Abstract
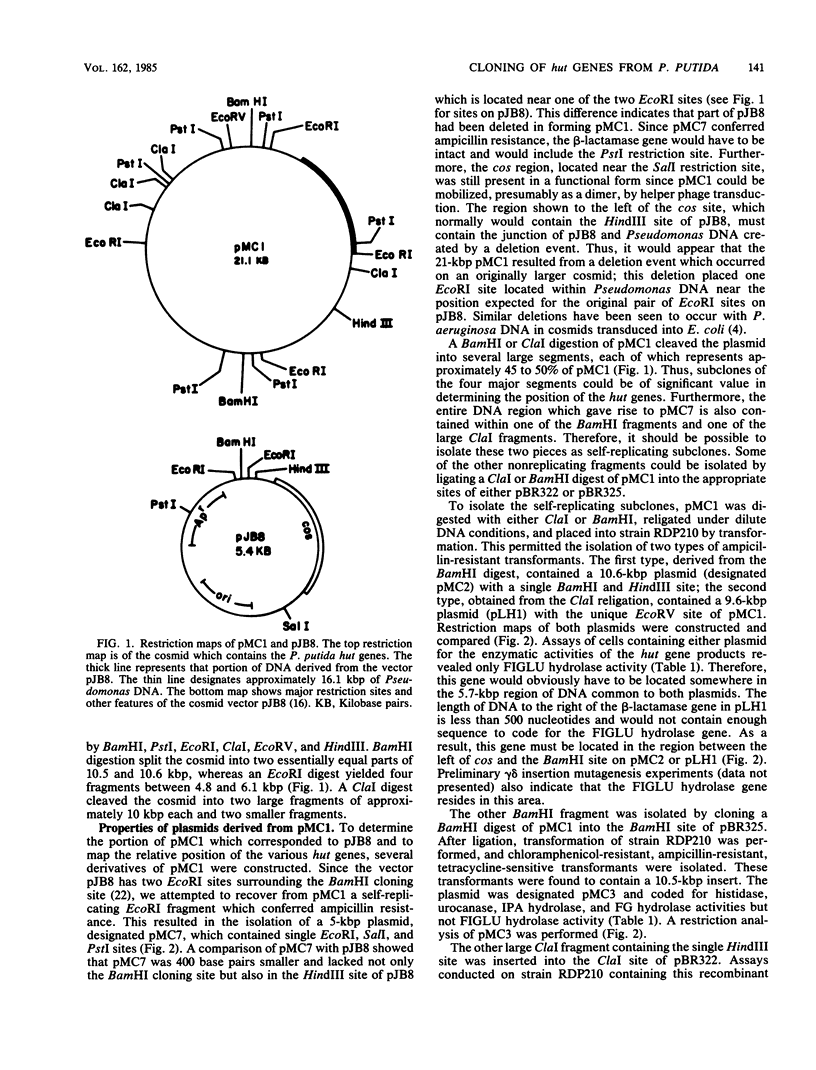
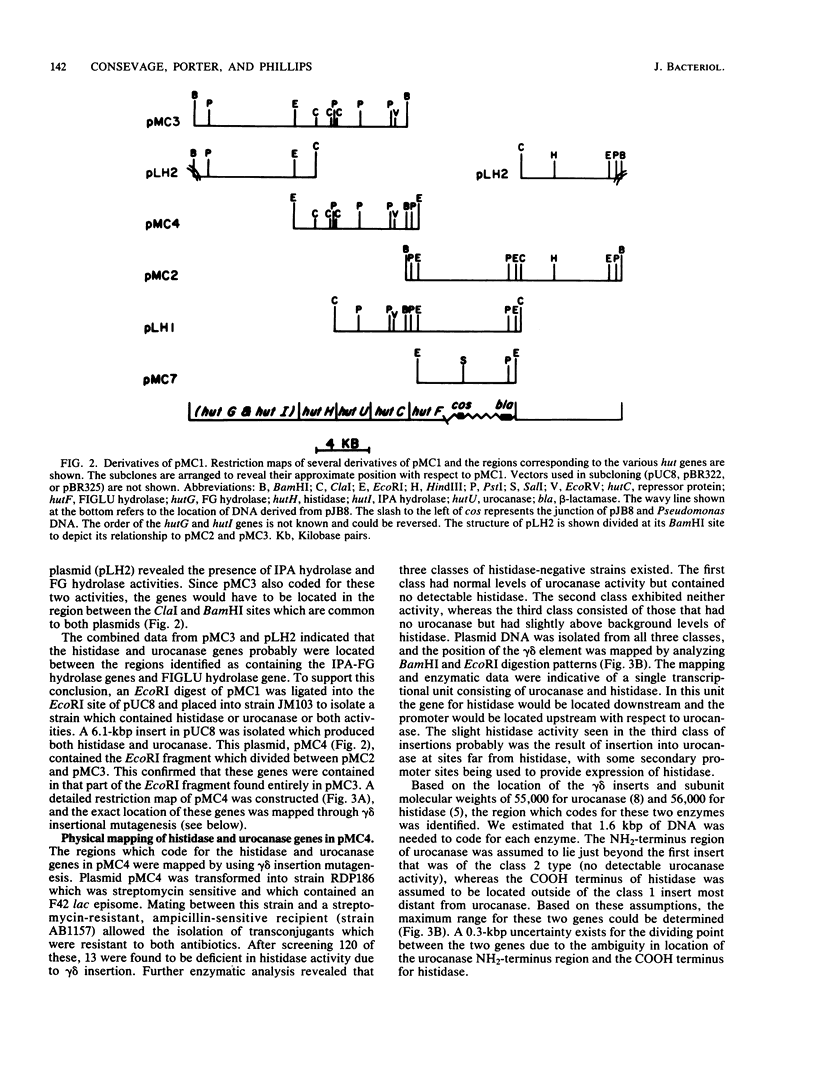
A library of the Pseudomonas putida chromosome, prepared through the use of the cosmid pJB8 ligated to a partial Sau3A digest of bacterial DNA, followed by in vitro packaging into bacteriophage lambda particles, was used to construct a strain of Escherichia coli which contained the genes for histidine utilization. This isolate produced a repressor product and all five enzymes required in Pseudomonas spp. for histidine dissimilation, whereas none of these could be detected in the nontransduced parent E. coli strain. When this transductant was grown on various media containing histidine or urocanate as the inducer, it was observed that production of the cloned histidine degradative enzymes was influenced somewhat by the choice of nitrogen source used but not by the carbon source. The recombinant cosmid was isolated and found to consist of 21.1 kilobase pairs of DNA, with approximately 16 kilobase pairs derived from Pseudomonas DNA and the remainder being from the pJB8 vector. Digestion of this insert DNA with EcoRI provided a 6.1-kilobase-pair fragment which, upon ligation in pUC8 and transformation into an E. coli host, was found to encode histidine ammonia-lyase and urocanase. The inducible nature of this production indicated that the hut repressor gene also was present on this fragment. Insertional inactivation of the histidine ammonia-lyase and urocanase genes by the gamma-delta transposon has permitted location of these structural genes and has provided evidence that transcription proceeds from urocanase through histidine ammonia-lyase. Mapping of the 16-kilobase-pair Pseudomonas DNA segment with restriction enzymes and subcloning of additional portions, one of which contained the gene for formiminoglutamate hydrolase and another that could constitutively express activities for both imidazolone propionate hydrolase and formylglutamate hydrolase, has provided evidence for the organization of all hut genes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boylan S. A., Bender R. A. Genetic and physical maps of Klebsiella aerogenes genes for histidine utilization (hut). Mol Gen Genet. 1984;193(1):99–103. doi: 10.1007/BF00327421. [DOI] [PubMed] [Google Scholar]
- Boylan S. A., Eades L. J., Janssen K. A., Lomax M. I., Bender R. A. A restriction enzyme cleavage map of the histidine utilization (hut) genes of Klebsiella aerogenes and deletions lacking regions of hut DNA. Mol Gen Genet. 1984;193(1):92–98. doi: 10.1007/BF00327420. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Coleman K., Dougan G., Arbuthnott J. P. Cloning, and expression in Escherichia coli K-12, of the chromosomal hemolysin (phospholipase C) determinant of Pseudomonas aeruginosa. J Bacteriol. 1983 Feb;153(2):909–915. doi: 10.1128/jb.153.2.909-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consevage M. W., Phillips A. T. Presence and quantity of dehydroalanine in histidine ammonia-lyase from Pseudomonas putida. Biochemistry. 1985 Jan 15;24(2):301–308. doi: 10.1021/bi00323a010. [DOI] [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The control of the enzymes degrading histidine and related imidazolyl derivates in Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):423–433. doi: 10.1042/bj1320423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L., Sternberg N. In vitro packaging of lambda Dam vectors and their use in cloning DNA fragments. Methods Enzymol. 1979;68:281–298. doi: 10.1016/0076-6879(79)68020-5. [DOI] [PubMed] [Google Scholar]
- George D. J., Phillips A. T. Identification of alpha-ketobutyrate as the prosthetic group of urocanase from Pseudomonas putida. J Biol Chem. 1970 Feb 10;245(3):528–537. [PubMed] [Google Scholar]
- Gerson S. L., Magasanik B. Regulation of the hut operons of Salmonella typhimurium and Klebsiella aerogenes by the heterologous hut repressors. J Bacteriol. 1975 Dec;124(3):1269–1272. doi: 10.1128/jb.124.3.1269-1272.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., Bloom F. R., Magasanik B. Regulation of histidase synthesis in intergeneric hybrids of enteric bacteria. J Bacteriol. 1976 Jul;127(1):114–119. doi: 10.1128/jb.127.1.114-119.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., Magasanik B. Gene order of the histidine utilization (hut) operons in Klebsiella aerogenes. J Bacteriol. 1975 Jun;122(3):1025–1031. doi: 10.1128/jb.122.3.1025-1031.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Magasanik B. Isolation of the self-regulated repressor protein of the Hut operons of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1973 Mar;70(3):808–812. doi: 10.1073/pnas.70.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug D. H., Roth D., Hunter J. Regulation of histidine catabolism by succinate in Pseudomonas putida. J Bacteriol. 1968 Aug;96(2):396–402. doi: 10.1128/jb.96.2.396-402.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen D. B., Herst P. M., Joosten H. M., van der Drift C. Nitrogen control in Pseudomonas aeruginosa: a role for glutamine in the regulations of the synthesis of nadp-dependent glutamate dehydrogenase, urease and histidase. Arch Microbiol. 1981 Feb;128(4):398–402. doi: 10.1007/BF00405920. [DOI] [PubMed] [Google Scholar]
- Kimhi Y., Magasanik B. Genetic basis of histidine degradation in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3545–3548. [PubMed] [Google Scholar]
- Leidigh B. J., Wheelis M. L. Genetic control of the histidine dissimilatory pathway in Pseudomonas putida. Mol Gen Genet. 1973 Feb 2;120(3):201–210. doi: 10.1007/BF00267152. [DOI] [PubMed] [Google Scholar]
- Lessie T. G., Neidhardt F. C. Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol. 1967 Jun;93(6):1800–1810. doi: 10.1128/jb.93.6.1800-1810.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matherly L. H., DeBrosse C. W., Phillips A. T. A covalent nicotinamide adenine dinucleotide intermediate in the urocanase reaction. Biochemistry. 1982 May 25;21(11):2789–2794. doi: 10.1021/bi00540a033. [DOI] [PubMed] [Google Scholar]
- Pahel G., Rothstein D. M., Magasanik B. Complex glnA-glnL-glnG operon of Escherichia coli. J Bacteriol. 1982 Apr;150(1):202–213. doi: 10.1128/jb.150.1.202-213.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada J. L., Magasanik B. Expression of the hut operons of Salmonella typhimurium in Klebsiella aerogenes and in Escherichia coli. J Bacteriol. 1975 Dec;124(3):1263–1268. doi: 10.1128/jb.124.3.1263-1268.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I., Adhya S. Cyclic adenosine 5'-monophosphate in Escherichia coli. Bacteriol Rev. 1976 Sep;40(3):527–551. doi: 10.1128/br.40.3.527-551.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A. T., Mulfinger L. M. Cyclic adenosine 3',5'-monophosphate levels in Pseudomonas putida and Pseudomonas aeruginosa during induction and carbon catabolite repression of histidase synthesis. J Bacteriol. 1981 Mar;145(3):1286–1292. doi: 10.1128/jb.145.3.1286-1292.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prival M. J., Magasanik B. Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J Biol Chem. 1971 Oct 25;246(20):6288–6296. [PubMed] [Google Scholar]
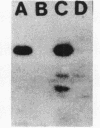
- Renart J., Sandoval I. V. Western blots. Methods Enzymol. 1984;104:455–460. doi: 10.1016/s0076-6879(84)04114-8. [DOI] [PubMed] [Google Scholar]
- Rothman N., Rothstein D., Foor F., Magasanik B. Role of glnA-linked genes in regulation of glutamine synthetase and histidase formation in Klebsiella aerogenes. J Bacteriol. 1982 Apr;150(1):221–230. doi: 10.1128/jb.150.1.221-230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K. Vectors for gene cloning in Pseudomonas and their applications. Curr Top Microbiol Immunol. 1982;96:31–45. doi: 10.1007/978-3-642-68315-2_3. [DOI] [PubMed] [Google Scholar]
- Schlesinger S., Scotto P., Magasanik B. Exogenous and endogenous induction of the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4331–4337. [PubMed] [Google Scholar]
- Smith G. R., Magasanik B. Nature and self-regulated synthesis of the repressor of the hut operons in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1493–1497. doi: 10.1073/pnas.68.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Goldberg R. B. Transduction of chromosomal genes between enteric bacteria by bacteriophage P1. J Bacteriol. 1976 Mar;125(3):1105–1111. doi: 10.1128/jb.125.3.1105-1111.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]