Abstract
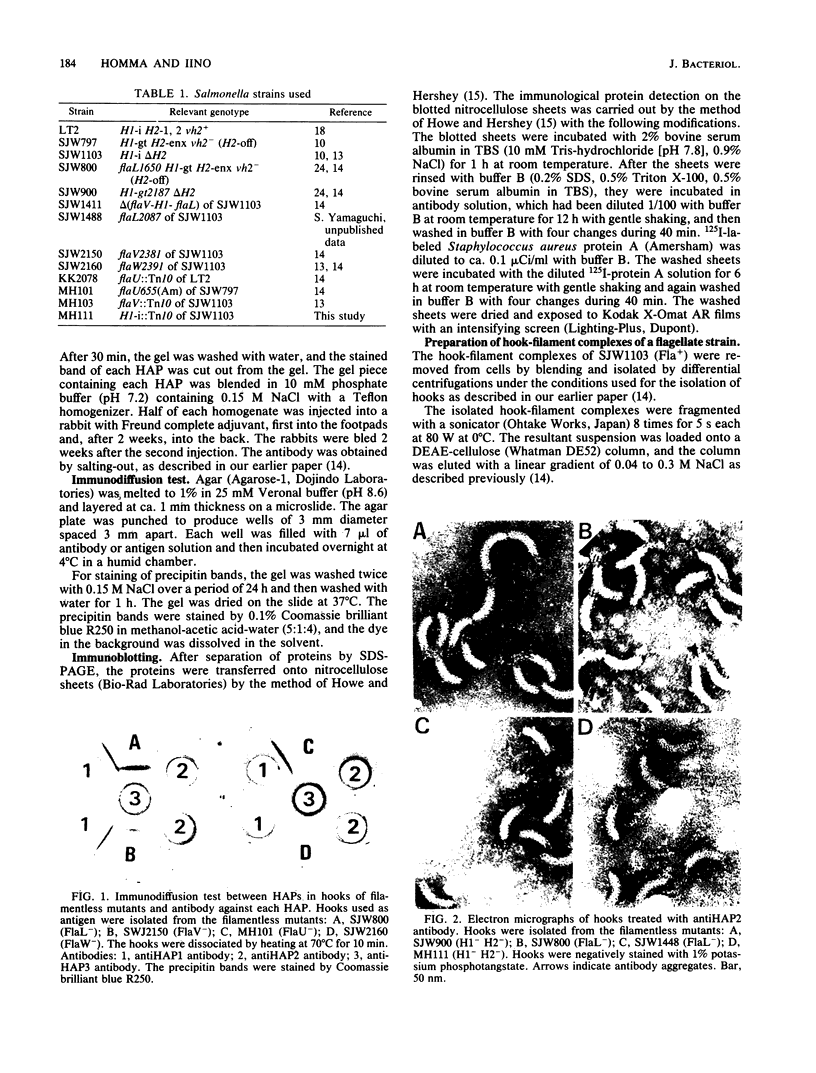
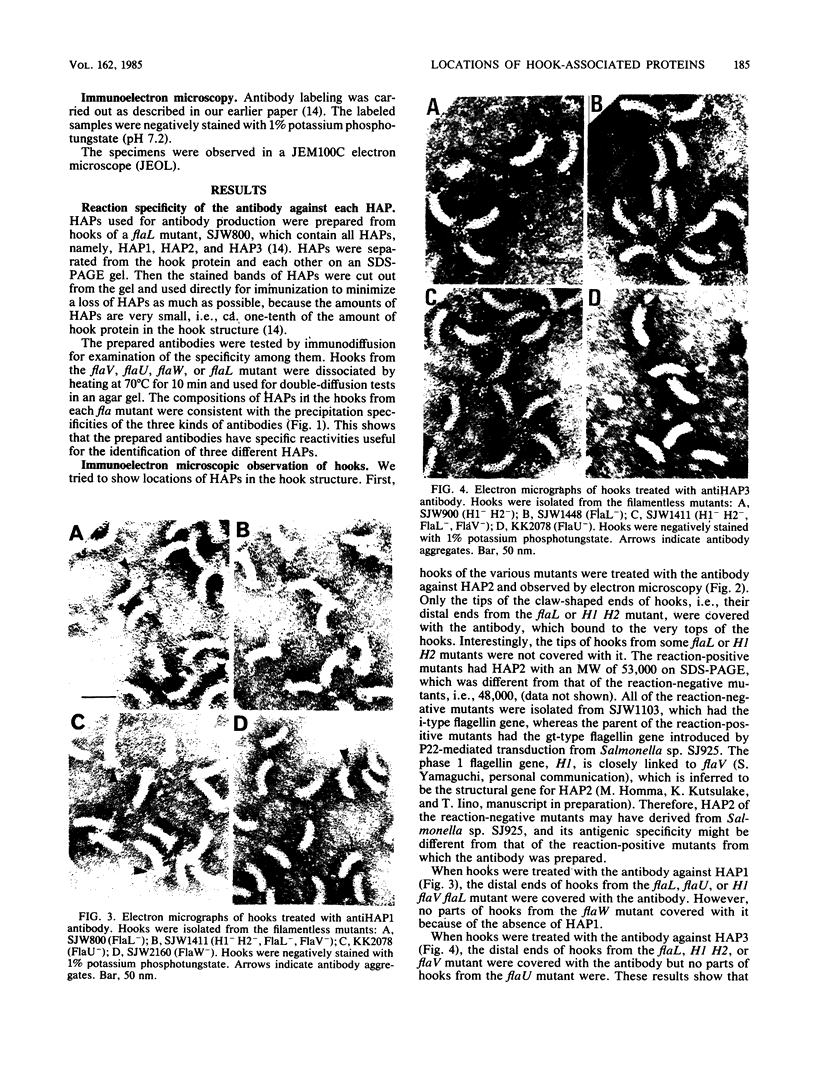
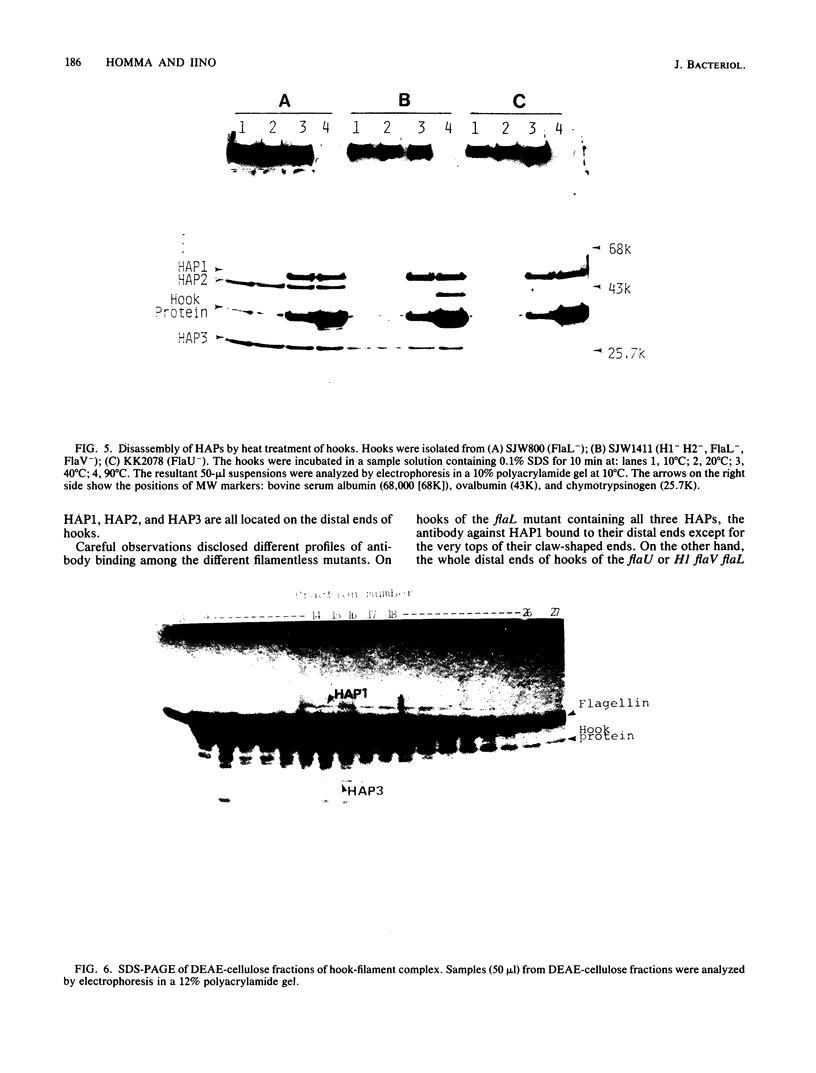
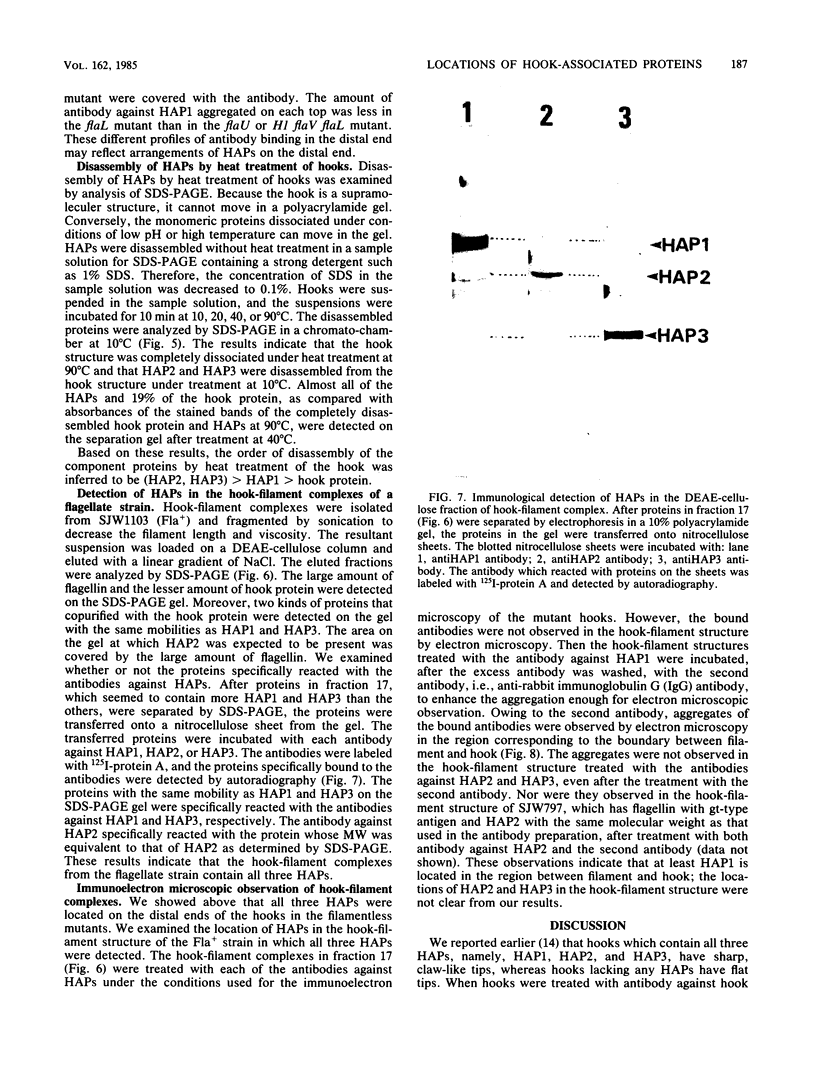
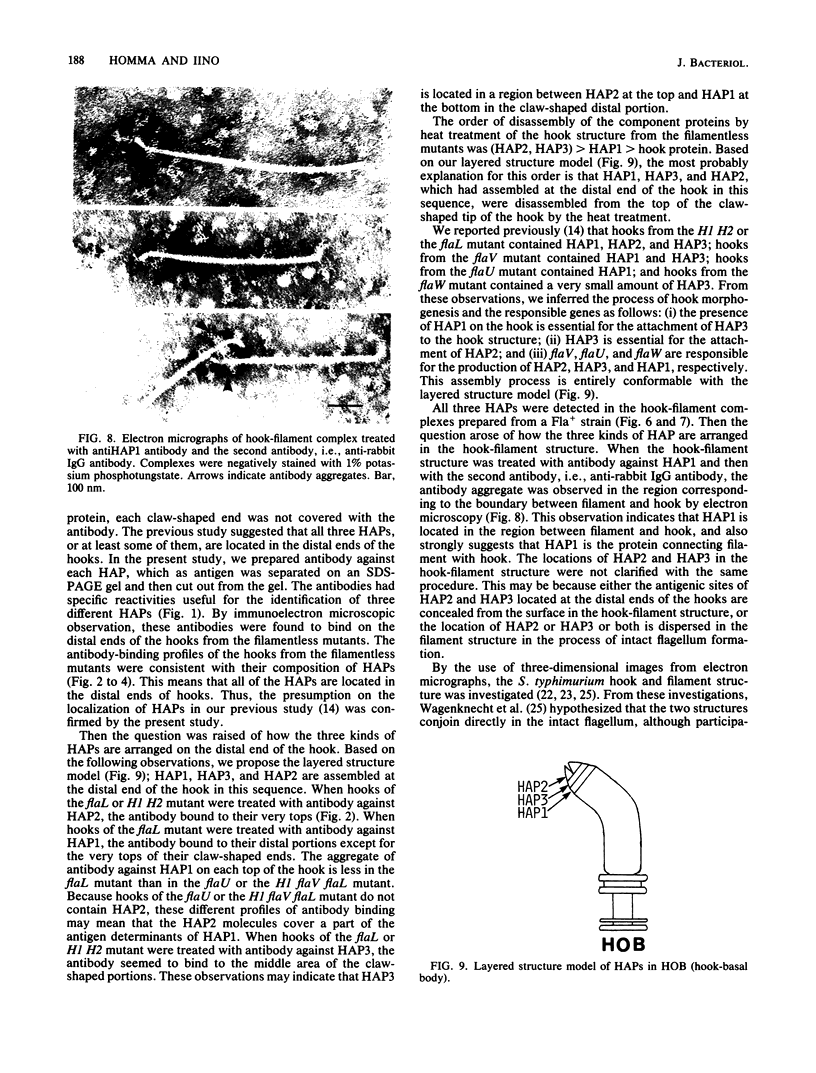
Hooks of the flagella of Salmonella typhimurium were purified from an flaL mutant. Hook-associated proteins, namely HAP1, HAP2, and HAP3, were separated from them, and the antibody against each HAP was prepared. By immunoelectron microscopic observation, these three kinds of antiHAP antibodies were found to bind on the distal ends of hooks of filamentless mutants consistently with their composition of HAPs. The antiHAP2 antibody bound to the very tops of the claw-shaped ends of the hooks which contain all three HAPS. The antibodies against HAP1 and HAP3 bound to the basal areas and the middle areas, respectively, of the claw-shaped ends. The order of disassembly of the component proteins by heat treatment of the hook structure from the filamentless mutants was (HAP2, HAP3) greater than HAP1 greater than hook protein. These observations were consistent with our layered structure model: HAP1, HAP3, and HAP2 are assembled at the distal end of the hook in this sequence. All three HAPs were detected in the hook-filament complexes prepared from a flagellate strain. When the hook-filament structure was treated with antibody against HAP1 and with the anti-rabbit immunoglobulin G antibody, the antibody aggregate was observed in the region corresponding to the boundary between filament and hook. This observation strongly suggests that HAP1 is the protein connecting filament with hook. The locations of HAP2 and HAP3 in the hook-filament structure were not clarified with the same procedure.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abram D., Koffler H., Vatter A. E. Basal structure and attachment of flagella in cells of Proteus vulgaris. J Bacteriol. 1965 Nov;90(5):1337–1354. doi: 10.1128/jb.90.5.1337-1354.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram D., Mitchen J. R., Koffler H., Vatter A. E. Differentiation within the bacterial flagellum and isolation of the proximal hook. J Bacteriol. 1970 Jan;101(1):250–261. doi: 10.1128/jb.101.1.250-261.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abram D., Vatter A. E., Koffler H. Attachment and structural features of flagella of certain bacilli. J Bacteriol. 1966 May;91(5):2045–2068. doi: 10.1128/jb.91.5.2045-2068.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura S. Polymerization of flagellin and polymorphism of flagella. Adv Biophys. 1970;1:99–155. [PubMed] [Google Scholar]
- Cohen-Bazire G., London J. Basal organelles of bacterial flagella. J Bacteriol. 1967 Aug;94(2):458–465. doi: 10.1128/jb.94.2.458-465.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cyoplasmic membrane. J Bacteriol. 1971 Jan;105(1):396–407. doi: 10.1128/jb.105.1.396-407.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):384–395. doi: 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch R. N., Sjoblad R. D. Flagellar structure and function in eubacteria. Annu Rev Microbiol. 1980;34:69–108. doi: 10.1146/annurev.mi.34.100180.000441. [DOI] [PubMed] [Google Scholar]
- Emerson S. U., Tokuyasu K., Simon M. I. Bacterial flagella: polarity of elongation. Science. 1970 Jul 10;169(3941):190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- Fujita H., Yamaguchi S., Taira T., Iino T. A simple method for the isolation of flagellar shape mutants in Salmonella. J Gen Microbiol. 1981 Jul;125(1):213–216. doi: 10.1099/00221287-125-1-213. [DOI] [PubMed] [Google Scholar]
- Hoeniger J. F., Van Iterson W., Van Zanten E. N. Basal bodies of bacterial flagella in Proteus mirabilis. II. Electron microscopy of negatively stained material. J Cell Biol. 1966 Dec;31(3):603–618. doi: 10.1083/jcb.31.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. C., Canale-Parola E. Fine structure of Spirochaeta stenostrepta, a free-living, anaerobic spirochete. J Bacteriol. 1968 Sep;96(3):822–835. doi: 10.1128/jb.96.3.822-835.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Fujita H., Yamaguchi S., Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984 Sep;159(3):1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T., Yamaguchi S. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium. J Bacteriol. 1984 Jan;157(1):100–108. doi: 10.1128/jb.157.1.100-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. A sensitive immunoblotting method for measuring protein synthesis initiation factor levels in lysates of Escherichia coli. J Biol Chem. 1981 Dec 25;256(24):12836–12839. [PubMed] [Google Scholar]
- Iino T. Assembly of Salmonella flagellin in vitro and in vivo. J Supramol Struct. 1974;2(2-4):372–384. doi: 10.1002/jss.400020226. [DOI] [PubMed] [Google Scholar]
- Iino T., Enomoto M. Genetical studies of non-flagellate mutants of Salmonella. J Gen Microbiol. 1966 Jun;43(3):315–327. doi: 10.1099/00221287-43-3-315. [DOI] [PubMed] [Google Scholar]
- Iino T. Polarity of flagellar growth in salmonella. J Gen Microbiol. 1969 May;56(2):227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- Kamiya R., Asakura S., Yamaguchi S. Formation of helical filaments by copolymerization of two types of 'straight' flagellins. Nature. 1980 Aug 7;286(5773):628–630. doi: 10.1038/286628a0. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Iino T., Komeda Y., Yamaguchi S. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1980 Apr;178(1):59–67. doi: 10.1007/BF00267213. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- O'Brien E. J., Bennett P. M. Structure of straight flagella from a mutant Salmonella. J Mol Biol. 1972 Sep 14;70(1):133–152. doi: 10.1016/0022-2836(72)90168-4. [DOI] [PubMed] [Google Scholar]
- Shirakihara Y., Wakabayashi T. Three-dimensional image reconstruction of straight flagella from a mutant Salmonella typhimurium. J Mol Biol. 1979 Jul 5;131(3):485–507. doi: 10.1016/0022-2836(79)90004-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T., DeRosier D. J., Aizawa S., Macnab R. M. Flagellar hook structures of Caulobacter and Salmonella and their relationship to filament structure. J Mol Biol. 1982 Nov 25;162(1):69–87. doi: 10.1016/0022-2836(82)90162-0. [DOI] [PubMed] [Google Scholar]