Abstract
Axis formation in Drosophila depends on correct patterning of the follicular epithelium and on signaling between the germ line and soma during oogenesis. We describe a method for identifying genes expressed in the follicle cells with potential roles in axis formation. Follicle cells are purified from whole ovaries by enzymatic digestion, filtration, and fluorescence-activated cell sorting (FACS). Two strategies are used to obtain complementary cell groups. In the first strategy, spatially restricted subpopulations are marked for FACS selection using a green fluorescent protein (GFP) reporter. In the second, cells are purified from animals mutant for the epidermal growth factor receptor ligand gurken (grk) and from their wild-type siblings. cDNA from these samples of spatially restricted or genetically mutant follicle cells is used in differential expression screens employing PCR-based differential display or hybridization to a cDNA microarray. Positives are confirmed by in situ hybridization to whole mounts. These methods are found to be capable of identifying both spatially restricted and grk-dependent transcripts. Results from our pilot screens include (i) the identification of a homologue of the immunophilin FKBP-12 with dorsal anterior expression in egg chambers, (ii) the discovery that the ecdysone-inducible nuclear hormone receptor gene E78 is regulated by grk during oogenesis and is required for proper dorsal appendage formation, and (iii) the identification of a Drosophila homologue of the human SET-binding factor gene SBF1 with elevated transcription in grk mutant egg chambers.
Recent years have seen an explosion in tools for analyzing differential gene expression. The development of a PCR-based differential display method by Liang et al. (1, 2) has quickly been followed by array-based methods for monitoring the expression of thousands of defined genes simultaneously (3, 4). Developmental biology, a field whose progress depends heavily on understanding regulated gene expression, stands to benefit from these methods. However, there are technical hurdles to be overcome before molecular screening methods can be widely applied to problems in development. Chief among these hurdles is purification of the tissue of interest from the complex mixture of cell types typically present in a developing system. Whereas some tissues are amenable to microdissection, others will require more sophisticated techniques such as fluorescence-activated cell sorting (FACS) of dissociated tissue (5–7). The molecular screens reported herein rely on a FACS-based approach to purify specialized follicle cells from Drosophila ovaries. The system we have chosen is particularly illustrative of the need for tissue purification: the Drosophila ovary is composed of egg chambers in which a cyst of large germ-line cells is surrounded by a thin epithelium of highly differentiated follicle cells. Patterning of subregions of the follicular epithelium involves intermingled cell groups, each comprising only a tiny fraction of the volume (and mRNA content) of the ovary. Isolation of follicle cell subgroups allows us to pursue direct molecular screening methods that complement genetic methods for studying follicle cell patterning and should provide insights into the key biological process of axis formation.
MATERIALS AND METHODS
Stocks.
Drosophila melanogaster stocks were raised on standard cornmeal/yeast/agar medium at 25°C. 55B (8) and A62 (9) both have insertions of the yeast GAL4 gene on the third chromosome. The green fluorescent protein (GFP) with a Ser-65 → Thr alteration under the control of the GAL4 upstream activating sequence (UAS-GFPS65T) was deposited in the Drosophila stock center by B. Dickson and is described in FlyBase (http://flybase.harvard.edu:7081). The following stocks were used: grk2B6/Cyo, grkDC9/Cyo (10); cn; In(3L)8h, pur2 ry506; Dd(3L)ME915/TM3, ryRK; Df(3L)ME5345 e/TM3, ryRK; w; P[hs-E78B-11]/CyO; w; P[hs-E78–13]/TM2 and w; P[hs-E78B-92] (gifts from Carl Thummel, Univ. of Utah, Salt Lake City). Heat shocks and egg collection were done as before (11).
Preparation of Follicle Cells.
Ovaries were dissected in S2 medium [Schneider’s insect medium supplemented with 10% fetal calf serum, 50 units/ml penicillin, 50 mg/ml streptomycin, and 0.25 mg/ml Fungizone (Sigma)]. Ovaries (75–150 pairs) were washed three times in calcium-free phosphate-buffered saline (PBS) and incubated at room temperature for 15 min with intermittent vigorous shaking in 0.7 ml of 0.5% trypsin in PBS. Supernatant was removed, filtered through a 40 μm nylon mesh into tubes containing 1 ml of S2 medium, pelleted at 1000 × g for 7 min in a Microcentrifuge, and washed in 1 ml of S2. Trypsin incubation and all subsequent steps were then repeated twice more with the remaining undissociated tissue. Cells were resuspended in 0.3 ml of S2 medium containing 2 μg/ml Hoechst 33342 and 10 μg/ml propidium iodide, incubated for 30 min at room temperature to allow dyes to equilibrate, and stored on ice.
Flow Cytometry.
FACS analysis and cell sorting were performed on the MoFlo high-speed molecular flow cytometer constructed by G.v.d.E. and co-workers. A laser tuned to 360 nm (50 mW) was used to excite Hoechst 33342 and propidium iodide. Hoechst signal was detected by collecting emitted light reflected off a 610-nm dichroic mirror through a 430-nm band-pass filter; the light passing the dichroic mirror was measured to determine propidium iodide fluorescence. A second laser was tuned to 488 nm (135 mW) to excite GFPS65T, whose fluorescence was detected through a 488-nm rejection band filter in combination with a 520-nm band-pass filter. Cells were sorted into 0.5 ml of S2 medium, spun at 1,000 × g for 7 min, and inspected and/or snap frozen in liquid nitrogen for subsequent RNA preparation.
Preparation of Amplified cDNA.
Poly(A)+ RNA was isolated from 1,000,000 A62− and 150,000 A62+ cells by using the PolyATract System 1000 (Promega), following the manufacturer’s recommendations for an extraction volume of 80 μl. RNA was resuspended in 75 μl (A62−) or 35 μl (A62+) of distilled H2O. Then 28.75 μl of A62− and 22.75 μl of A62+ RNA were each reverse transcribed in a 50 μl reaction volume containing 1× First Strand Buffer (BRL), 0.01 M DTT, 0.5 mM dNTPs, 0.2 mg/ml acrylamide (Ambion), 0.05 mg/ml of the primer TP1 (sequence: CGAAACGACGGCCAGTGAATTGTAATACGACTCACTATAGGCGCTTTTTTTTTTTTTTTTTTTTTTTT), and 400 units of SuperScript II (BRL), incubated for 90 min at 42°C. Second-strand cDNA synthesis was performed in a 250-μl reaction volume using 1× Second Strand Buffer (BRL), 0.2 mM dNTPs, 17 units of Escherichia coli DNA ligase (NEB), 67 units of E. coli DNA polymerase I (NEB), 3.4 units of RNase H (Promega), and 50 μl of the first-strand reaction products. The reaction mixtures were then extracted twice with phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and resuspended in 25 μl of distilled H2O. A 6.9-μl sample of cDNA was amplified by PCR by using the procedure of Brady et al. (12), as modified by Dulac and Axel (13), with the following further modifications: double-stranded cDNA was used in place of the first-strand reaction mixture, the primer TP1 was used for PCR amplification, and 0.2 mM dATP was used in the polyadenylation reaction.
Slot Blots.
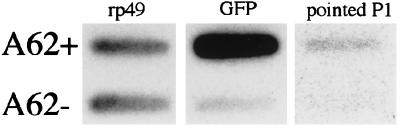
Ten nanograms each of the amplified A62+ and A62− cDNA product was re-amplified by 25 rounds of PCR with the TP1 primer. One-tenth of this reaction mixture was blotted to a nylon membrane in each of the spots shown in Fig. 3. Radiolabeling of probes and Southern hybridization were performed according to standard protocols (14). The probes used (given in the order that the hybridizations were performed) were GFP (15), rp49 (16), and pointed P1 (17).
Figure 3.
Autoradiographs showing that GFP and pointed P1 are enriched in the GFP-selected, posterior-derived A62+ cells. Amplified cDNA was prepared from sorted A62+ and A62− cells and blotted onto a nylon membrane. This membrane was hybridized to three successive radioactive oligonucleotides: the universally expressed ribosomal protein rp49, the reporter transgene GFP, and the posterior-expressed transcript pointed P1.
Microarray Hybridization and Analysis.
Six micrograms each of the original amplified A62+ and A62− cDNA was used as a template for fluorescent labeling by random hexamer priming. Microarray hybridization and scanning was performed by Ken Burtis as described in ref. 18 by using a microarray (constructed by Ken Burtis, Kevin White, David Hogness, and co-workers, Stanford Univ., Stanford, CA) of 4,500 unique Drosophila cDNAs obtained from the Berkeley Drosophila Genome Project/Howard Hughes Medical Institute EST project (Ken Burtis, personal communication). Quantitation of array data was performed by integrating pixel values over spots, subtracting background, and normalizing the average signal of the two probes to each other. Eleven clones enriched for one of the two probes were obtained from Research Genetics (Huntsville, AL) and processed for in situ hybridization. (Typical enrichment values were approximately 2-fold; our quantitation method tends to underestimate signal ratios.)
Differential Display.
Total RNA was isolated from sorted cells by using a hot phenol method (19). About 0.5–1.0 μg of nucleic acid was isolated per 100,000 sorted cells. Differential display was performed as described in ref. 2, with the following modifications: Silane-treated tubes were used to maximize recovery of nucleic acid. DNase treatment was performed with 0.1 unit of DNase I and 0.1 unit of human placental RNase per microgram of nucleic acid and a 15-min incubation time. Reverse transcription was performed with 0.5 μg of total RNA and an anchored primer (T12MA, T12MT, T12MC, or T12MG, where M is a mixture of all four nucleotides). PCR amplification was conducted using the anchored primer and one of 18 random 10-mers, under the following conditions: 40 cycles of 94°C for 30 sec, 40°C for 2 min, 72°C for 30 sec. For the comparison of A62+ with 55B+ cells, 5 primer pairs were used for a total of approximately 250 displayed bands. For the comparison of FACS-purified grk+ and grk− cells, 10 primer pairs were used for a total of 312 displayed bands; a screen using grk+ and grk− cells that skipped the FACS step employed 6 primer pairs for 181 displayed bands. Selected bands were extracted, amplified by PCR, purified from agarose gels, reamplified, and repurified to provide sufficient probe for in situ hybridization. All bands selected for further analysis were present in at least two PCRs per sample.
Molecular Cloning.
DNA cloning and library screening were performed according to ref. 14. A 4.4-kb DSbf1 cDNA was recovered from an ovarian gt22A library (library generated by P. Tolias, Public Health Research Institute, New York). The N-terminal region of the gene was sequenced from the P1 clone DS00597 (86F3–10; Berkeley Drosophila Genome Project).
In Situ Hybridization.
Hybridization with DNA probes was performed as described in ref. 20. Hybridization with RNA probes was performed as above, with the following modifications: Digoxigenin (DIG)-labeled sense and antisense RNA probes were transcribed from linearized pGEM-T vector (Promega) containing the insert of interest, using the DIG RNA Labeling Kit (Boehringer Mannheim), following the recommendations of the manufacturer. Hybridization to ovaries was carried out at 55°C. Plasmids containing E78A and E78B cDNA, from which probes were made, are described in ref. 21 and were a gift of Carl Thummel.
Fluorescent Staining of Ovaries and Microscopy.
A62 GAL4/UAS-GFP and 55B GAL4/UAS-GFP ovaries were fixed for 1 hr in 4% paraformaldehyde/1× PBS, washed three times in 1× PBS, incubated 30 min in 10 μg/ml RNase H at 37°C, stained for 10 min in 10 μg/ml propidium iodide, washed in PBS, and mounted in 70% glycerol. Light and fluorescence microscopy were performed on a Leitz DMRB with Nomarski differential interference contrast. Confocal microscopy was performed on a Bio-Rad confocal microscope.
RESULTS AND DISCUSSION
Marking of Follicle Cell Subgroups.
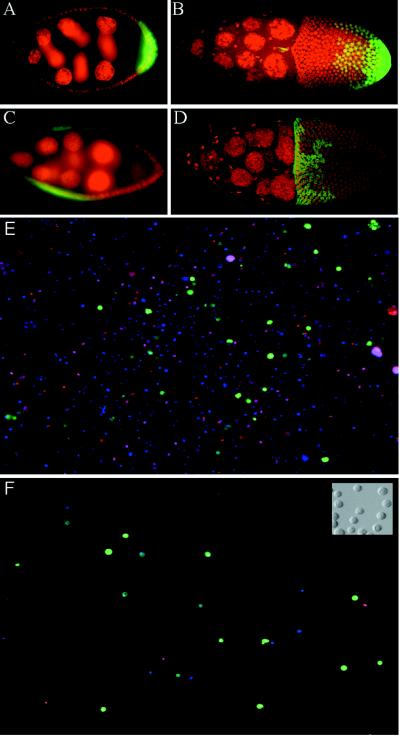
Follicle cell subgroups were labeled by using the GAL4 system of Brand and Perrimon (8) to drive expression of a UAS-GFPS65T reporter. Two GAL4 insertions were chosen for this study. A62 GAL4 (9) drives expression of GFP in the follicle cells covering the posterior of the oocyte (Fig. 1 A and B). 55B GAL4 (9) drives expression only in the lateral follicle cells, which cover the nurse cells in early stages (Fig. 1C) and later migrate to cover the anterior of the oocyte (Fig. 1D). The posterior follicle cells, which respond to grk signaling and whose differentiation is required for correct anterior/posterior axis formation (22), are included in the expression pattern of A62 but not of 55B. Likewise, the grk-sensitive dorsal anterior triangle of cells implicated in dorsal/ventral patterning (22, 23) is included in the expression pattern of 55B but not of A62. Thus, transcripts restricted to either marked subpopulation should include molecules involved in axis formation.
Figure 1.
Marking and purification of follicle cell subgroups. (A–D) GAL4 expression patterns in egg chambers. Posterior is to the right. Nuclei are stained with propidium iodide, shown as red. Green fluorescence reflects GAL4-activated transcription of GFP. (A and B) A62 GAL4/UAS-GFP. (A) Stage 7 egg chamber showing GFP expression in posterior follicle cells. (B) Stage 10 egg chamber showing GFP expression in posterior follicle cells and in border cells (small anterior group). (C and D) 55B GAL4/UAS-GFP. Expression is seen in lateral follicle cells in stage 8 (C) through stage 10 (D) egg chambers. (E and F) Fluorescence images of cells released from A62 GAL4/UAS-GFP ovaries, before (E) and after (F) purification of GFP-positive cells by FACS. All nuclei stain blue with Hoechst 33342; the nuclei of dead cells stain red with propidium iodide (cells were restained after sorting). GFP fluorescence is shown in green. (Inset) Nomarski image of A62− cells after sorting.
Characterization of Released Cells.
Follicle cells were separated from germ-line cells by treatment with trypsin followed by filtration (see Materials and Methods). Examination of follicle cell preparations showed single dissociated round cells. Staining with the DNA dye Hoechst 33332 (Fig. 1E) revealed a wide range of ploidies; this result was expected, since follicle cells cease division at stage six of oogenesis and enter several rounds of endoreplication. Between 60% and 80% of released cells were found to be living on the basis of staining with propidium iodide, which is excluded from cells with intact membranes. GFP-fluorescing cells were observed in suspensions derived from the ovaries of A62 GAL4/UAS-GFP (henceforth A62/GFP) or 55B/GFP flies, demonstrating that the marked subpopulations were included in the released cell preparations.
FACS Analysis and Sorting of Follicle Cell Subgroups.
Cells were stained with Hoechst 33332 and propidium iodide (PI) and analyzed by flow cytometry (Fig. 2). Cells from A62/GFP or 55B/GFP ovaries were sorted to collect cells that satisfied three criteria: their Hoechst fluorescence fell within a range empirically determined to include single DNA-containing cells; their green fluorescence was indicative of GFP expression; and their membranes were uncompromised on the basis of low PI fluorescence. Inspection of sorted cells showed >90% purity of visibly GFP-positive cells (Fig. 1F). GFP-negative cells were also collected separately. Cells from grk2B6/grkDC9 ovaries, as well as from their grk/+ siblings, were sorted solely on the basis of PI and Hoechst staining.
Figure 2.
FACS analysis of follicle cells. Cells are plotted as fluorescence events on the axes shown. (A) Wild-type follicle cells. There are very few events in the region of high GFP fluorescence. Arrows indicate concentrated regions along the Hoechst axis roughly corresponding to the four major ploidies in the population. (B) A62 GAL4/UAS-GFP. A population of GFP-positive cells can clearly be distinguished. The rectangle represents the approximate sort window used to select GFP-positive cells. (C) Live/dead selection. The approximate sort window used to select live cells is overlaid on the plot. The upper streak, which is high in propidium iodide fluorescence, represents dead cells, which are excluded from the sorted population. The separation of cells of different ploidies can most easily be seen in the dead cells (arrows), whose exposed nuclei can readily equilibrate with the Hoechst dye.
To assay the suitability of the sorted cells for differential screening, poly(A)+ RNA was prepared from A62+ and A62− cells (i.e., GFP+ and GFP− cells purified from A62/GFP ovaries). cDNA was prepared from these samples, amplified by PCR using a modification of the procedure of Brady et al. (12) (see Materials and Methods), blotted onto a nylon membrane, and analyzed by hybridization to radioactive probes for expected transcripts (Fig. 3). When the uniformly expressed ribosomal protein rp49 was used as a normalizing control, A62+ cells were found to have GFP transcripts enriched at least 14× over A62− cells. pointed P1, a transcript expressed in the posterior of developing egg chambers (24), was found to be highly enriched in the posterior-derived A62+ cells, as expected; it was essentially undetectable in A62− cells.
Molecular Screens.
The amplified cDNA samples tested above were used to screen a microarray of 4,500 Drosophila cDNAs for genes preferentially expressed in or excluded from the posterior follicle cells (see Materials and Methods). In addition, differential display (1, 2) was used to compare RNA from posterior (A62+) and lateral (55B+) follicle cells, as well as to screen for differences in expression between wild-type follicle cells and cells purified from grk2B6/grkDC9 ovaries, which are defective in follicle cell differentiation mediated by the epidermal growth factor receptor (EGFr). Differentially expressed genes identified in these screens are summarized in Table 1 and discussed below.
Table 1.
Results of expression screens
| cDNA comparison method | Cell types compared | No. of probes selected for secondary screening | No. of probes showing anticipated follicle cell patterns | Differentially expressed genes of interest | Enriched in | Novel follicle cell gene? | Fig. 4 ref. |
|---|---|---|---|---|---|---|---|
| Microarray | A62+ vs. A62− | 11 | 4* | GM07659 (FKBP-12 homologue) | A62− | Yes | A–C |
| GM04985 (novel E75 transcript) | A62+ | Yes | D | ||||
| Differential display | A62+ vs. 55B+ | 23 | 1 | VM34C | 55B+ | No | E and F |
| grk/grk vs. grk/+ | 16 | 1 | E78B | grk/+ | Yes | G–J | |
| grk/grk vs. grk/+ (no FACS) | 11 | 1 | DSbf1 | grk/grk | Yes | K–N |
Includes two clones of lesser interest because they show temporal but not spatial regulation, reflecting the stage specificity of A62 GAL4.
An FKBP-12 Homologue Expressed in Dorsal Anterior Follicle Cells May Help Elucidate a Link Between FKBP-12 and TGF-β Signaling.
The cDNA clone GM07659 showed greater signal on microarray hybridization with the A62− probe than with the posterior-derived A62+ probe. It was therefore expected to be expressed in the follicle cells and selectively excluded from the posterior of the egg chamber. Consistent with this expectation, it has a dorsal anterior in situ hybridization pattern in stage 9–10 egg chambers (Fig. 4 A and B); this expression depends on grk (Fig. 4C). The predicted gene product of the clone (Berkeley Drosophila Genome Project/Howard Hughes Medical Institute EST Project, unpublished: clot 5249) shows homology to the human immunophilin FKBP-12 and related proteins, which have prolyl isomerase activity in vitro and are responsible for binding and mediating the activity of the immunosuppressant drug FK506 (25). FKBP-12 has been shown to bind to TGF-β type I receptors (26) and has been proposed, on the basis of cell culture studies, to act as a regulated inhibitor of TGF-β type I signaling (27). The Drosophila TGF-β homologue dpp is expressed in anterior follicle cells and is required for the formation of anterior chorion structures (28). Follicle cell patterning may provide an instructive system in which to study the interactions between TGF-β signaling, FKBP-12-like proteins, and the EGFr pathway, which is required for the induction of GM07659 and other genes in the dorsal anterior follicle cells. Consistent with a role for GM07659 in modulating dpp signaling, preliminary overexpression studies show defects in anterior chorion structures (M.T. and H.R.-B., unpublished data).
Figure 4.
In situ hybridization patterns of microarray (A–D) and differential display positives (E–L). Egg chambers are shown with posterior to the right. (A–C) GM07659, a gene with homology to human FKBP-12. (A) Lateral view of a stage 10A egg chamber, showing expression restricted to a dorsal anterior triangle of follicle cells. (B) Dorsal view of a stage 10B egg chamber, showing dorsal anterior staining. (C) No staining is seen in grk2B6/grkDC9 egg chambers. (D) GM04985, a splice variant of the E75 nuclear hormone receptor gene, hybridizes to follicle cells over the posterior (arrows) of stage 6–9 egg chambers. (E and F) The vitelline membrane protein VM34C. Stage 8 (E) and stage 10 (F) egg chambers, showing expression in the columnar follicle cells over the oocyte. Staining is excluded from the posterior (arrows). (G–J) The The ecdysone-responsive nuclear hormone receptor E78. (G) Hybridization of an RNA antisense probe to a stage 12 egg chamber, showing expression in the dorsal anterior follicle cells (bracket). No staining in the follicle cells is seen with an RNA sense probe (H) nor in grk egg chambers (I). (J) A stage 14 egg chamber showing expression in the follicle cells covering the dorsal appendages (bracket). (K–N) Drosophila SET-binding factor 1. In grk2B6/grkDC9 flies, staining can be detected in follicle cells over the oocytes of stage 8 through stage 11 egg chambers (K), with the highest concentration in the posterior (arrow). (L) Close-up of staining in the posterior follicle cells (stage 10). (M) In the wild-type egg chambers of grk/CyO flies, hybridization is faint or undetectable in the follicle cells surrounding the oocyte, although staining is evident in the nurse cells. (N) Stage 9 grk2B6/grkDC9 egg chamber
A Transcript of the Ecdysone Response Gene E75 Is Expressed in the Posterior of Stage 6–9 Egg Chambers.
The cDNA clone GM04985 showed greater signal on microarray hybridization with the posterior-derived A62+ probe than with the A62− probe. It was therefore predicted to be preferentially expressed in the posterior follicle cells; this prediction was borne out by in situ hybridization (Fig. 4D). GM04985 hybridizes to the posterior follicle cells during stages 6–9. Sequence from the 5-′ end [Berkeley Drosophila Genome Project(BDGP)/Howard Hughes Medical Institute EST Project, unpublished data] shows that the clone is a novel splice variant of the ecdysone-inducible nuclear hormone receptor superfamily gene E75 (29, 30), in which a unique region (base pairs 1–344 of the BDGP sequence) is spliced into the exon 2/3 junction of the E75A/B common region (31). We have sequenced the remainder of the clone, and found it to match the common exons 3, 4, and 5 of E75A/B, with a poly(A) region beginning at base pair 4574 of the E75A sequence. Thus, the variant appears to encode a receptor that lacks the N-terminal DNA-binding zinc fingers encoded by the unique regions and exon 2 of E75A/B. A probe specific to this variant reproduces the hybridization pattern shown. E75 transcripts have also been observed to have dorsal anterior expression in stage 10 egg chambers (William Segraves, personal communication); in agreement with this observation, GM04985 hybridizes to the dorsal anterior of stage 10 chambers (not shown).
A Vitelline Membrane Protein Expressed Only in Lateral Follicle Cells.
A fragment of the vitelline membrane protein gene VM34C (31) was amplified in differential display reactions from 55B+ lateral follicle cells but not from A62+ posterior follicle cells. Consistent with this display pattern, it hybridizes only to lateral follicle cells in stages 8–10 (Fig. 4 E and F), as previously reported by its discoverers. While it encodes a known structural gene product, this band demonstrates the ability of the differential display screen to identify spatially restricted transcripts according to desired criteria.
The Nuclear Hormone Receptor Superfamily Gene E78 Is Transcriptionally Regulated by grk.
A fragment of a second ecdysone-responsive nuclear hormone receptor superfamily gene, E78 (21), was amplified in differential display reactions from sorted grk/+ but not from grk2B6/grkDC9 cells. The fragment hybridizes to dorsal anterior follicle cells and is absent from the dorsal follicle cells of grk− ovaries (Fig. 4 G–J). The use of specific probes shows that only one of the two E78 transcripts, E78B, is expressed in these cells. Staining for E78B is detected from stage 10B onward. This gene was hitherto unknown to be under the control of the EGFr pathway, and demonstrates that our method can correctly identify follicle cell transcripts whose expression is regulated by grk. Two other ecdysone-induced genes have recently been observed to be under the control of grk during oogenesis: E75 (William Segraves, personal communication) and BRC (32).
DSbf1 Is a Gene with High Homology to Human Sbf1 and Elevated Transcription in grk− Follicle Cells.
In a separate differential display screen comparing follicle cells that were isolated by enzymatic digestion and filtration (excluding the FACS step) from grk2B6/grkDC9 and grk/+ ovaries, a band was found to be present in grk2B6/grkDC9 lanes but absent or faint in grk/+ lanes. This band was cloned and found to encode a fragment of a previously undescribed gene, DSbf1, with high homology to the human gene SBF1 (SET binding factor 1) (33). A partial cDNA for DSbf1 was isolated, and the gene was localized to the 86F region of the 3rd chromosome. The cDNA and N-terminal genomic DNA were sequenced; an alignment of the predicted Drosophila gene product with human Sbf1 is published as supplemental data on the PNAS web site (www.pnas.org). In situ hybridization shows elevated Dsbf1 expression in the follicle cells over the oocyte with the highest concentration in the posterior follicle cells of grk2B6/grkDC9 ovaries (Fig. 4 K–N). Expression in the corresponding anterior follicle cells of grk and WT ovaries is difficult to assess because of staining in the underlying nurse cells. Human SBF1 has been shown to bind to SET domains, found in genes, such as trithorax, involved in epigenetic regulatory mechanisms, and prevent their dephosphorylation by myotubularin-related dual-specificity phosphatases (33). An attractive speculation is that DSbf1 forms a link between the grk-activated EGFr pathway and epigenetic gene regulation.
E78 Mutants Have Defective Dorsal Appendages.
Because our method does not directly select for functional genes, we wished to assay one of the genes retrieved from our differential expression screens for function in the expected tissue. Having determined that E78B was expressed under EGFr control in the follicle cells that form the dorsal appendages, we asked whether it had a function in these cells. Eggs were collected from viable E78 mutant flies and examined for defects in the chorion. Eggs were found that had shortened and/or broadened dorsal appendages (Fig. 5 A–C). The eggshells of flies expressing E78B under control of a heat shock promoter (34) were also examined, and they were found to have fused dorsal appendages (Fig. 5D). E78 has previously been shown to be inessential for viability and fertility (34). Our observation of an incompletely penetrant defect in dorsal appendage morphogenesis suggests that E78 has a partially redundant role in follicle cell migration or patterning. Its role in larval chromosome puffing is similarly redundant (34): mutants reduce the size of late puffs but do not abolish them. The identification of a role for E78 illustrates an advantage of molecular expression screening over loss-of-function female sterile screens, which are unable to detect genes with redundant requirements.
Figure 5.
(A–C) Chorions of eggs laid by E78 mutant flies. Dorsal side is facing the camera; anterior is to the left. Twenty percent of eggshells have broadened and/or shortened dorsal appendages (A and B); the remainder have wild-type appearance (C). (D) Eggs laid by hs-E78B flies have fused appendages.
Our Expression Screens Have Uncovered Tools for Analyzing the Determination of the Dorsal Anterior Follicle Cells.
These cells are responsible for both the formation of dorsal chorion structures and the establishment of the dorsal–ventral axis of the embryo (35). Their fate is influenced by at least three known signaling pathways: the EGFr pathway (22, 23, 36), the TGF-β pathway (28, 32), and the ecdysone response pathway (ref. 32; William Segraves, personal communication). The identification of EGFr-dependent dorsal anterior expression of both a potential player in TGF-β signaling (an FKBP-12 homologue) and a known ecdysone response gene (E78B) provides new markers and functional tools for assessing the interactions between these three pathways.
Evaluation of a Strategy for Cell Type-Specific Developmental Analysis.
A major aim of this pilot study was to a establish a tissue purification method for preparing samples of follicle cells that are suitable for use in cDNA preparation and differential expression screens. When enzymatic digestion is used to dissociate cells, as was done in this study, there is a concern that the released cells may not maintain their original fates, because of generalized trauma or activation of signaling by cleavage of surface proteins. We have addressed this concern by confirming via slot blots that a reporter construct (GFP) and an endogenous cell-specific transcript (pointed P1) were enriched, as expected, in cDNA amplified from posterior GFP-expressing follicle cells. We have further addressed the concern by retrieving confirmed positives from our microarray and differential display screens (see above). The transcripts we retrieved were largely previously unknown or unknown in the follicle cells, excluding only the vitelline membrane protein VM34C. It is unsurprising that our screens did not retrieve more of the known follicle cell components, as the differential display screens were on a scale considerably below saturation of the transcript pool, and the microarray used contained only one cDNA with a known polarized follicle cell pattern, pointed P1. (Hybridization to this clone was below the detection limit for both cell groups used. This likely reflects low sensitivity in our microarray analysis, as pointed P1 was detected on slot blots in cDNA amplified from A62+ cells.)
As compared with our differential display screens, our microarray screen produced a considerably smaller proportion of false positives (see Table 1). This apparent greater efficiency, combined with the rapidity of cDNA microarray screening, will likely make it the method of choice for future screens; it is hoped that the sensitivity of the method can be improved. Future screens could also combine the strategies of genetic mutation and spatial selection of follicle cells, for example by crossing GAL4 lines and UAS-GFP into a grk− background and analyzing the effect of grk on the expression profiles of specific subregions of the follicular epithelium. This type of targeted mutant analysis will likely be the most useful future application of these methods—screens for purely spatial selection may be supplanted by large-scale in situ screening (37), but it is unlikely to become feasible to perform many thousands of in situ hybridizations for every mutant situation a researcher wishes to analyze. These methods can also be readily applied to other systems; Drosophila imaginal discs are an obvious candidate, and as Krasnow et al. (5) have previously pointed out, FACS purification of cells marked with cell type-specific reporter constructs can in principle be applied to any organism receptive to germ-line transformation. As microarrays for higher organisms become more comprehensive and more readily available, experiments that probe the intricate regulation of transcription on a genomic scale will become routinely feasible in developmental systems. If these experiments are to provide targeted results, many of them will rely on cell type-specific purification schemes like the one described here.
Supplementary Material
Acknowledgments
We thank Ken Burtis, Kevin White, and the laboratories of Pat Brown and David Hogness for supplying the microarray and performing the array hybridization; Celeste Berg for locating the cytological position of DSbf1 by in situ hybridization; Kristen Holder for technical assistance; Gabrielle Boulianne, Gertrud Schüpbach, and Carl Thummel for supplying Drosophila stocks and plasmids; Steve Hauschka and David Baker for critical readings of the manuscript; and William Segraves for sharing results prior to publication. This work was supported by an Established Investigatorship Award from the American Heart Association (AHA 95002990), a grant from the March of Dimes Birth Defect Foundation, a Basil O’Conner Starter Research Grant (5-FY94-0929), a National Institutes of Health grant (R01 HD 32464), and by the Pew Memorial Trust, in the form of a Pew Scholarship in the Biomedical Sciences to H.R.-B.
ABBREVIATIONS
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- EGFr
epidermal growth factor receptor
- TGF-β
tumor growth factor β
Footnotes
References
- 1.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 2.Liang P, Bauer D, Averboukh L, Warthoe P, Rohrwild M, Muller H, Strauss M, Pardee A B. Methods Enzymol. 1995;254:304–321. doi: 10.1016/0076-6879(95)54022-9. [DOI] [PubMed] [Google Scholar]
- 3.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 4.Ramsay G. Nat Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 5.Krasnow M A, Cumberledge S, Manning G, Herzenberg L A, Nolan G P. Science. 1991;251:81–85. doi: 10.1126/science.1898782. [DOI] [PubMed] [Google Scholar]
- 6.Cumberledge S, Krasnow M A. Methods Cell Biol. 1994;44:143–159. doi: 10.1016/s0091-679x(08)60911-5. [DOI] [PubMed] [Google Scholar]
- 7.Amrein H, Axel R. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 8.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Yeh E, Gustafson K, Boulianne G L. Proc Natl Acad Sci USA. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neuman-Silverberg F S, Schüpbach T. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- 11.Ruohola-Baker H, Grell E, Chou T B, Baker D, Jan L Y, Jan Y N. Cell. 1993;73:953–965. doi: 10.1016/0092-8674(93)90273-s. [DOI] [PubMed] [Google Scholar]
- 12.Brady G, Barbara M, Iscove N N. Methods Mol Cell Biol. 1990;2:17–25. [Google Scholar]
- 13.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Maniatis T, Fritsch E F. Molecular Cloning: A Laboratory Manual. 2nd. Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 16.Kongsuwan K, Yu Q, Vincent A, Frisardi M C, Rosbash M, Lengyel J A, Merriam J. Nature (London) 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- 17.O’Neill E M, Rebay I, Tjian R, Rubin G M. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 18.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 19.Jowett T. In: Drosophila: A Practical Approach. Roberts B, editor. Oxford: Information Printing; 1986. pp. 275–286. [Google Scholar]
- 20.Ruohola H, Bremer K A, Baker D, Swedlowe J R, Jan L, Jan Y N. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- 21.Stone B L, Thummel C S. Cell. 1993;75:307–320. doi: 10.1016/0092-8674(93)80072-m. [DOI] [PubMed] [Google Scholar]
- 22.Ray R, Schüpbach T. Genes Dev. 1996;10:1711–1723. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- 23.Wasserman J D, Freeman M. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto A M, Jordan K C, Tietze K, Britton J S, O’Neill E M, Ruohola-Baker H. Development (Cambridge, UK) 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- 25.Fruman D A, Burakoff S J, Bierer B E. FASEB J. 1994;8:391–400. doi: 10.1096/fasebj.8.6.7513288. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Donahoe P K, Zervos A S. Science. 1994;265:674–676. doi: 10.1126/science.7518616. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Li B Y, Danielson P D, Shah P C, Rockwell S, Lechleider R J, Martin J, Manganaro T, Donahoe P K. Cell. 1996;86:435–444. doi: 10.1016/s0092-8674(00)80116-6. [DOI] [PubMed] [Google Scholar]
- 28.Twombly V, Blackman R K, Jin H, Graff J M, Padgett R W, Gelbart W M. Development (Cambridge, UK) 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- 29.Feigl G, Gram M, Pongs O. Nucleic Acids Res. 1989;17:7167–7178. doi: 10.1093/nar/17.18.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segraves W A, Hogness D S. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- 31.Mindrinos M N, Scherer L J, Garcini F J, Kwan H, Jacobs K A, Petri W H. EMBO J. 1985;4:147–153. doi: 10.1002/j.1460-2075.1985.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W M, Bownes M. Development (Cambridge, UK) 1997;124:4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 33.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary M L. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 34.Russell S R H, Heimbeck G, Goddard C M, Carpenter A T C, Ashburner M. Genetics. 1996;144:159–170. doi: 10.1093/genetics/144.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schüpbach T, Roth S. Curr Opin Genet Dev. 1994;4:502–507. doi: 10.1016/0959-437x(94)90064-a. [DOI] [PubMed] [Google Scholar]
- 36.Queenan A M, Ghabrial A, Schüpbach T. Development (Cambridge, UK) 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 37.Kopczynski C C, Noordermeer J N, Serano T L, Chen W-Y, Pendleton J D, Lewis S, Goodman C S, Rubin G M. Proc Natl Acad Sci USA. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.