or CD1FOP oligonucleotides were used as competitors. The protein–DNA complexes were analyzed as in Fig. 4A.
Transactivation of the Cyclin D1 Promoter in Colon Cancer Cells and Its Inhibition by APC, Axin, and the Cadherin Cytoplasmic Domain.
To analyze effects on transcription of the cyclin D1 promoter in a cellular system where endogenous β-catenin is elevated, we employed SW480 colon carcinoma cells in which the level of β-catenin is increased because of inactivating mutations in APC (37). Nuclear extracts from these cells could specifically bind to and shift the electrophoretic mobility of the LEF-1 binding sequence in the cyclin D1 promoter (Fig. 5A, lane 3), similar to in vitro translated LEF-1 (Fig. 5A, lane 1). An additional shifted band (Fig. 5A, marked X) bound nonspecifically; it also was observed when the mutant LEF-1 binding site (CD1FOP) was used (Fig. 5A, lanes 6–8).
Figure 5.
Electrophoretic mobility-shift assays of the cyclin D1 promoter with SW480 nuclear lysates and regulation of cyclin D1 promoter transcriptional activity by APC and axin. (A) Oligonucleotides labeled with [32P]dATP (CD1, lanes 1–5; CD1FOP, lanes 6–8) were incubated with in vitro translated LEF-1 (lanes 1 and 2) or with nuclear extracts from SW480 cells (lanes 3–8) in the presence of 100-fold excess of cold competitor oligonucleotides. (B) SW480 cells were transiently transfected with 1 μg of −163CD1LUC or −163ΔLefCD1LUC and 2 μg of either GFP or APC plus axin expression vectors. The bars represent luciferase activity in the transfected cells after normalizing for transfection efficiency with β-galactosidase. Note that, in A, an additional, faster-migrating band was obtained with CD1 and the SW480 nuclear extract (X). This band also was seen with the mutant CD1FOP and therefore was considered nonspecific.
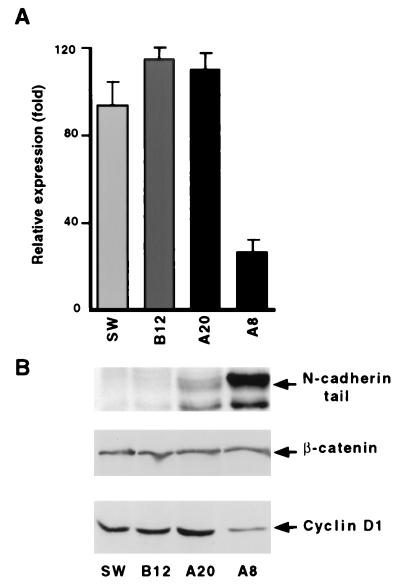
When the cyclin D1 promoter, in the context of the −163CD1LUC reporter plasmid, was transfected into SW480 cells, transcription from this plasmid was active without cotransfection of exogenous β-catenin (Fig. 5B). This transcription was reduced by cotransfection of wild-type APC or a mixture of APC and axin (Fig. 5B), which act together to decrease levels of β-catenin by enhancing its degradation (15, 16). In contrast, APC and axin did not affect the basal transcription observed in the deletion mutant of this cyclin D1 promoter construct (−163ΔLefCD1LUC; Fig. 5B). We have shown previously that either full-length N-cadherin or its cytoplasmic domain can block the constitutive β-catenin-mediated transactivation in SW480 cells by competing with LEF-1 for β-catenin binding (7, 32). We therefore analyzed the effect of a stably overexpressed N-cadherin cytoplasmic tail in SW480 cells on the transcriptional activity of the cyclin D1 promoter and the level of the cyclin D1 protein. In SW480 cells stably expressing high levels of the cadherin cytoplasmic tail (Fig. 6B, clone A8) but not in clones expressing low levels of the cadherin tail (Fig. 6B, clone A20), β-catenin-mediated transactivation of the cyclin D1 promoter was inhibited (Fig. 6A; compare clones A8 and A20), and the level of cyclin D1 protein was reduced specifically (Fig. 6B; compare clones A8 and A20).
Figure 6.
Decreased transcription from the cyclin D1 promoter and cyclin D1 protein in SW480 cells expressing the N-cadherin cytoplasmic domain. (A) Individual SW480 cell clones stably expressing different levels of the N-cadherin cytoplasmic domain were transiently transfected with 1 μg of −1745CD1LUC. The bars represent luciferase activity in cells transfected with −1745CD1LUC divided by luciferase activity in cells transfected with the empty pCIneo vector. (B) Total cellular proteins from the cells described in A were separated by SDS/PAGE and analyzed by Western blotting with antibodies that recognize the N-cadherin tail, β-catenin, and cyclin D1.
DISCUSSION
Elevated β-catenin expression in colorectal cancer caused by inactivating mutations in APC or in the GSK-3β phosphorylation sites of β-catenin results in the accumulation of β-catenin in the nucleus and the uncontrolled activation of target gene expression—a process believed to contribute to tumor progression. The nature of these target genes is still largely unknown, but their involvement in the control of cell proliferation is suggested. In this study, we have identified the cyclin D1 gene as a target for the β-catenin/LEF-1 complex. We have shown that the LEF-1 binding consensus sequence in the cyclin D1 gene promoter can bind to nuclear extracts from human colon cancer cells and is transcriptionally active when linked to reporter plasmids and introduced into human colon cancer cells expressing elevated wild-type β-catenin. In such cells expressing inactive mutant APC, cotransfection with wild-type APC inhibited transactivation of the cyclin D1 gene. Based on these results, we suggest that increased β-catenin levels in colon cancer cells result in the activation of the cyclin D1 gene promoter by the heterodimeric complex formed between β-catenin and LEF-1, which, in turn, results in the elevation of cyclin D1 gene expression and protein level. This elevation, in turn, may lead to uncontrolled stimulation of the cells into the proliferative stage of the cell cycle. In the presence of wild-type APC, the level of β-catenin is kept in check, and the formation of the β-catenin/LEF-1 complex is prevented. The reduction in the transcriptional activity of the cyclin D1 promoter, resulting most probably from competition between the cytoplasmic tail of cadherin and LEF-1 for β-catenin binding (32), and the concomitant decrease in cyclin D1 protein support this view (Figs. 5 and 6). These results also show the potential usefulness of the cadherin cytoplasmic domain (and its derivatives) in the attempt to antagonize the possibly oncogenic transcription driven by the β-catenin/LEF-1 complex (32).
Elevated transcription from the cyclin D1 gene induced by the β-catenin/LEF-1 complex may also explain the notion that, although alterations in the cyclin D1 gene were detected in many types of human cancer (38–40), no mutations in the cyclin D1 gene were identified in colorectal cancer, whereas ≈30% of primary human colon tumors display increased expression of cyclin D1 (28, 29). This increase in cyclin D1 in only some colon cancers may result from the heterogeneity in β-catenin expression in the tumor and from the fact that many colon cancer cell lines with mutant APC do not display increased levels of β-catenin (41).
The involvement of cyclin D1 in human colon cancer also is supported by experiments showing that expression of an antisense cyclin D1 construct in SW480 colon cancer cells (also used in this study) results in decreased cyclin D1, inhibition of growth, and suppression of their tumorigenicity in nude mice (30).
β-Catenin and cyclin D1 apparently also share another component of the Wnt pathway, GSK-3β. A recent study showed that cyclin D1 is a target for GSK-3β, which, by regulating cyclin D1 phosphorylation, may control the subcellular localization and proteolysis of cyclin D1 by the ubiquitin–proteasome system (42). This pathway may constitute another means by which the Wnt signaling pathway can target cyclin D1 in colon cancer.
The link between β-catenin-mediated transactivation of the cyclin D1 promoter and neoplastic transformation also may be relevant to intestinal and mammary epithelial cells transformed by the elevated expression of integrin-linked kinase (ILK) (43). ILK-transfected cells were shown to become neoplastic and to display high levels of cyclin D1 protein constitutively (44). In addition, the level of extrajunctional β-catenin in such cells is increased, concomitant with nuclear translocation of β-catenin and activation of β-catenin-mediated LEF-1-directed transcription (45). Cyclin D1 elevation in these ILK-transfected cells may result from a direct activation of cyclin D1 transcription by the β-catenin/LEF-1 complex.
In a recent study, the c-MYC promoter was identified also as a target of the β-catenin/LEF-1 complex in human colorectal cancer (26). In other types of tumors, overexpression of either c-myc or cyclin D1 were shown to induce cell-cycle progression in breast cancer cell lines (46), whereas, in fibroblasts, myc induction of cell-cycle progression seems to be linked to the induction of cyclin E, independently of cyclin D1 (47). Together, these results are consistent with a model in which myc and cyclin D1 can induce S phase entry in parallel pathways, and the elevation of cyclin D1 in colorectal cancer, by inactivation of APC or mutations in β-catenin, may promote uncontrolled proliferation and thus contribute to the neoplastic transformation of cells.
Acknowledgments
We are grateful to the following colleagues for sending reagents: S. Sokol, R. Kemler, H. Clevers, M. van de Wetering, M. Wheelock, and R. Grosschedl. These studies were supported by grants from the USA–Israel Binational Foundation, the German–Israeli Foundation for Scientific Research and Development, the Forchheimer Center for Molecular Genetics, the Cooperation Program in Cancer Research between the German Cancer Research Center and the Israel Ministry of Science (to A.B.-Z.), and from the National Institute of Health (to R.G.P.). A.B.-Z. holds the Lunenfeld–Kunin Chair in Cell Biology and Genetics, and R.G.P. is a recipient of the Ira T. Hirschl Award and the Susan G. Komen Breast Cancer Foundation Award.
ABBREVIATIONS
- APC
adenomatous polyposis coli
- GSK-3β
glycogen synthase kinase 3β
- GFP
green fluorescent protein
References
- 1.Ben-Ze’ev A, Geiger B. Curr Opin Cell Biol. 1998;10:629–639. doi: 10.1016/s0955-0674(98)80039-2. [DOI] [PubMed] [Google Scholar]
- 2.Kemler R. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 3.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 4.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 5.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–11. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 6.Salomon D, Sacco P A, Guha Roy S, Simcha I, Johnson K R, Wheelock M J, Ben-Ze’ev A. J Cell Biol. 1997;139:1325–1335. doi: 10.1083/jcb.139.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simcha I, Shtutman M, Salomon D, Zhurinsky J, Sadot E, Geiger B, Ben-Ze’ev A. J Cell Biol. 1998;141:1433–1448. doi: 10.1083/jcb.141.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S-C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Tpma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 10.Cavallo R, Rubenstein D, Peifer M. Curr Opin Genet Dev. 1997;7:459–466. doi: 10.1016/s0959-437x(97)80071-8. [DOI] [PubMed] [Google Scholar]
- 11.Bullions L C, Levine A. Curr Opin Oncol. 1998;10:81–87. doi: 10.1097/00001622-199801000-00013. [DOI] [PubMed] [Google Scholar]
- 12.Willert K, Nusse R. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 13.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 14.Rubinfeld B, Souza B, Albert I, Müller O, Chamberlain S F, Masiarz R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 15.Hart M, de los Santos R, Albert I, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 16.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 17.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 18.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1372–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 22.Korinek V, Backer N P, Morin J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 23.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 24.Peifer M. Science. 1997;275:1752–1753. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 25.Gumbiner B M. Curr Biol. 1997;7:R443–R446. doi: 10.1016/s0960-9822(06)00214-4. [DOI] [PubMed] [Google Scholar]
- 26.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 27.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 28.Bartkova J, Lukas J, Strauss M, Bartek J. Int J Cancer. 1994;58:568–573. doi: 10.1002/ijc.2910580420. [DOI] [PubMed] [Google Scholar]
- 29.Arber N, Hibshoosh H, Moss S F, Sutter T, Zhang Y, Begg M, Wang S, Weinstein I B, Holt P R. Gastroenterology. 1996;110:669–674. doi: 10.1053/gast.1996.v110.pm8608874. [DOI] [PubMed] [Google Scholar]
- 30.Arber N, Doki Y, Han E K-H, Sgambato A, Zhou P, Kim N-H, Delohery T, Klein M G, Holt P R, Weinstein I B. Cancer Res. 1997;57:1569–1574. [PubMed] [Google Scholar]
- 31.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 32.Sadot E, Simcha I, Shtutman M, Ben-Ze’ev A, Geiger B. Proc Natl Acad Sci USA. 1998;95:15339–15344. doi: 10.1073/pnas.95.26.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiber E, Matthias P, Müller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe G, Albanese C, Lee J R, Reutens A, Vairo G, Henglein B, Pestell R G. Mol Cell Biol. 1998;18:3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavallo R A, Cox R T, Moline M M, Roose J, Polevoy G A, Clevers H, Peifer M, Bejsovec A. Nature (London) 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 36.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. Nature (London) 1998;395:607–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 37.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter T, Pines J. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 39.Motokura T, Arnold A. Curr Opin Genet Dev. 1993;3:5–10. doi: 10.1016/s0959-437x(05)80334-x. [DOI] [PubMed] [Google Scholar]
- 40.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 41.Polakis P. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 42.Diehl A J, Cheng M, Rousell M F, Sherr C J. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanigan G E, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino M G, Radeva G, Filmus J, Bell J C, Dedhar S. Nature (London) 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 44.Radeva G, Petrocelli T, Behrend E, Leung-Hagelsteijn C, Filmus J, Slingerland J, Dedhar S. J Biol Chem. 1997;272:13937–13944. doi: 10.1074/jbc.272.21.13937. [DOI] [PubMed] [Google Scholar]
- 45.Novak A, Hsu S-C, Leung-Hagelsteijn C, Radeva G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prall O W, Rogan E M, Musgrove E A, Watts C K, Sutherland R L. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vlach J, Hennecke S, Amati B. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]