Abstract
Yeast two-hybrid (Y2H) has been successfully used for genome-wide screens to identify protein–protein interactions for several model organisms. Nonetheless, the logistics of pair-wise screening has resulted in a cumbersome and incomplete application of this method to complex genomes. Here, we develop a modification of Y2H that eliminates the requirement for pair-wise screening. This is accomplished by incorporating lox sequences into Y2H vectors such that cDNAs encoding interacting partners become physically linked in the presence of Cre recombinase in vivo. Once linked, DNA from complex pools of clones can be processed without losing the identity of the interacting partners. Short linked sequence tags from each pair of interacting partner (binary interaction Tags or BI-Tags) are then recovered and sequenced. To validate the approach, comparisons between interactions found using traditional Y2H and the BI-Tag method were made, which demonstrate that the BI-Tag technology accurately represents the complexity of the interaction partners found in the screens. The technology described here sufficiently improves the throughput of the Y2H approach to make feasible the generation of near comprehensive interaction maps for complex organisms.
INTRODUCTION
Knowledge of specific protein–protein interactions is an important component in understanding biological processes and disease mechanisms. The yeast two-hybrid (Y2H) method was developed by Fields and Song (1) to detect protein–protein interactions. Recently, genome-wide data for several model organisms has been generated by high-throughput Y2H screening. Uetz et al. (2) and Ito et al. (3) have both systematically assayed each of the 6000 full length open reading frames (ORFs) from Saccharomyces cerevisiae for interactions with each other and identified 957 and 4549 interactions, respectively. Other large-scale interaction maps include: for Drosophila, 20 000 interactions (4) and for Caenorhabditis elegans, 4000 interactions (5). Two independent human interaction maps were produced by testing of ∼5000 protiens, yielding ∼3200 interactions (6) and testing ∼8000, resulting in 2800 interactions (7). There were two basic strategies for screening used for all of the interaction maps: individual pair-wise testing and library screening. In the first strategy, pair-wise or array testing, a single ‘bait’ test protein is tested with each ‘prey’ test protein individually, i.e. one assay for each pair of proteins to be tested. The second strategy, library screening, tests a single bait-test-protein-containing strain for interactions with a mixed pool of ORF- or cDNA-prey-test-protein-containing strains. Following two-hybrid selection, prey proteins must be identified by sequencing the ORF or cDNA. The common aspect of all current strategies is the use of large arrays, in order to maintain the identity of one or both of the tested proteins; even with current pooling strategies this is limiting. In addition, when using the library screening strategies there is a requirement for sequencing of thousands to hundreds of thousands of cDNAs.
Surprisingly, there is very little overlap between the two-yeast interaction map data sets (3). The most significant reason for the lack of concordance is that the screens were not saturated due to incomplete mating of a single DNA binding domain (DBD) to the activation domain (AD) library and because, in the Uetz et al. screen, a limit was set for the total number of interactions that would be identified for each DBD by sequencing. Consequently, the most important solutions to reduce the number of false negative results are to increase the number of potential interactions screened, including several different clones for each cDNA, and to increase the number of positive colonies analyzed.
To improve the efficiency of the Y2H method, we have developed a modification referred to as binary interaction tag Y2H analysis, or BI-Tag Y2H. In this modification, interacting partner cDNAs are physically linked in those yeast cells selected for a two-hybrid interaction. This physical linkage then maintains the association between cDNAs encoding interacting proteins even when cDNAs from large numbers of clones surviving selection are pooled. These pooled linked DNAs can be handled as a single sample and processed to identify interaction partners by use of short sequence tags (generated by the type IIs restriction enzyme MmeI).
As proof of principle, first, the mouse protein HoxA1 was used as a bait fusion protein and screened for interaction partners from an E12.5 mouse embryo AD fusion protein cDNA library. In a second proof of principle experiment, a library of DBD fusion proteins was screened against a library of AD fusion proteins. This screen demonstrates the utility of BI-Tag Y2H in improving the efficiency of library × library screens.
MATERIALS AND METHODS
Plasmid construction
The Lox71 sequence was added to the plasmid, pC-Act.2 (8), by adding double stranded oligonucleotides to create pC-Act.2lox71. The promoter driving the GAL4 AD coding sequence, lox71 and the transcription terminator were inverted with respect to the rest of the vector at AatII and SacI resulting in pC-Act.2lox71rev. The pGADt7Lox71 was created by cloning the EcoRV and PvuII fragment of pC-Act.2revlox71 into SphI-, BsrGI-digested, blunt-ended pGADt7 (Clontech).
The pCDlox66HoxA1 was cloned full length from pKS-HoxA1 (9), an MmeI site and lox66 sequence were included in the 3′ PCR primer and cloned into AvrII- and PstI-digested pCD.2 (8). The pGBKt7lox66MmeI was constructed by ligating dsDNA containing lox66 and MmeI into pGBKt7 (Clontech) between EcoRI and SalI sites.
The SalI fragment of pCreERt2 (10) containing CreERt2 was cloned into pFA6a-KanMX6 (11) to make pFA6a-KanMX6-CreERt2. The 1600 bp fragment of pGBKt7 (Clontech) containing the 2 μ replication origin was cloned into SacII-digested pFA6a-KanMX6-CreERt2 to create pFA6a-KanMX6-CreERt2-2 μ. The Adc1 promoter was cloned from pCAct.2 as a NotI, ApaI fragment into pFA6a-KanMX6-CreERt2-2 μ. The resulting vector is pFA6a-KanMX6-CreERt2-Adc1-2 μ. Cre was inserted under the control of the Adc1 promoter by gap-repair cloning with KpnI-linearized pFA6a-KanMX6-CreERt2-Adc1-2 μ and a PCR product templated by pBS185 (12). Finally, CreERt2 was removed by ApaI and AscI digestion and religation, resulting in the final construct, pFA6a2 μ-Adc1Cre.
Yeast strains and library construction
A+ RNA was purified from 12.5 dpc C57Bl/6J mouse by the guanidine isothiocyanate (GITC) method and oligo dT-cellulose. The RNA was reverse transcribed with primers (for prey libraries, SMART pCAct: 5′-TGGCCATGGA CCTAGGCAGA TCTGATCAAG GGATCCGGG-3′ and CDS-52 lox71: 5′-GCTGCAGATA ACTTCGTATA ATGTATGCTA TACGAACGGT ATCCAACNNN NN-3′; for bait libraries, SMART pGBK47: 5′-GAGCAGAAGC TGATCTCAGA GGAGGACCTG CATATGGCCA TGGAGGG-3′ and CDS-54 lox66: 5′-GGCTGCAGCA TAACTTCGTA TAGCATACAT TATACGAACG GTATCCAACN NNNN-3′) adapted from the Clontech SMART cDNA synthesis protocol. Primary cDNA was amplified by PCR and cloned by gap-repair cloning with pGADt7Lox71 or pGBKt7lox66 vector and AH109 (Clontech) or YD116 for prey libraries or MATα strain YD119cre for bait libraries. Prey library transformants were selected for on medium lacking leucine. Bait libraries were selected for on synthetic media based on glutamate (referred to as SE), (13) medium lacking tryptophan and containing G418. Negative selection was accomplished during library selection by adding 0.2% 5-FOA to the medium.
HoxA1 expressing bait strain
YD119Cre was transformed by pCDlox66HoxA1 and selected on SE medium containing G418 lacking tryptophan.
Y2H assay
The Y2H library E12.5-AH109-pGADt7lox71MmeI and YD119Cre-HoxA1 were mated, and were selected on SE medium (lacking leucine, tryptophan, adenine and histidine) and containing G418. Two-hybrid positive colonies from library screening were selected on SE medium (lacking leucine, tryptophan and uracil) containing G418.
Retesting
Retesting was performed by generating two PCR amplicons from each pick and recloning them by gap-repair cloning with pGBKt7lox66MmeI and pGADt7lox71 into YD119 and Yd116, respectively. Fusion proteins were tested for auto-activation by assaying for growth on medium lacking tryptophan and uracil or leucine and uracil. To retest interaction partners, corresponding interaction partner strains were mated and tested for URA3 gene expression. To test the ability of a protein to bind the GAL4 protein to activate the GAL4 responsive promoter, bait fusion proteins were mated with a strain carrying pGADt7lox71 and prey fusion proteins were mated with a strain carrying pGBKt7 and assayed for growth on SE medium -Trp, -Leu and -Ura.
BI-Tag Y2H analysis
DNA was purified from the pool of two-hybrid positive colonies as described previously (14). PCR amplification of the linked cDNAs was performed with primers that anneal in the GAL4 DBD and GAL4 AD cDNA. The product was purified by phenol : chloroform : isoamyl alcohol (PCIA) extraction and EtOH precipitation. It was then digested by MmeI New England Biolabs (NEB) and purified by 6% PAGE and the excised band was eluted into TE. Linkers (NotI linker t3: 5′-GCGGGATAGC GTGCCAGCGA GTGACGTTGC GGCCGCNN-3′, NotI linker b3: 5′-GCGGCCGCAA CGTCACTCGC TGGCACGCTA TCCCGC-3′; NotI linker t4: 5′-GGTATAGCCC GGCAGTTGCG CTGACGAGCA GCGGCCGCNN-3′, NotI linker b4: 5′-GCGGCCGCTG CTCGTCAGCG CAACTGCCGG GCTATACC-3′) were ligated to the BI-Tags, then gel purified on PAGE followed by elution into TE. This DNA was used as template for PCR. The resultant 160 bp band was purified by PCIA extraction and EtOH precipitation and digested by NotI, generating a 94-bp band. The 94 bp band was gel purified by PAGE, elution and EtOH precipitated. The pellet was dissolved in 6 μl of H2O, and concatenation was performed in a 10 μl total volume with T4 DNA liga0se (Invitrogen). DNA >500 bp was purified from a 1.5% agarose gel and cloned into NotI-digested pBluescriptKS(+). Inserts were then amplified by PCR and sequenced.
Southern blot
Southern blotting was performed using yeast total DNA prepared as described previously (14) and digested by PstI and HindIII. The probe was a HindIII, EcoRI fragment of pC-Act.2, which contains the GAL4 AD cDNA.
RESULTS
Vector construction and Cre-mediated recombination between AD and DBD Y2H vectors in vivo
To produce Y2H libraries configured for use in the BI-Tag Y2H method, Y2H vectors were modified to contain mutant lox sequences (15) adjacent to the 3′ end of the cDNA insertion site (Figure 1). Lox sites are recognized by the site-specific recombinase, Cre (16). The lox66 and lox71 sites can recombine with each other to form a wild-type loxP site and double mutant lox66/71 site for which Cre has a very low affinity, suppressing recombination at the double mutant site (15). In the present application, the lox66/71 site is between the linked cDNAs.
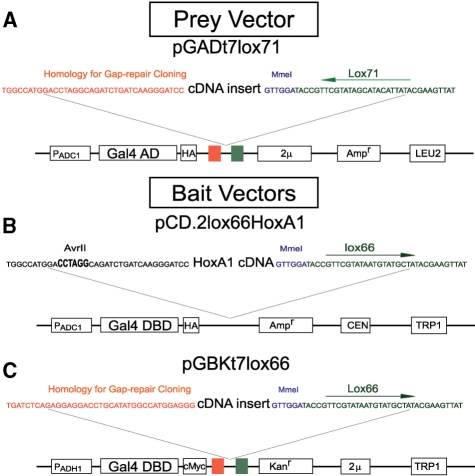
Figure 1.
BI-tag Y2H vectors. The prey vector (panel A) pGADt7lox71 includes an Adc1 promoter, GAL4 AD cDNA, a hemagglutinin epitope, a gap-repair cloning sequence that includes the lox71 sequence, the 2 μ ori, an ampicillin resistance gene for bacterial selection and the LEU2 gene for yeast selection. The figure also shows a cDNA molecule flanked by vector homology, an MmeI RE binding site, and a lox71 site. The bait vector (panel B) pCD.2lox66HoxA1 includes an Adc1 promoter, GAL4 DBD cDNA, a hemagglutinin epitope, the HoxA1 sequence, an ampicillin resistance gene for bacterial selection, a CEN sequence for low copy number replication, and a TRP1 gene for yeast selection. A bait vector pGADt7lox71 for library creation (panel C) includes an ADH1 promoter, GAL4 DBD cDNA, a cMyc epitope, a gap-repair cloning sequence that includes the lox66 sequence, the 2 μ ori, a kanamycin resistance gene for bacterial selection and the TRP1 gene for yeast selection.
Y2H vectors were transformed into yeast in the presence or absence of the Cre expression vector, pFA6a2 μ-Adc1Cre, and assessed for recombination (Figure 2). Recombination occurs between lox71 AD vectors and lox66 DBD vectors in the presence of Cre, while no recombination is detected in its absence. It also should be noted that recombination between lox66 and lox71 creates a loxP site in addition to the lox66/71 site and that the loxP site will recombine with other lox sites forming higher order plasmids. These molecules are likely not stable and were not assayed for in the Southern blot or in downstream applications (e.g. BI-Tag purification).
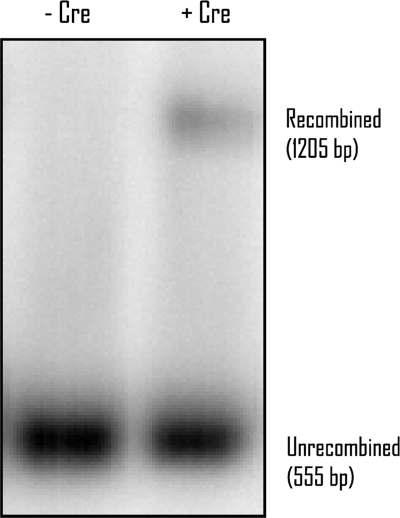
Figure 2.
Cre-mediated recombination in vivo. A Southern blot of HindIII and PstI-digested yeast DNA probed with a GAL4 AD fragment is shown. Lane 1: Bait and prey vectors in yeast strain AH109 in the absence of Cre expression vector (AH109-pCDlox66, pGADt7lox71). Lane 2: Bait and prey vectors in yeast strain AH109 in the presence of Cre expression vector (AH109-pCDlox66, pGADt7lox71, pFA6a2 μ-Adc1Cre).
Library preparation and screening
To generate a library of cDNAs in the BI-Tag activation-domain vector, pGADt7lox71, cDNAs were prepared using poly-A positive RNA isolated from E12.5 day embryos. First strand synthesis was conducted by random priming with a primer containing five random nucleotides at the 3′ end, followed by an MmeI restriction enzyme site and ∼30 nts of vector-homologous sequence. Moloney Murine Leukemia Virus (MMLV) reverse transcriptase was utilized to generate the first-strand cDNA. This enzyme has the property of incorporating several 3′ non-templated C residues following completion of first-strand synthesis. Second-strand cDNA synthesis was then accomplished by SMART technology (Clontech), which takes advantage of these C residues to prime second-strand synthesis using a second-strand primer that contains three G's at its 3′ end. Additionally, the 5′ end of the second-strand primer is homologous to the vector. These steps result in cDNAs with an MmeI site adjacent to the gene-specific DNA sequence and flanked by vector-homologous sequence that can be used for PCR amplification and gap-repair cloning in yeast. The MmeI site is used in subsequent steps to generate 20 bp tags for cDNA identification.
The cDNAs prepared as mentioned earlier were used in gap-repair cloning in AH109 yeast (Clontech) with the pGADt7Lox71 vector to generate a library of 2.1 × 106 total individual transformants and an average insert size of ∼500 bp (called E12.5-AH109-pGADt7lox71MmeI). An insert size of 500 bp will produce multiple fragments from most genes, which is anticipated to be advantageous since it has been shown that random-primed libraries detected valid two-hybrid interactions that were not seen when using full length ORFs (17).
A full length HoxA1 gene was cloned into pCDlox66, a centromere (CEN)-based vector, as a GAL4 DBD fusion protein. CEN-based vectors are carried by yeast in one to three copies per cell, which eliminates the toxicity observed for some fusion proteins when they are expressed at high levels (8). The bait vector, pCDlox66HoxA1 was transformed into the Y2H yeast strain YD119Cre, which carries a plasmid that expresses Cre. This line was mated with the E12.5 library and selected for two-hybrid interactions, resulting in ∼1000 colonies.
Comparison of interaction partners identified by individual clone analysis and BI-Tag methodologies
For comparison with the BI-Tag method, the library of clones selected for HoxA1 interactions was first characterized using standard methods. Eighteen individual Y2H positive fusion proteins were identified using a PCR-based strategy similar to that described previously (2), and BLAST searches of NCBI's nucleotide database (Table 1).
Table 1.
Results Of HoxA1 Y2H screen generated by individual sequences and BI-Tag method
| BI-Tag IDs | No. | Individual IDs | No. |
|---|---|---|---|
| Uhrf1 | 36 | Uhrf1 | 2 |
| Lamc1 | 12 | Lamc1 | 4 |
| Col4a2 | 5 | Col4a2 | 3 |
| Hand2 | 10 | Sema3G | 4 |
| Psmd7 | 9 | Sdhb | 2 |
| eIF3s3 (or similar to eIF3s3) | 7 | Atp2a2 | 1 |
| Mtvr2 | 5 | Cpox | 1 |
| Gnb2 | 4 | Snrp1c | 1 |
| Sh3bp1 | 2 | ||
| Anapc7 | 1 | ||
| Arih2 | 1 | ||
| HnrnpA1a | 1 | ||
| Rprc1 | 1 | ||
| Pfn1 | 1 | ||
| Tubb5 | 1 | ||
| Total | 95 | Total | 18 |
a100% match at other non-gene genomic location.
Comparison of BI-Tag and traditional Y2H analysis, HoxA1 screen: in the first column is a list of interaction partners that were identified by the BI-Tag method. The second column shows the number of times that BI-Tags were sequenced for each cDNA. The third column is the names of the cDNAs that were identified by traditional analysis followed by the number of times that each was identified.
The BI-Tag method was then used (diagrammed in Figure 3). First, DNA, which includes recombined plasmid DNA (Figure 3A and B), was isolated from a pool of the ∼1000-colony HoxA1 interaction library. Amplicons across interacting cDNAs were generated using PCR with GAL4 DBD- and GAL4 AD-specific primers (Figure 3C). This reaction resulted in DNA fragments ranging from 1500 bp and larger in length when assessed on a 1% agarose gel (Figure 4A). Each amplicon includes the HoxA1-DBD fusion cDNA, an MmeI site, the lox66/71 double mutant recombination product, a second MmeI site, and the interacting AD-cDNA fusion. MmeI digestion was then used to excise an ∼86 bp fragment from each amplicon (Figure 3C). These fragments are visible on a PAGE gel (Figure 4B). These DNA fragments contain lox66/71 flanked by MmeI sites and the 19–21 bp BI-Tags used to identify the two interaction partners. Linkers with NotI cleavage sites were ligated to each end and used as primer binding sites in PCR amplification (Figures 3D and 4C, left lane). NotI digestion, which results in ∼94 bp fragments with complementary overhangs for concatenation, are gel purified by 6% PAGE (Figure 4C, lane center lane). Purified DNA was ligated (Figure 3E), and the resulting concatamers >500 bp were purified and cloned into a NotI-digested cloning vector. Figure 4D shows amplicons across concatenated BI-Tags with size distributions between ∼300 and 700 bp. DNAs recovered from the clones were sequenced.
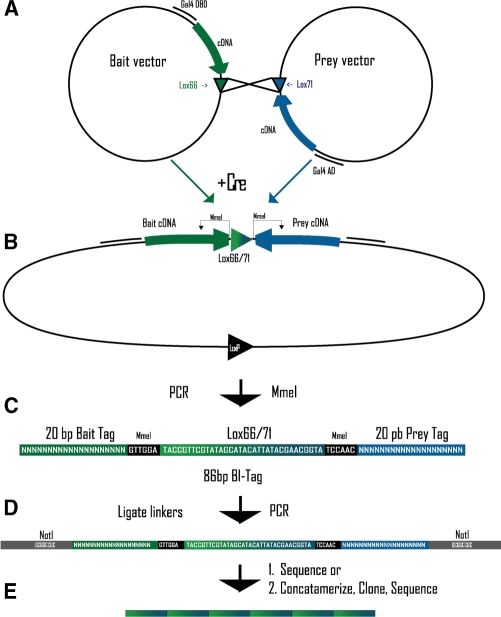
Figure 3.
BI-Tag method diagram. (A) Bait and prey BI-Tag Y2H vectors with lox66 and lox71, respectively. (B) Cre induced recombination at lox sites causes linkage of the BI-Tag Y2H vectors. (C) MmeI digestion generates the BI-Tag-containing lox66/71 flanked by MmeI and 20 bp sequence tags. (D) NotI linkers are ligated to the BI-Tag, and PCR is performed. (E) After NotI digestion the BI-Tags are ligated to form concatamers.
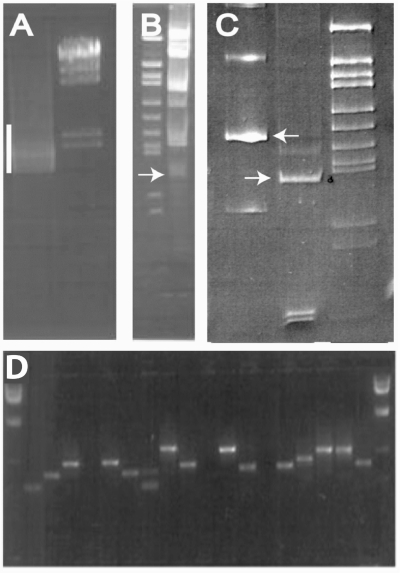
Figure 4.
BI-Tag method. (A) PCR amplification (with primers that anneal to GAL4 AD and DBD cDNAs) across linked cDNAs and lox sequence of the HoxA1 Y2H positive colony DNA (bar). (B) MmeI digestion of the PCR product to produce the 86 bp BI-Tag (arrow). (C) Left lane: 160 bp PCR product that includes the BI-Tag and 40 bp linkers (arrow), Middle lane: 94 bp BI-Tag (arrow) generated by NotI digestion with NotI compatible overhangs for concatenation. (D) Amplicons of BI-Tag concatamer inserts in a cloning vector.
Unexpectedly, we found that in all but one case, all of the BI-Tags in each vector were orientated in the same direction. That is, that the bait (HoxA1) tag was always on the left of the prey tag or vice-versa. BI-Tags were expected to have no preferred orientation within the cloning vector since each one has a NotI site on each end, which allows concatenation. This head to tail orientation could be a result of homologous recombination and/or hairpin formation within the bacteria during the BI-Tag cloning step, which either deletes sequence or causes a selection against these clones. This potential artifact does not affect the present screen but will be a concern for a library by library screen as the recombination could disconnect interaction partners.
BI-Tags were identified from sequence data by BLAST of the NCBI nucleotide database. A total of 95 tags representing 15 different genes were identified. A comparison of putative HoxA1 interacting proteins identified in the BI-Tag analysis described here with results from traditional individual clone analysis is shown in Table 1.
DNA binding domain, fusion protein library construction and screening for interaction partners in an activation domain library
A bait library was constructed similar to the E12.5 prey library described earlier, using pGBKt7lox66MmeI and YD119cre. Additionally, the medium contained 0.2% 5-FOA, which is used to select against the presence of auto-activating bait fusion proteins (8). Previous studies have shown that ∼4–20% of all DBD-cDNA fusion proteins are auto-activating (2,4,6), i.e. are able to activate a GAL4 responsive promoter in the absence of a prey fusion protein. The resultant library, E12.5-YD119cre-pGBKt7lox66MmeI, had 5 × 105 independent transformants.
A prey library was prepared as previously described, except the strain YD116 was used for negative selection against auto-activating prey fusion proteins. This library, E12.5-YD116-pGADt7lox71MmeI, contains 3 × 105 transformants.
The libraries were mated and selected on two-hybrid selection medium (SE—Leu, -Trp, -Ura, 200 μg/ml G418). Thirty of these colonies were picked to a new plate, subjected to standard PCR amplification of cDNA inserts and sequenced for the identification of interaction partners. The result of this analysis is summarized in Table 2. A total of seventeen different proteins were found from 54 cDNAs that were successfully sequenced. Based on analysis of the sequences, we found that a bias was present in the cDNA synthesis step of the SMART library creation protocol that we used, which significantly limited the diversity of bait and prey libraries. The SMART primer failed to prime correctly at the cytosine nucleotides added at the end of first strand by MMLV reverse transcriptase's terminal deoxycytodine transferase activity. Rather, priming occurred at short regions of homology (∼8–15 bp) within cDNA molecules resulting in inclusion of only a subset of cDNAs within the library.
Table 2.
Results of library by library Y2H screen generated by individual sequences and BI-Tag method
| BI-Tag Ids | Individual IDs | ||
|---|---|---|---|
| DBD cDNA | AD cDNA | DBD cDNA | AD cDNA |
| Arid1a | Npc22,1 | Arid1a | Npc22 |
| Arid1a | Pcbp313,11 | Arid1a | Pcbp33 |
| Sf1 | Ttll125,2 | Sf1 | Ttll122 |
| Sf1 | Pcbp3 | Sf1 | Pcbp3 |
| Sf1 | Dpysl22,1 | Sf1 | Dpysl2 |
| Sf1 | Tubb5 | Sf1 | Tubb5 |
| 3100004P22Rik | Pcbp4 | Sf1 | Falz2 |
| 4921511K06Rik | Mast2 | Aprt | Npc2 |
| Arid1a | Bnc2 | Arid1a | Khsrp |
| Arid1a | D530005L17Rik4,3 | Arid1a | Ttll12 |
| Arid1a | Itsn2 | Arid1a | Prmt7 |
| Arid1a | Numa1 | Arid1a | Ucp2 |
| Arid1a | Pcbp43,1 | Arid1a | Dpys12 |
| Arid1a | Pfn13,3 | Cugbp1 | Pcbp3 |
| Arid1a | Tubb510,5 | Hmmr | Npc2 |
| Arid1a | Tubgcp2 | Rai17 | Npc2 |
| Arid1a | Ubl4 | Rai17 | Tln1 |
| BC021381 | Cfl1 | Sf1 | fblimp1 |
| Gm1302 | Zfp2192,1 | ||
| H2afv | Anxa66,1 | ||
| Mrpl17 | Pfn1 | ||
| Myst4 | Pcbp3 | ||
| Ncoa2 | Atn1 | ||
| Palm | Fkbp8 | ||
| Plagl1 | Pcbp3 | ||
| Psmd8 | Ap2m1 | ||
| Rai17 | Ttll122,2 | ||
| Sf1 | Hnrpab4,1 | ||
| Sf1 | Npc2 | ||
| Sf1 | Numa1 | ||
| Sf1 | Pcbp4 | ||
| Sf1 | Upc2 | ||
| Ss18 | Col1a1 | ||
| Ss18 | D530005L17Rik2,1 | ||
| Ss18 | Pcbp32,1 | ||
| Ss18 | Scarb1 | ||
| Ss18 | Tubb52,1 | ||
| Vim | Khsrp | ||
Comparison of BI-Tag and traditional Y2H analysis, library by library screen: The first two columns show a list of interaction partners that were identified by the BI-Tag method. The third and fourth columns show the names of the cDNAs that were identified by traditional analysis. Interactions found in both screens are indicated in bold font. The numbers in superscript are the number of times a protein pair was found, the 1st number is total times, and 2nd number is pairs with unique junctions (ensuring that it is from a unique clone).
All of the colonies were collected, pooled and processed as previously described for BI-Tag identification with the exception that BI-Tags were not concatenated. The results of this analysis are summarized in Table 2. From a total of 83 BI-Tags that were sequenced and contained one bait cDNA and one prey cDNA, each in the correct orientation, we found a total of 61 unique cDNA pairs which can be collapsed into 39 protein pairs.
In order to further characterize this library of Y2H positive interaction partners, we have subjected the 30 picks that have been identified by individual sequencing to several different tests. These picks include protein pairs, which were also identified by the BI-Tag method. They were all retested for two-hybrid interactions (in the absence of Cre), tested for auto-activation, and tested for their ability to bind to the GAL4 protein. Of the 10 protein pairs that repeated in the retest experiments, six were also found in the BI-Tag data (Sf1 : Ttll12 twice in retest data, Sf1 : Dpysl2, Arid1a : Pcbp3, Sf1 : Pcbp3 and Sf1 : Tubb5). Also, in the retest data there were two protein pairs that retested positively but the cDNAs were unable to be identified based on individual sequence reads because the PCR product was a doublet. Two retested protein pairs were not found in the BI-tag set (Cugbp1 : Pcbp3 and Rai17 : Tln1). Of the protein pairs that did not pass the retest, some failed to activate the two-hybrid promoter and other fusion proteins were able to activate the reporter in the absence of an interaction partner (by autoactivation or by binding the complementing Gal4 fragment). It should be noted that the Arid1a : Pcbp3 interaction retested positive only one time and failed two other times to retest, which may indicate that one or both proteins may be somewhat promiscuous. Other high-throughput Y2H screening projects have reported that 55% (7), and about 20% (2) of first round two-hybrid positive protein pairs were reproducible. In all, we found that 10 of 23 interactions (43%) retested successfully and many of these protein pairs were also found in the BI-Tag data set.
In high throughput Y2H interaction testing, multiple positive results with the same protein pair increases the likelihood that that protein pair represents a true interaction and is usually used as a criteria for confidence scoring of data (2–4, and others). Protein pairs from BI-Tag Y2H are not easily recovered for retesting but they can occur multiple times in a data set and this criterion can be used as a surrogate for a retest. There were 14 protein pairs identified by the BI-Tag method multiple times and three of these were retested. Sf1 : Ttll12 and Sf1 : Dpysl2 had positive retests and Arid1a : Pcbp3 had a positive retest one of three times. Taken together, two of three protein pairs that were identified multiple times by BI-Tag Y2H were confirmed by retesting. This supports the notion that protein pairs found multiple times by BI-Tag Y2H are of higher confidence.
As shown in Table 2, one-third of the individually identified pairs were also identified by the BI-Tag method, including five protein pairs that were shown to retest successfully. Additionally, all but one of the individually identified protein pairs that were identified multiple times were also identified by BI-Tag Y2H and the one case in which the protein pair was not represented (Sf1/Falz) could be explained by the occurrence of a mutation in the MmeI site.
DISCUSSION
Defining protein–protein interactions is essential for understanding the functions of both individual proteins and larger regulatory networks. Several large-scale Y2H projects to define interactions across the proteomes of model organisms, including Helicobacter pylori (18), S. cerevisiae (2,3) C. elegans (5) and Drosophila melanogaster (4), and humans (6,7) have been conducted. Although the two human screens are the two largest array-based Y2H screens, together they cover only a fraction of the potential human proteins due to the large complexity of the human proteome. Rual et al. (7) suggest that by using 8100 bait and prey fusion proteins, they have assayed 10% of the total protein pairs based on the most minimal estimate (one protein per gene). Despite the enormous scale of all of these studies, there is a significant lack of concordance between data sets when different studies (even those conducted for the same organism) are compared. In part, this is considered to be a consequence of the low coverage of interaction data sets, since current methods are capable of assaying only a fraction of the potential interactions (3).
In the present study, a method for improving the efficiency of Y2H interaction screening using Cre-mediated recombination to physically link, within the yeast cell, cDNAs encoding interacting bait and prey proteins was developed and tested using mouse HoxA1 as the bait protein for interaction partners in a library and by performing a library by library screen with mouse proteins.
Efficiency of BI-Tag Y2H in defining Hoxa1 interacting proteins
The BI-Tag screen conducted in the present study demonstrates that physical linkage between interacting bait and prey cDNAs can be accomplished using Cre-mediated recombination. Additionally, sequence tags of 19–21 nucleotides generated using MmeI as shown in Figure 4 were sufficient to identify 95 of the 97 tags that localized within genes to either a unique gene (14 cases) or one of two closely related family members (two cases). Comparison of the spectrum of prey molecules identified by individual sequencing from selected clones with those identified from BI-Tags showed that many of the more abundant clones (Uhrf1, Lamc1 and Col4a2-b) were identified in both data sets. However, a number of cDNAs were identified only in the BI-Tag (13) or individual clone (6) data sets, suggesting that each method may incorporate steps that result in a bias to the clones that are represented. By concatenating BI-Tags, we were able to clone and sequence an average of five BI-Tags per sequencing run, resulting in a 5-fold reduction in sequencing requirements when one bait is used and prey proteins are identified. However, the unexpected head to tail orientation of all of the BI-Tags in these studies suggests the possibility that recombination is occurring within concatamers during amplification in bacteria. While this would not affect the identification of interacting partners when single bait is utilized, as is the case here, in a library × library screen recombination between BI-Tags would destroy the association between interacting partners.
The most prominent interaction partner identified in this screen (in both BI-Tag and individual clone data sets) is Uhrf1 (also referred to as Np95, ICBP90). Uhrf1 as well as several other potential interaction partners identified in the HoxA1 screen have been shown to be involved in the ubiquitin–proteasome pathways (Arih2, Psmd7, Anapc7). The ubiquitin–proteasome system has been shown to regulate transcription by several different mechanisms involving histone ubiquitination, transcription factor degradation or transcription activation by the proteasome in a ubiquitin-dependant manner [for a review see (19)]. Uhrf1, Arih1 and the APC/C (of which Anapc7 is a component) have E3 ubiquitin ligase activity and Uhrf1 has been shown to ubiquitinate histones (20). Some of the other interactions are consistent with known functions of the Hox family of proteins and suggest potential regulatory mechanisms. For example, Hand2 is structurally related to the Twist gene, which has been shown to interact with a domain on HoxA5 (21). As with previous Y2H studies, several of the other interactions detected appear unlikely to be biologically relevant and all putative interactions discovered in such studies require validation by alternative methods.
Application of the BI-Tag Y2H technology to high-throughput protein–protein interaction screening
Here we describe a proof of principle library by library screen of mouse proteins for protein-protein interactions where, based on the retest fidelity, our data is of comparable quality to that of other high-throughput Y2H studies (2). The sample of BI-Tags that were sequenced have a similar level of redundancy to the individually identified colonies, suggesting that the BI-Tag procedure is able to accurately represent the members of the original two-hybrid positive library. Furthermore, one-third of the individually identified pairs were also identified by BI-Tags. Five out of six of the protein pairs that were individually identified multiple times were also identified by BI-Tag Y2H. Based on these data, we find no obvious bias in the representation of cDNAs identified by the BI-Tag method relative to the interactions defined by traditional YTH methods, and suggest that application of the BI-Tag method to complex libraries is likely to be efficient.
We anticipate that the BI-Tag Y2H technology described here can, in conjunction with high-throughput (HTP) parallel sequencing technologies, provide the means to assay a number of interactions sufficient to allow the generation of near comprehensive interaction maps for one or even many mammalian tissues. Although cloning artifacts unrelated to the BI-Tag technology limited the complexity of the DBD cDNA library used here, generation of bait and prey libraries of a size sufficient to represent near to the total complexity of a cDNA from any given tissue (∼100 000 independent transformants from a normalized cDNA pool) is possible. In addition, methodologies allowing the generation of a sufficient number of yeast zygotes (1 × 1010) to assay all of the potential interactions between bait and prey libraries of this size by mating have been described (22). However, a screen of this size could result in as many as 300 000 interactions which, in a standard Y2H approach, would need to be defined by individual sequencing reactions. Advances in DNA sequencing technology now allow the parallel generation of ∼250 000 unique 100-nucleotide-long sequence reads within a few hours using a single instrument (23), providing the capacity to efficiently generate the requisite amount of sequence data. However, using the standard YTH method associations between interacting bait and prey cDNAs would be lost in this format. The contribution that the BI-Tag technology described here can make is to allow the associations between individual interacting bait and prey sequences to be maintained such that one interaction is represented in each 100-nucleotide long sequence read. Specifically, direct parallel sequencing of the ∼86 bp MmeI cleavage products from the linked interacting bait-prey cDNA sequences (as shown schematically in Figure 3C and in Figure 4B and omitting the concatenation step), each containing the lox66/71 sequence flanked by MmeI sites and 19–21 bp tags that identify interaction partners could identify on the order of 250 000 protein interaction partners per sequencing run. The number of interactions identified from even a few such sequencing runs could, theoretically, allow redundant coverage of all of the potential interactions occurring between the proteins encoded by cDNA derived from a mammalian tissue. From this information, a near comprehensive protein interaction map could be established. Further, confidence levels for specific interactions could be estimated based on repeated representation of specific protein pairs.
ACKNOWLEDGEMENTS
The authors are grateful to Melanie Ruszczyk and Amy Freeland for technical assistance and Drs Lawrence Mielnicki, Kimberly Bailey and Joel Huberman for helpful suggestions to the article. This work was supported by a grant from the NIH (R21GM068856) to S.C.P. and a Cancer Center Support Grant (P30CA016056). Funding to pay the Open Access publication charges to this article was provided by a grant from the Roswell Park Alliance Foundation.
Conflict of interest statement. S.C.P. is an owner of and an officer in the Buffalo Molecular Target Laboratories LLC.
REFERENCES
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 8.Durfee T, Draper O, Zupan J, Conklin DS, Zambryski PC. New tools for protein linkage mapping and general two-hybrid screening. Yeast. 1999;15:1761–1768. doi: 10.1002/(SICI)1097-0061(199912)15:16<1761::AID-YEA494>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Pruitt SC, Bussman A, Maslov AY, Natoli TA, Heinaman R. Hox/Pbx and Brn binding sites mediate Pax3 expression in vitro and in vivo. Gene Expr. Patterns. 2004;4:671–685. doi: 10.1016/j.modgep.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 11.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 12.Sauer B, Henderson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 13.Cheng TH, Chang CR, Joy P, Yablok S, Gartenberg MR. Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res. 2000;28:e108. doi: 10.1093/nar/28.24.e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 15.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 16.Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 17.Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 18.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 19.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 20.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P, Jr, Raman V. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J. Biol. Chem. 2005;280:2294–2299. doi: 10.1074/jbc.M411018200. [DOI] [PubMed] [Google Scholar]
- 22.Soellick TR, Uhrig JF. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-research0052. RESEARCH0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]