Abstract
Hormone-sensitive lipase (HSL) is a cytosolic neutral lipase that functions as the rate-limiting enzyme for the mobilization of free fatty acids in adipose tissue. By using the yeast two-hybrid system to examine the potential interaction of HSL with other cellular proteins, evidence is provided to demonstrate a direct interaction of HSL with adipocyte lipid-binding protein (ALBP), a member of the family of intracellular lipid-binding proteins that binds fatty acids, retinoids, and other hydrophobic ligands. The interaction was demonstrated in vitro by the binding of ALBP to HSL translated in vitro, to HSL in extracts of HSL overexpressing Chinese hamster ovary (CHO) cells, and to HSL in extracts of rat adipose tissue. Finally, the presence of ALBP was documented in immune complexes from rat adipose tissue immunoprecipitated with anti-HSL antibodies. The HSL–ALBP interaction was mapped to an N-terminal 300-aa region of HSL that is distinct from the C-terminal catalytic domain. These results suggest that HSL-derived fatty acids are bound by ALBP to facilitate intracellular trafficking of hydrophobic lipids.
Free fatty acids (FFA) derived from the breakdown of stored triacylglycerols in adipose tissue are a major energy source for most tissues. The critical enzyme that catalyzes the hydrolysis of stored triacylglycerols in adipose tissue is hormone-sensitive lipase (HSL) (1). HSL has broad substrate specificity; in addition to triacylglycerol, HSL can also catalyze the hydrolysis of diacylglycerol, 1(3) monoacylglycerol, cholesteryl esters, lipoidal esters of steroid hormones, and retinyl esters in adipose tissue (2–5). In the rat, HSL is a 767-aa protein that has a molecular mass of 84 kDa on SDS/PAGE (2, 6). Its primary sequence is unrelated to any of the other known mammalian lipases; however, within its catalytic domain, it shares some sequence similarity with liver arylacetamide deacetylase (7) as well as with esterases isolated from several bacteria (8–11). The secondary structure of the C-terminal portion of HSL has been demonstrated to possess homology with some features of the secondary structure of acetylcholinesterase and of fungal lipases from Geotrichum candidum and Candida rugosa (12, 13), whereas the N-terminal 320 aa have no primary or secondary structural similarity with any known proteins.
The activity of HSL against triacylglycerol and cholesteryl ester substrates is regulated acutely via phosphorylation–dephosphorylation reactions. The activation of HSL by fast-acting lipolytic hormones (catecholamines, glucagon, corticotropin) involves a hormone/receptor-induced increase in the cellular concentration of cAMP, which then activates cAMP-dependent protein kinase, resulting in the phosphorylation of HSL and an increase in hydrolytic activity. However, phosphorylation increases HSL activity in vitro only ≈2- to 4-fold compared with the up to 50-fold stimulation of lipolysis observed in intact fat cells. To explain this discrepancy, it has been proposed that the catecholamine-induced stimulation of lipolysis in vitro is caused by the translocation of phosphorylated HSL from an aqueous cytosolic compartment to the lipid droplet (14, 15). It would be expected that this translocation is facilitated through the interaction of HSL with specific cellular proteins. HSL could interact with structural or other proteins in the cell that facilitate the exposure of the lipid droplet. Perilipin, a protein that is located on the surface of the lipid droplet and is the major substrate in adipose cells of cAMP-dependent protein kinase (16, 17), as well as adipocyte differentiation-related protein (18, 19), are potential candidates for such a function. Conversely, HSL might be tethered in the cytoplasm in the basal state while droplet-associated proteins prevent its interaction with the lipid droplet. After phosphorylation, HSL could be released, while droplet-associated proteins fall off the lipid droplet (20, 21), thus allowing access to HSL. The interaction of HSL with other cellular proteins has not been reported. In the present study, we used the yeast two-hybrid assay system to identify specific HSL binding partners and have identified one such protein as the major intracellular lipid binding protein in adipose tissue.
MATERIALS AND METHODS
Yeast Two-Hybrid System.
Full-length rat HSL cDNA was cloned into the SmaI site of the pAS1-CYH2 yeast two-hybrid vector (22) to yield an HSL-GAL4 DNA-binding domain vector; and the correct clone was confirmed by sequencing. Adipose tissue was obtained from adult, male Sprague–Dawley rats and was used to generate a yeast two-hybrid library (HybriZAP 2.1, Stratagene) that was constructed by using directional cloning into EcoRI/XhoI sites in the pAD-GAL4–2.1 vector (titer: 2.9 × 1010 plaque-forming units/ml). YRG-2 yeast competent cells were transformed with 10 μg each of pAS1-CYH2-HSL and the HybriZAP 2.1 adipose cDNA library (23). After heat shock for 5 min, the cells were grown in synthetic minimal medium (SD) without Leu, Trp, and His for 3–4 h and replated on similar SD plates. The plates were incubated at 30°C for 3–4 d. Colonies that grew on the selective plates were regrown in SD media for 2 days and assayed for β-galactosidase activity (23). To eliminate false positives, the colonies were regrown on SD plates without Leu, but with 10 μg/ml cyclohexamide to remove the pAS1-CYH2-HSL plasmid and then reassayed for β-galactosidase activity. Plasmids were isolated from the colonies that showed no β-galactosidase activity under these conditions. The isolated library plasmids were then retransformed along with pAS1-CYH2-HSL cDNA, and the colonies were again assayed for β-galactosidase activity to confirm a positive interaction. Isolated plasmids were sequenced bidirectionally by using an ABI Prism 377 DNA Sequencer (Perkin–Elmer).
In Vitro Protein–Protein Binding.
Full-length rat HSL was cloned into pcDNA3 vector (Invitrogen) and in vitro translated with [35S]methionine by using the TNT transcription/translation system (Promega). Mouse adipocyte lipid-binding protein (ALBP) was inserted into the pGEX vector (Amersham Pharmacia), and glutathione S-transferase (GST) fusion proteins were produced in Escherichia coli (24). Extracts (400 μg) of E. coli transformed with pGEX-ALBP or with pGEX alone were incubated with glutathione-agarose beads in buffer B (20 mM Tris, pH 8.0/0.15M NaCl/1 mM EDTA/0.5% Nonidet P-40). After 1 h incubation at room temperature, the beads were washed three times in buffer B, and then incubated with [35S]methionine-labeled HSL. After 1 h incubation at room temperature, the beads were washed five times in buffer B, and proteins that bound to the beads were eluted in SDS/PAGE sample buffer, separated on SDS/10% PAGE, and visualized by using a PhosphorImager (Molecular Dynamics). In other experiments, extracts of E. coli transformed with pGEX-ALBP or with pGEX alone were incubated with glutathione-agarose beads and washed, as above, and the beads were then incubated either with extracts from Chinese hamster ovary (CHO) cells stably transfected with pCEP4-HSL (25) prepared by brief sonication in buffer A (0.15 M NaCl/3% Triton X-100/0.1% lauryl sarcosyl, pH 7.4/1 unit/ml leupeptin) or with the 10,000 × g supernatant of extracts prepared from epididymal fat pads isolated from male Sprague–Dawley rats that were homogenized in buffer A. Proteins that bound to the beads were then eluted in SDS/PAGE sample buffer, separated on SDS/10% PAGE, immunoblotted with anti-rat HSL fusion protein antibodies, and visualized by using enhanced chemiluminescence (26).
Truncations of HSL.
Truncations of HSL were generated by cutting pcDNA3-HSL with EcoRI to produce HSL 1–625, with BamHI to produce HSL 1–469, and with NheI to produce HSL 1–300. HSL 300–767 was produced by removing the HinD III–NheI fragment from pcDNA3-HSL, followed by religation by using a HinD III–ATG–NheI adaptor (5′ → 3′, AGCTTCCATGCCTGTG; 3′ → 5′, AGGTACGGACACGATC).
Immunoprecipitation and Immunoblotting.
Immunoprecipitation of HSL was performed by using anti-rat HSL fusion protein antibodies as described (26), with some modifications. Epididymal fat pads isolated from male Sprague–Dawley rats were homogenized in buffer A and centrifuged at 10,000 × g for 15 min. The infranatant below the fat and oil cake was used for immunoprecipitation and protein determination. An aliquot (250 μg) was precleared with Protein A beads and then incubated with an immunomatrix consisting of rabbit polyclonal anti-HSL/fusion protein IgG and protein A. The immunomatrix was prepared with minor modifications as described by Doolittle et al. (27). Briefly, 200 μl of protein A beads were washed three times in PBS followed by two washes in PBS with 0.05% BSA. The beads were resuspended in 200 μl of PBS with 0.05% BSA and incubated with 10 μg of anti-rat HSL fusion protein IgG for 90 min at 4°C. After this, the protein A beads were pelleted, washed two times in PBS with 0.05% BSA, washed two times with 0.2 M sodium borate buffer (pH 9.0), and incubated at room temperature for 90 min with 20 mM dimethylpimelimidate⋅2HCl, after readjusting the pH to 9.0 with NaOH. The cross linking reaction was stopped by adding 0.2 M ethanolamine. Uncrosslinked antibodies were removed by suspending the immunomatrix in 0.2 M glycine (pH 2.5), and the immunomatrix was then washed two times in 0.1 M Tris (pH 8.0) and three times in PBS with 0.05% BSA. The adipose tissue extract and freshly prepared immunomatrix (50 μl) were incubated overnight at 4°C. After incubation, the immune complex was centrifuged at 10,000 × g for 15 min and washed two times in PBS with 0.05% BSA and two times in PBS. The pellet was resuspended in 0.063 M Tris⋅HCl (pH 6.8) containing 1% 2-mercaptoethanol, 1% SDS, and 13% (vol/vol) glycerol, boiled for 5 min, electrophoresed on 15% polyacrylamide gels containing 0.1% SDS, transferred to nitrocellulose paper, and immunoblotted with rabbit anti-mouse ALBP IgG (28).
RESULTS AND DISCUSSION
To study the regulation of HSL and to identify protein(s) that interact with HSL, we used the yeast two-hybrid system (29) to screen a rat adipocyte expression library for proteins that would interact with a full-length rat HSL used as bait. Seven positive clones were identified of the 1 × 107 colonies that were screened. When sequenced, one of the clones that demonstrated positive interaction with the HSL bait showed a 93% identity to mouse ALBP (also known as aP2, Fig. 1), a member of the family of intracellular fatty acid-binding proteins. The sequence determined for ALBP from the Sprague–Dawley rat is 97% identical to a sequence from a Norway rat that was recently deposited in GenBank (accession no. 2494405). Differences in sequence were seen at position 47, where an Asp is found in the Norway rat and commonly in other fatty acid-binding proteins. In addition, position 106 was reported to be a Lys in the Norway rat, as opposed to the Arg in the current and other fatty acid-binding proteins; this Arg participates in forming a hydrogen bond with the bound fatty acid carboxylate (30). It is not clear whether these sequence variations are polymorphisms or real differences. Fatty acid-binding proteins are low-molecular-weight, single-chain polypeptides that generally form 1:1 complexes with fatty acids, retinoids, or other hydrophobic ligands (30). As revealed by x-ray crystallography, the lipid-binding proteins fold into β-barrels, forming a large internal water-filled cavity. Ligands, such as fatty acids, are bound within the cavity, occupying about one-third of the accessible volume. The bound fatty acid is stabilized via a combination of enthalpic and entropic forces that govern ligand affinity and selectivity. It has been suggested that one of the roles for ALBP is sequestration of fatty acids within the cytosol, serving as an intracellular buffer for fatty acids, thus protecting cells from the deleterious effects of excess free fatty acids by binding the fatty acid inside the ligand cavity. The sequestered fatty acids may also be shuttled by ALBP within the cytosol, delivering them to specific enzymatic systems. ALBP-null mice fail to express tumor necrosis factor α and fail to develop obesity-linked insulin resistance when maintained on high fat diets (31).
Figure 1.
Comparison of the amino acid sequence of Sprague–Dawley rat ALBP (rALBP) with mouse ALBP (mALBP). The amino acids from rat ALBP that are different from mouse ALBP are underlined.
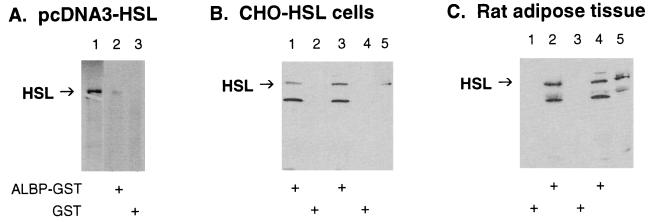
To confirm the interaction of HSL with ALBP, the ability of mouse ALBP–GST fusion protein to interact with HSL derived from three different sources was examined (Fig. 2). Fig. 2A shows results with HSL that was [35S]methionine-labeled by in vitro translation; lane 1 shows the migration of the ≈84-kDa in vitro translation product of pcDNA3-HSL alone. When [35S]methionine-labeled HSL and ALBP–GST were incubated with glutathione-agarose beads and the proteins that bound to the beads were washed, eluted, and separated on SDS/PAGE, labeled HSL was seen (lane 2). However, when GST was substituted for ALBP–GST, no labeled HSL was pulled down by the glutathione-agarose beads (lane 3). Fig. 2B shows the results of an experiment in which extracts from HSL-overexpressing CHO cells and ALBP–GST (lanes 1 and 3) or GST alone (lanes 2 and 4) were incubated with glutathione-agarose beads. When the proteins that bound to the beads were washed, eluted, separated on SDS/PAGE, and then immunoblotted with anti-HSL antibodies, only incubation with ALBP–GST allowed HSL to be pulled down by the glutathione-agarose beads. The identity of the lower molecular mass (≈55 kDa) band recognized by the anti-HSL antibodies is not known. Fig. 2C displays the results of a similar experiment in which extracts of rat adipose tissue and GST alone (lanes 1 and 3) or ALBP–GST (lanes 2 and 4) were incubated with glutathione-agarose beads. When the proteins that bound to the beads were washed, eluted, separated on SDS/PAGE, and then immunoblotted with anti-HSL antibodies, only incubation with ALBP–GST allowed HSL to be pulled down by the glutathione-agarose beads. Therefore, under each of the incubation conditions, ALBP–GST specifically interacted with HSL, whereas GST alone showed no interaction with HSL. Thus, both genetic and biochemical experiments have confirmed the specific interaction of HSL with ALBP.
Figure 2.
Interaction of HSL with ALBP in vitro. (A) Interaction of ALBP–GST with in vitro-translated HSL. pcDNA3–HSL was in vitro translated with [35S]methionine by using the TNT transcription/translation system (lane 1). Extracts (400 μg) of E. coli transformed with pGEX–ALBP (ALBP–GST, lane 2) or with pGEX alone (GST, lane 3) were incubated with glutathione-agarose beads and with [35S]methionine-labeled HSL as described in Materials and Methods. Proteins that bound to the beads were eluted, separated on SDS/10% PAGE, and visualized by using a PhosphorImager. (B) Interaction of ALBP–GST with CHO cells overexpressing HSL. ALBP–GST (lanes 1 and 3) or GST (lanes 2 and 4) was incubated with glutathione beads as above. The beads were then incubated with extracts from CHO cells stably overexpressing HSL. Proteins that bound to the beads were eluted, separated on SDS/10% PAGE, immunoblotted with anti-rat HSL fusion protein antibodies, and visualized by using enhanced chemiluminescence. Lane 5 contains extracts (25 μg) of CHO cells overexpressing HSL that were used in the incubations. (C) Interaction of ALBP–GST with extracts of rat adipose tissue. ALBP–GST (lanes 2 and 4) or GST (lanes 1 and 3) was incubated with glutathione beads as above. The beads were then incubated with extracts prepared from rat epididymal fat pads. Proteins that bound to the beads were eluted, separated on SDS/10% PAGE, immunoblotted with anti-rat HSL fusion protein antibodies, and visualized by using enhanced chemiluminescence. Lane 5 contains extracts (25 μg) of adipose tissue homogenate that were used in the incubations.
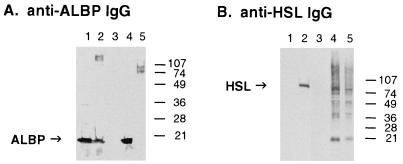
To document the interaction of HSL with ALBP in vivo, anti-HSL antibodies were used to immunoprecipitate HSL from rat adipose tissue and the immune complexes were electrophoresed and immunoblotted with anti-ALBP antibodies to detect the presence of ALBP in the immunoprecipitated HSL complexes. As shown in Fig. 3A, anti-ALBP antibodies recognize purified ALBP and ALBP in extracts of adipose tissue as a 15-kDa protein. Importantly, the 15-kDa ALBP also is detected in adipose tissue immunoprecipitated with anti-HSL antibodies, substantiating the interaction of HSL with ALBP in vivo. In contrast, adipose tissue immunoprecipitated with preimmune antibodies or CHO cells overexpressing HSL that were immunoprecipitated with anti-HSL antibodies do not have any detectable ALBP, establishing that ALBP was not found in the anti-HSL immune complex because of nonspecific interactions during the immunoprecipitation and confirming that CHO cells do not express the adipose specific ALBP. Fig. 3B displays an immunoblot of these same samples by using anti-HSL antibodies that verifies the presence of the 84-kDa HSL in extracts of adipose tissue, in the anti-HSL immunoprecipitated complexes, and in the CHO cells overexpressing HSL.
Figure 3.
Interaction of HSL with ALBP in vivo. Extracts of rat epididymal fat (lane 3) were immunoprecipitated with preimmune antibodies, or extracts of rat epididymal fat (lane 4) or extracts of CHO cells stably overexpressing HSL (lane 5) were immunoprecipitated with anti-HSL antibodies, as described in Materials and Methods. The immunoprecipitated pellets were separated on SDS/15% PAGE, immunoblotted with anti-mouse ALBP antibodies (A) or anti-rat HSL fusion protein antibodies (B), and visualized by using enhanced chemiluminescence. Lane 1 contains purified ALBP (0.8 μg) and lane 2 contains extracts (10 μg) of adipose tissue homogenate before immunoprecipitation.
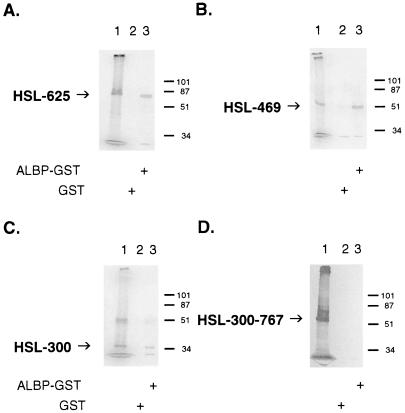
To determine the region within HSL that is important for its interaction with ALBP, we performed a series of deletions of HSL and tested the ability of the [35S]methionine-labeled in vitro translation products of the truncated HSLs to interact with ALBP. As shown in Fig. 4, [35S]methionine-labeled truncations of HSL and either GST or ALBP–GST were incubated with glutathione-agarose beads, and the proteins that bound to the beads were washed, eluted, and separated on SDS/PAGE. In each panel, lane 1 shows the migration of the in vitro translation product alone, lane 2 shows the results with GST, and lane 3 shows the results with ALBP–GST. As the C-terminal portion of HSL is removed from the 767-aa intact protein to HSL 1–625 (Fig. 4A, translation product ≈68 kDa), to HSL 1–469 (Fig. 4B, translation product ≈51 kDa), and to HSL 1–300 (Fig. 4C, translation product ≈33 kDa), the truncated portion of HSL continues to be able to interact specifically with ALBP–GST, whereas no interaction is observed with GST alone. From these data, it appears that the first 300 aa of HSL are responsible for the interaction of HSL with ALBP. To confirm the importance of the initial 300 aa of HSL in interacting with ALBP, a form of HSL was generated that lacked the initial 300 aa, HSL 300–767 (Fig. 4D, translation product ≈51 kDa). This truncated HSL no longer displayed the ability to form an interacting complex with ALBP. These results demonstrate that the region of HSL responsible for interacting with ALBP resides within the first 300 aa of the protein. This region of HSL has no homology with any known protein, nor has any function previously been shown to be associated with this portion of the protein because the catalytic and regulatory regions of the enzyme have been localized within the C-terminal region (8, 12, 13, 32–34). We propose that the N-terminal 300-aa region represents a docking domain for the interaction with ALBP.
Figure 4.
Identification of the region of HSL that interacts with ALBP. Truncations of HSL were produced as described in Materials and Methods. The truncated HSL constructs were in vitro-translated with [35S]methionine and incubated with GST (lane 2) or ALBP–GST (lane 3) and with glutathione beads, as described in Fig. 2. Proteins that bound to the beads were eluted, separated on SDS/10% PAGE, and visualized as described in Fig. 2. (A) HSL 1–625; lane 1, in vitro translation product of pcDNA3–HSL 1–625. (B) HSL 1–469; lane 1, in vitro translation product of pcDNA3–HSL 1–469. (C) HSL 1–300; lane 1, in vitro translation product of pcDNA3–HSL 1–300. (D) HSL 300–767; lane 1, in vitro translation product of pcDNA3–HSL 300–767. Each of the gels presented is representative of three separate experiments.
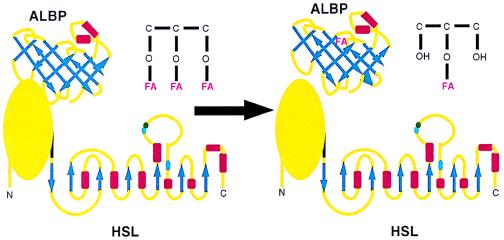
The current observations of a specific interaction between HSL and ALBP provide a basis for a new framework whereby lipolysis is controlled within adipose cells. Based on these findings, it is proposed that HSL and ALBP constitute a lipolytic complex, with ALBP binding to the N-terminal portion of HSL in the basal state (Fig. 5). When lipolytic signals activate HSL, hydrolysis of triacylglycerols is mediated, resulting in the generation of FFA and monoacylglycerol; the close physical association of ALBP with HSL within this complex would enable FFA released to be efficiently and rapidly sequestered by ALBP. This sequestration would prevent the high concentrations of FFA produced locally at the site of hydrolysis from causing any deleterious effects. Moreover, because fatty acids have been reported to inhibit HSL activity (35), this sequestration of FFA by ALBP might potentially prevent feedback inhibition of HSL by excessive concentrations of FFA. We propose a mechanism by which (as depicted in Fig. 5), on binding of released FFA by ALBP, the HSL–ALBP complex dissociates. This would allow the fatty acid sequestered by ALBP to be shuttled out of the cell, whereas HSL-mediated hydrolysis would continue, because an apo-ALBP could now bind to HSL, thus repeating the process.
Figure 5.
Proposed effects of the interaction of HSL and ALBP. The HSL diagram (Lower) depicts the C-terminal portion of HSL as α/β-hydrolase folds based on models of its secondary structure, whereas the N-terminal portion is depicted as a globular protein because its structure does not fit any current predicted models. The ALBP diagram (Upper) depicts a β-barrel. β-sheets are depicted as blue arrows and α-helices as red rectangles. See text for a description of the interaction of HSL and ALBP.
Consistent with the observation that ALBP forms a complex with HSL, mice null for the ALBP allele exhibit diminished basal and isoproterenol-stimulated lipolysis in isolated fat cells (36). In such ALBP-null mice, fatty acids accumulate intracellularly, concomitant with the decrease in lipolysis. These findings strongly suggest that the ALBP–HSL interaction is functional and results in the facilitation of fatty acid efflux from the cell. In the absence of the ALBP–HSL interaction, fatty acids are not efficiently released from the adipocyte and accumulate intracellularly. Overall, the results suggest that fatty acid trafficking during lipolysis is mediated by ALBP and that the complex with HSL is the first step in an organized lipid transfer process. Whether the interaction of HSL with ALBP influences the function of perilipin or other proteins potentially involved in lipolysis is not known, but additional characterization of the interaction of HSL with ALBP will provide further insight into the regulation of intracellular lipid metabolism within adipose cells.
Acknowledgments
This work was supported in part by Grants DK 46942 and DK 49705 from the National Institutes of Health (F.B.K.), by the Research Service of the Department of Veterans Affairs (F.B.K.), and by Grants MCB 9506088 and MCB 9816575 from the National Science Foundation (D.A.B.). K.S. was supported by a postdoctoral training grant (DK 07217) from the National Institutes of Health.
ABBREVIATIONS
- HSL
hormone-sensitive lipase
- FFA
free fatty acids
- ALBP
adipocyte lipid-binding protein
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF144756).
References
- 1.Yeaman S J. Biochim Biophys Acta. 1990;1052:128–132. doi: 10.1016/0167-4889(90)90067-n. [DOI] [PubMed] [Google Scholar]
- 2.Fredrikson G, Stralfors P, Nilsson N O, Belfrage P. J Biol Chem. 1981;256:6311–6320. [PubMed] [Google Scholar]
- 3.Cook K G, Yeaman S J, Stralfors P, Fredrikson G, Belfrage P. Eur J Biochem. 1982;125:245–249. doi: 10.1111/j.1432-1033.1982.tb06675.x. [DOI] [PubMed] [Google Scholar]
- 4.Lee F T, Adams J B, Garton A J, Yeaman S J. Biochim Biophys Acta. 1988;963:258–264. doi: 10.1016/0005-2760(88)90289-5. [DOI] [PubMed] [Google Scholar]
- 5.Wei S, Lai K, Patel S, Piantedosi R, Shen H, Colantuoni V, Kraemer F B, Blaner W S. J Biol Chem. 1997;272:14159–14165. doi: 10.1074/jbc.272.22.14159. [DOI] [PubMed] [Google Scholar]
- 6.Holm C, Kirchgessner T G, Svenson K L, Fredrikson G, Nilsson S, Miller C G, Shively J E, Heinzmann C, Sparkes R S, Mohandas T, et al. Science. 1988;241:1503–1506. doi: 10.1126/science.3420405. [DOI] [PubMed] [Google Scholar]
- 7.Probst M R, Beer M, Beer D, Jenö D, Meter U A, Randolfo G. J Biol Chem. 1994;269:21650–21656. [PubMed] [Google Scholar]
- 8.Hemilä H, Koivula T T, Palva I. Biochim Biophys Acta. 1994;1210:249–253. doi: 10.1016/0005-2760(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 9.Kanaya S, Koyanagi T, Kanaya E. Biochem J. 1998;332:75–80. doi: 10.1042/bj3320075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manco G, Adinolfi E, Pisani F M, Ottolina G, Carrea G, Rossi M. Biochem J. 1998;332:203–212. doi: 10.1042/bj3320203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo D-W, Kurihara T, Suzuki T, Soda K, Esaki N. Appl Environ Microbiol. 1998;64:486–491. doi: 10.1128/aem.64.2.486-491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterlund T, Danielsson B, Degerman E, Contreras J A, Edgren G, Davis R C, Schotz M C, Holm C. Biochem J. 1996;319:411–420. doi: 10.1042/bj3190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contreras J A, Karlsson M, Osterlund T, Laurell H, Svensson A, Holm C. J Biol Chem. 1996;271:31426–31430. doi: 10.1074/jbc.271.49.31426. [DOI] [PubMed] [Google Scholar]
- 14.Egan J J, Greenberg A S, Chang M-K, Wek S A, Moos M C, Jr, Londos C. Proc Natl Acad Sci USA. 1992;89:8537–8541. doi: 10.1073/pnas.89.18.8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie A W, McCormick D K T, Emmison N, Kraemer F B, Alberti K G M M, Yeaman S J. Diabetologia. 1996;39:45–53. doi: 10.1007/BF00400412. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg A S, Egan J J, Wek S A, Garty N B, Blanchette-Mackie E J, Londos C. J Biol Chem. 1991;266:11341–11346. [PubMed] [Google Scholar]
- 17.Greenberg A, Egan J, Wek S, Moos M C, Jr, Londos C, Kimmel A. Proc Natl Acad Sci USA. 1993;90:12035–12039. doi: 10.1073/pnas.90.24.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H P, Serrero G. Proc Natl Acad Sci USA. 1992;89:7856–7860. doi: 10.1073/pnas.89.17.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasaemle D L, Barber T, Wolins N E, Serrero G, Blanchette-Mackie E J, Londos C. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 20.Londos C, Gruia-Gray J, Brasaemle D L, Rondinone C M, Takeda T, Dwyer N K, Barber T, Kimmel A R, Blanchette-Mackie E J. Int J Obesity Relat Metab Disord. 1996;20, Suppl. 3:S97–S101. [PubMed] [Google Scholar]
- 21.Clifford G M, McCormick D K, Vernon R G, Yeaman S J. Biochem Soc Trans. 1997;25:S672. doi: 10.1042/bst025s672. [DOI] [PubMed] [Google Scholar]
- 22.Elledge S J, Mulligan J T, Ramer S W, Spottswood M, Davis R W. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nawaz Z, Baniahmad C, Burris T P, Stillman D J, O’Malley B W, Tsai M J. Mol Gen Genet. 1994;245:724–733. doi: 10.1007/BF00297279. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z H, Buelt M K, Banaszak L J, Bernlohr D A. J Biol Chem. 1991;266:14367–14370. [PubMed] [Google Scholar]
- 25.Kraemer F B, Fong L, Patel S, Komaromy M C. J Lipid Res. 1997;38:1553–1561. [PubMed] [Google Scholar]
- 26.Kraemer F B, Patel S, Saedi M S, Sztalryd C. J Lipid Res. 1993;34:663–671. [PubMed] [Google Scholar]
- 27.Doolittle M H, Ben-Zeev O, Briquet-Laugier V. J Lipid Res. 1998;39:934–942. [PubMed] [Google Scholar]
- 28.Bernlohr D A, Doering T L, Kelly T J, Jr, Lane M D. Biochem Biophys Res Commun. 1985;132:850–855. doi: 10.1016/0006-291x(85)91209-4. [DOI] [PubMed] [Google Scholar]
- 29.Bai C, Elledge S J. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- 30.Bernlohr D A, Simpson M A, Vogel Hertzel A, Banaszak L J. Annu Rev Nutr. 1997;17:277–303. doi: 10.1146/annurev.nutr.17.1.277. [DOI] [PubMed] [Google Scholar]
- 31.Hotamisligil G S, Johnson R S, Distel R J, Ellis R, Papaioannou V E, Spiegelman B M. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 32.Osterlund T, Contreras J A, Holm C. FEBS Lett. 1997;403:259–262. doi: 10.1016/s0014-5793(97)00063-x. [DOI] [PubMed] [Google Scholar]
- 33.Anthonsen M W, Rönnstrandt L, Wernstedt C, Degerman E, Holm C. J Biol Chem. 1998;273:215–221. doi: 10.1074/jbc.273.1.215. [DOI] [PubMed] [Google Scholar]
- 34.Shen W-J, Patel S, Natu V, Kraemer F B. Biochemistry. 1998;37:8973–8979. doi: 10.1021/bi980545u. [DOI] [PubMed] [Google Scholar]
- 35.Jepson C A, Yeaman S J. FEBS Lett. 1992;310:197–200. doi: 10.1016/0014-5793(92)81328-j. [DOI] [PubMed] [Google Scholar]
- 36.Coe, N. R., Simpson, M. A. & Bernlohr, D. A. (1999) J. Lipid Res., in press. [PubMed]