Abstract
Background and purpose:
Genistein, a tyrosine kinase inhibitor used to block caveolae dependent endocytosis, reduces the cellular uptake of anandamide in RBL2H3 basophilic leukaemia cells. However, genistein is also a competitive inhibitor of fatty acid amide hydrolase, the enzyme responsible for anandamide hydrolysis. Here we have investigated whether inhibition of fatty acid amide hydrolase rather than inhibition of endocytosis is the primary determinant of genistein actions upon anandamide uptake.
Experimental approach:
Cellular uptake of anandamide, labelled in the arachidonoyl part of the molecule was assessed in four different cell lines using a standard method. Fatty acid amide hydrolase activity in homogenates and intact cells was measured using anandamide labelled in the ethanolamine part of the molecule.
Key results:
The fatty acid amide hydrolase inhibitor URB597 inhibited anandamide uptake into RBL2H3 cells and R3327 AT-1 prostate cancer cells, but not into 3T3-L1 preadipocytes or PC-3 prostate cancer cells. An identical pattern was seen with genistein. The related compound daidzein inhibited anandamide hydrolysis in homogenates and intact cells, and reduced its uptake into RBL2H3 and R3327 AT-1, but not PC-3 cells. Anandamide hydrolysis by cell homogenates was in the order RBL2H3 > R3327 AT-1 > PC-3 ≈3T3-L1.
Conclusions and implications:
The ability of genistein to inhibit anandamide uptake is mimicked by daidzein (which does not affect tyrosine kinase), and is only seen in cells that show sensitivity to URB597. This indicates that blockade of fatty acid amide hydrolase is the primary determinant of the effects of genistein on cellular anandamide uptake.
Keywords: endocannabinoid, anandamide, cellular uptake, fatty acid amide hydrolase, tyrosine kinase, genistein, daidzein
Introduction
An important component in any signalling process is the ability of the organism to terminate the signal. In the case of the endogenous cannabinoid compound anandamide (arachidonoylethanolamide, AEA), this is brought about by a process of cellular uptake followed by metabolism, the latter primarily by the enzyme fatty acid amide hydrolase (FAAH) (Deutsch and Chin, 1993; Di Marzo et al., 1994). FAAH has been well characterized, and selective inhibitors, such as (3′-(aminocarbamoyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate (URB597) (Kathuria et al., 2003), are currently being used to explore the physiological importance of this enzyme. One particular role that FAAH undertakes is the regulation of the uptake of AEA: thus, in cells like rat RBL2H3 basophilic leukaemia cells, inhibition of FAAH reduces the rate of uptake of AEA (Rakhshan et al., 2000; Kaczocha et al., 2006). In other cell types, however, FAAH inhibitors do not affect AEA uptake, despite expression of the enzyme (see Ruiz-Llorente et al., 2004 for an example using PC-3 human prostate adenocarcinoma cells).
In 2004, McFarland et al. demonstrated that inhibitors of caveolae-related endocytosis reduced the uptake of AEA into RBL2H3 cells and argued, on the basis of these and other data, that an endocytotic process was involved in the cellular uptake of AEA. In their model, (shown schematically by McFarland and Barker, 2004), AEA is anchored in caveolae by a putative binding protein, which, after endocytosis, is ‘cleared' from the binding protein by FAAH. We have recently questioned the evidence of an endocytotic process on the basis of the specificity of the compounds used (Thors et al., 2007). Thus, for example, genistein, used to prevent caveolae-related endocytosis (review, see Nichols, 2003), was found at a concentration of 200 μM to reduce by about half the uptake of AEA into RBL2H3 cells in the study of McFarland et al. (2004). Genistein was originally termed a ‘specific' inhibitor of tyrosine kinase (Akiyama et al., 1987), but has subsequently been found to have other actions over the same concentration range, including competitive inhibition of FAAH (Thors et al., 2007). Given that the effects of URB597 and genistein upon the uptake of AEA were not additive (Thors et al., 2007), we argued that the action of genistein upon AEA uptake could be ascribed to inhibition of FAAH. However, additivity would not be expected if effects of genistein upon FAAH and upon endocytosis occur at different points along the same pathway of cellular AEA removal, such as that postulated by McFarland and Barker (2004), and the possibility remains that the effects of genistein upon endocytosis are at least as important as those mediated by FAAH inhibition, particularly in cells that are rich in caveolae (such as fibroblasts, Couet et al., 1997), but with low expression of FAAH. This hypothesis has been tested by utilizing the mouse 3T3-L1 fibroblast line and PC-3 human prostate adenocarcinoma line. Both cell lines accumulate AEA (the latter in a manner not affected by FAAH inhibition) (Ruiz-Llorente et al., 2004; Gasperi et al., 2007) and show sensitivity to tyrosine kinase inhibitory effects of genistein in other biochemical systems (Roig and Traugh, 1999; Skogseth et al., 2005). The morphology of caveolae on the plasma membrane of 3T3-L1 fibroblasts has also been well described (Westermann et al., 2005).
Materials and methods
Culturing of cells
The cells were grown in 75 cm2 culturing flasks at 37°C with 5% CO2 in humidified atmospheric pressure. Cells were passaged twice a week and cell culture medium was changed every other day. The following cells were used: RBL2H3 cells (passage range 13–43) were obtained from American Type Culture Collection, Manassas, VA, USA. 3T3-L1 mouse fibroblast cells (passage range 24–42 (above those initially propagated before deposition)) were obtained from European Collection of Cell Cultures, Porton Down, UK. PC-3 cells (passage range 5–14) and R3327 AT-1 rat prostate cancer cells (passage range 44–51) were obtained from Professor Anders Bergh (Department of Medical Biosciences, Umeå University). Culturing conditions were as follows: RBL2H3, minimum essential medium with Earl's salts, 15% foetal bovine serum; 3T3-L1, Dulbecco's modified Eagle's medium, 10% foetal bovine serum; PC-3, Hams F-10, 2 mM L-glutamine, 10% foetal bovine serum; R3327 AT-1, RPMI 1640, 250 nM dexamethasone, 10% foetal bovine serum. In all cases, these media also contained 100 U ml−1 penicillin+100 μg ml−1 streptomycin.
Assay of [3H]AEA uptake
The assay was based on the method of Rakhshan et al. (2000), as modified by Sandberg and Fowler (2005). For experiments with uncoated wells, cells were plated at a density of 2 × 105 cells well−1 in 24-well plates and incubated overnight at 37°C in an atmosphere of 5% CO2. For experiments with coated wells, the wells were first incubated for at least 5 h at 37°C with phosphate-buffered saline containing either fibronectin (final concentration 10 μg ml−1), vitronectin (final concentration 0.5 μg ml−1), fibronectin-like protein polymer (final concentration 10 μg ml−1) or bovine serum albumin (BSA) (2% w/v), after which the cells (2 × 105 per well) were added and the 24-well plates incubated for a further 60–90 min. After these incubation phases, cells were washed once with Krebs–Ringer HEPES (KRH) buffer (120 mM NaCl, 4.7 mM KCl, 2.2 mM CaCl2, 10 mM 4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid (HEPES), 0.12 mM KH2PO4, 0.12 mM MgSO4 in milliQ deionised water, pH 7.4) containing 1% of BSA and once with KRH buffer alone. The cells were then preincubated with KRH buffer containing 0.1% of fatty acid-free BSA and selected compounds for the times shown in the figure legends. [3H-arachidonoyl]AEA (50 μl, final concentration 100 nM, in KRH buffer containing 0.1% (15 μM of fatty acid-free BSA)) was added to give a final volume of 400 μl, and the wells were incubated for the times shown. Solvent concentrations were kept as low as possible; the amount of solvent in the vehicles added per 400 μl assay were 0.8 μl ethanol for genistein, 0.04 μl dimethyl sulphoxide (DMSO) for URB597, 1.6 μl DMSO for daidzein and 0.72 μl ethanol for [3H-arachidonoyl]AEA unless otherwise stated. After incubation, the plates were placed on ice and each well was washed three times with 500 μl of ice-cold KRH buffer containing 1% (w/v) of BSA. After aspiration of the buffer, 500 μl of 0.2 M NaOH was added, and the plates were incubated for 15 min at 75°C before removal of aliquots (300 μl) and assessment of their tritium content by liquid scintillation spectroscopy with quench correction. Oleic acid (OA) uptake assays were undertaken in the same way. It should be noted that the use of fatty acid-free BSA means that the actual free concentration of AEA is lower than the added concentration of this substrate (100 nM) (for discussion, see Thors and Fowler, 2006). However, under the conditions used here, the total uptake was <10% of the added [3H-Ara]AEA, and thus had not saturated.
Assay of FAAH
FAAH activity was measured in homogenates from either cultured cells or rat brain (available at the Department) and intact cells as described by Boldrup et al. (2004) and Paylor et al. (2006), with [ethanolamine-1-3H]AEA as substrate. For the membranes, cells were cultured in culturing flasks and after two washes with cold phosphate-buffered saline, the cells were collected using a rubber policeman. The cells (on ice) were resuspended in cold phosphate-buffered saline, centrifuged for 5 min at 1000 g before being resuspended in 10 mM Tris buffer at the appropriate pH and frozen in aliquots. It should be noted that protease inhibitor cocktails were not used, simply because they contain sulphonyl fluoride compounds, which will inhibit FAAH directly (Deutsch and Chin, 1993). For homogenates, the assay concentrations of AEA are given in the figure legends. An assay concentration of 100 nM, a preincubation time of 10 min and an incubation time of 20 min were used for experiments with intact cells (2.5 × 105 seeded into 24-well plates the day before the experiment), and blank values defined as the recovery of tritium for assays conducted on the same assay plates in the absence of cells. In the experiments undertaken using intact cells, the total amount of solvent added in the vehicles (per 400 μl assay) was 1.2 μl ethanol for genistein, 1.2 μl DMSO for daidzein and 0.5 μl for the [ethanolamine-1-3H]AEA.
Statistical analyses
Curve fitting and statistical comparisons were undertaken utilizing the statistical package in the GraphPad Prism computer programme (GraphPad Software Inc., San Diego, CA, USA). Kmapp and Vmaxapp values were calculated from pooled data using the direct linear plot analysis (Eisenthal and Cornish-Bowden, 1974) built into the Enzyme Kinetics v1.4 computer programme (Trinity Software, Campton, NH, USA).
Compounds
AEA [arachidonoyl 5,6,8,9,11,12,14,15-3H] (specific activity 7.4 TBq mmol−1; for the uptake experiments), AEA [ethanolamine-1-3H] (specific activity 2.22 TBq mmol−1; for the FAAH assays) and OA [9,10-3H(N)] (specific activity 7.4 TBq mmol−1) were obtained from American Radiolabeled Chemicals Inc., St Louis, MO, USA. Rat fibronectin was obtained from EMD Biosciences, La Jolla, CA, USA. Genistein, daidzein, vitronectin, fibronectin-like protein polymer, dexamethasone, fatty acid-free and normal BSA were obtained from Sigma Aldrich Inc., St Louis, MO, USA. URB597 and unlabelled AEA were obtained from the Cayman Chemical Co., Ann Arbor, MI, USA.
Results
The effects of URB597 and genistein upon the uptake of AEA into RBL2H3 and 3T3-L1 cells
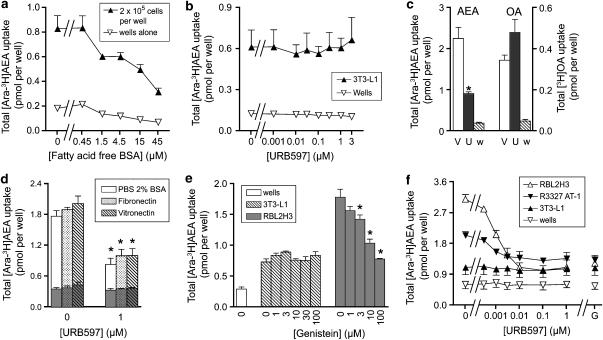
In our assays of AEA uptake, we use fatty acid-free BSA to stabilize the AEA and to minimize retention of the ligand by the wells (which can be as much as 40% of added AEA, see Karlsson et al., 2004) (for a review on the methodology of AEA uptake and the use of BSA in this manner, see Glaser et al., 2005). The sensitivity to fatty acid-free BSA of AEA uptake into 3T3-L1 cells and its retention by wells is shown in Figure 1a. A clear accumulation of AEA above that seen for wells alone was found at all concentrations of fatty acid-free BSA. The apparent reduction of uptake at increasing fatty acid-free BSA concentrations can be explained in terms of a reduction in the free substrate concentration available for uptake (see Thors and Fowler, 2006). Similar results were seen when OA was used as substrate (data not shown). In the remaining experiments, a fatty acid-free BSA concentration of 15 μM was used.
Figure 1.
Uptake of AEA by 3T3-L1, RBL2H3 and R3327 AT-1 cells. In (a–c) uncoated wells were used, whereas in (d–f) the wells were coated with fibronectin, unless otherwise shown. (a) Effect of fatty acid-free BSA upon the uptake by 3T3-L1 cells of 100 nM (added concentration) [3H]AEA labelled in the arachidonate part of the molecule (‘[3H-Ara]AEA'). The incubation time used was 4 min. (b) No effect of URB597 upon the uptake of 100 nM [3H-Ara] into 3T3-L1 cells or the retention of this substrate by wells alone. (c) Comparison of the effect of either vehicle (‘V', DMSO, final concentration 1.6 μl per assay) or 0.1 μM URB597 (‘U') upon the uptake of either AEA or OA into RBL2H3 cells. The cells were preincubated with URB597 for 20 min before addition of [3H-Ara]AEA or [3H]OA (100 nM) and incubation for a further 4 min. Data for vehicle-treated wells alone (‘w') are also shown. *Significantly different from the corresponding vehicle value (P<0.05, two-tailed paired t-test). (d) Comparison of the uptake of 100 nM [3H-Ara]AEA into RBL2H3 cells on either control (BSA), fibronectin or vitronectin-coated plates. After attachment and washing, the cells were preincubated with the indicated concentration of URB597 for 10 min before addition of AEA and incubation for a further 4 min. Values enclosed within the columns are the corresponding values for wells alone. One-way ANOVA for repeated measures indicated no significant differences between the uptakes for the different coatings; *significantly different (Tukey's test) from the corresponding value in the absence of URB597. (e) Effect of genistein upon the uptake of 100 nM [3H-Ara]AEA into 3T3-L1 and RBL2H3 cells adherent to fibronectin-coated plates. Preincubation and incubation times were 30 and 4 min, respectively. *Significantly different (Dunnett's test following significant one-way ANOVA for repeated measures) from the corresponding value in the absence of genistein. (f) Effect of URB597 or 100 μM genistein (‘G', in DMSO, assay concentration 0.04 μl per assay) upon the uptake of 100 nM [3H-Ara]AEA into 3T3-L1, RBL2H3, R3327 AT-1 cells adherent to fibronectin-coated plates. Preincubation times were 10 and 30 min for URB597 and genistein, respectively, and incubation times with AEA were 4 min. All data are means±s.e.m., n=4 except for (d) where n=3. AEA, anandamide (arachidonoylethanolamide); ANOVA, analysis of variance; OA, oleic acid; URB597, (3′-(aminocarbamoyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate.
The effects of URB597 upon the uptake of AEA into 3T3-L1 and RBL2H3 cells are shown in Figures 1b and c. In contrast to the robust effect of 0.1 μM URB597 upon the uptake of AEA into the RBL2H3 cells, this compound did not significantly affect AEA uptake into the 3T3-L1 cells. URB597 was also without significant effect upon OA uptake into either RBL2H3 cells (Figure 1c) or 3T3-L1 cells (data not shown), indicating that its ability to inhibit AEA uptake into the RBL2H3 cells is not a nonspecific effect upon the transport of lipophilic compounds. Initial experiments (n=2) indicated that genistein did not affect the uptake of AEA into 3T3-L1 cells at concentrations that reduced the uptake into RBL2H3 cells (data not shown). However, after these initial experiments, the 3T3-L1 cells no longer adhered satisfactorily to the uncoated wells, a prerequisite for the uptake assays. The problem recurred when a new batch of cells (from the same original batch) was thawed and used.
In order to circumvent the problems of adherence of the 3T3-L1 cells, coated wells were used. In order to be sure that the uptake assay behaved in the same manner for coated wells, RBL2H3 cells were tested in wells coated with either fibronectin, vitronectin or, as controls, with BSA (Figure 1d). The degree of uptake and its sensitivity were the same for all three conditions. A similar result was found for wells coated with fibronectin-like protein polymer (data not shown). In consequence, fibronectin-coated wells were used for the rest of the study. The 3T3-L1 cells were found to adhere well to the fibronectin-coated wells. In contrast to the situation for RBL2H3 cells cultured in the coated wells, the AEA uptake into the 3T3-L1 cells was not affected by either genistein or URB597 (Figures 1e and f). R3327 AT-1 rat prostate cancer cells were also investigated. These cells had a basal AEA uptake level in between that seen for the 3T3-L1 and RBL2H3 cells, and the uptake was inhibited both by URB597 and genistein (Figure 1f). For all three cell types, the residual activity in the presence of either URB597 or genistein were similar, and higher than the level of retention of AEA by the wells alone (Figures 1e and f).
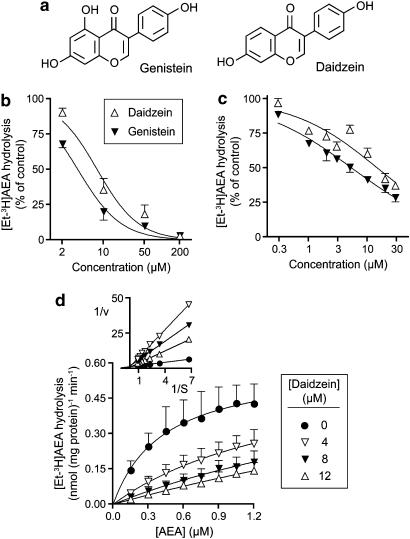
Daidzein inhibits the hydrolysis of AEA with a similar potency to that seen for genistein
Daidzein is a close structural analogue of genistein (Figure 2a) that lacks activity towards tyrosine kinase (Akiyama et al., 1987) and is often used as a negative control for genistein in this respect (see for example Harmon et al., 2002; Cayabyab and Schlichter, 2002; Jeong et al., 2005). The compound, however, was almost as effective as genistein as an inhibitor of AEA hydrolysis both in RBL2H3 cell homogenates (Figure 2b) and in intact RBL2H3 cells (Figure 2c). The pI50 values (with IC50 values in brackets) for daidzein and genistein, respectively, were: homogenates, 5.11±0.08 (7.8 μM) and 5.44±0.05 (3.6 μM); intact cells, 4.83±0.07 (15 μM) and 5.27±0.04 (5.4 μM). Kinetic experiments were undertaken using rat brain homogenates (Figure 2d). The Kmapp values calculated by direct linear plot of the mean data were 0.45, 1.67, 2.43 and 3.82 μM for 0, 4, 8 and 12 μM daidzein, respectively. The corresponding Vmaxapp values (nmol (mg protein)−1 min−1) were 0.60, 0.63, 0.53 and 0.58, respectively. Thus, daidzein inhibited [3H]AEA hydrolysis by rat brain homogenates in a competitive manner. A plot of Kmapp vs [daidzein] gave a Ki value of 1.7 μM.
Figure 2.
(a) Structures of genistein and daidzein. (b and c) Inhibition by genistein and daidzein of the hydrolysis of AEA labelled in the ethanolamine portion of the molecule (‘[3H-EA]AEA') by RBL2H3 cell homogenates ((b) AEA concentration 2 μM, 2 μg protein per assay, assay pH 7.4) and intact RBL2H3 cells ((c) AEA concentration 100 nM). The cells or homogenates were preincubated with the isoflavones for 10 min before addition of [3H-Et]AEA and incubation for a further 20 min. Note, that in (b) genistein was dissolved in DMSO, whereas in (c) it was dissolved in ethanol. In (c), the lowest four and the highest four concentrations were assayed on separate plates, and the data expressed as % of the values (after subtraction of blanks) for the vehicle controls on the same plates. The control activities (as pmol AEA hydrolysed per well for the 20 min incubation time were 4.81±0.31 and 4.64±0.21 for genistein and daizdein, respectively). (d) Inhibition of [3H-Et]AEA hydrolysis by rat brain homogenates at pH 7.4 and using an incubation time of 10 min and 1.5 μg protein per assay. The double reciprocal plot shown in the inset illustrates the competitive nature of the inhibition. Data are means±s.e.m., n=3 (b and d) or 4 (c). AEA, anandamide (arachidonoylethanolamide); DMSO, dimethyl sulphoxide.
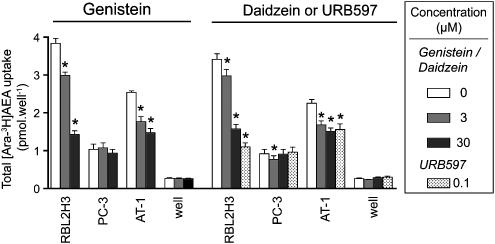
Comparison of the effects of genistein, daidzein and URB597 in RBL2H3, PC-1 and R3327 AT-1 cells
The effects of genistein (3 and 30 μM), daidzein (3 and 30 μM) and URB597 (0.1. μM) upon the uptake of AEA into the RBL2H3, PC-3 and R3327 AT-1 cells are shown in Figure 3. As with the experiments shown in Figure 1, the pattern of inhibition with genistein and URB597 was the same: thus, in the R3327-AT1 and RBL2H3 cells, both compounds reduced the uptake, whereas in the PC-3 cells, neither compound was effective. Once again, the residual activity seen after URB597 treatment was roughly the same for all three cells. The higher uptake for RBL2H3 cells in Figure 3 compared with Figure 1 is due to the use of a longer AEA incubation time (10 min vs 4 min). Daidzein gave a similar pattern of inhibition to that seen with genistein (Figure 3).
Figure 3.
Effects of genistein, daidzein or URB597 upon the uptake of 100 nM [Ara-3H]AEA into RBL2H3, PC-3 or R3327 AT-1 (‘AT-1') cells. Cells were preincubated with the compounds for 10 min before addition of AEA and incubation for a further 10 min. Data are means±s.e.m., n=4. Note that the DMSO concentration in the Daidzein/URB597 control is matched to the URB597 rather than the daidzein. *Significantly different (Dunnett's test following significant one-way ANOVA for repeated measures) from the corresponding control value. AEA, anandamide (arachidonoylethanolamide); ANOVA, analysis of variance; URB597, (3′-(aminocarbamoyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate.
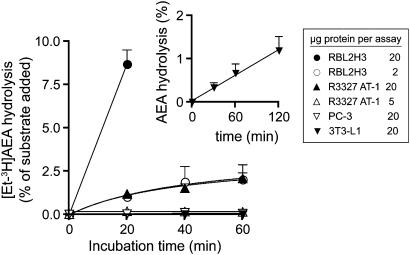
FAAH activities in homogenates of 3T3-L1, RBL2H3, PC-3 and R3327 AT-1 cells
FAAH has a pH optimum at pH 9 (Schmid et al., 1985), and the ability of the cell homogenates to hydrolyse 2 μM AEA was investigated at this pH value. The rates of hydrolysis of AEA by the homogenates of the four cell types were RBL2H3>R3327 AT-1>PC-3≈3T3-L1 (Figure 4). The rates of hydrolysis of the PC-3 and 3T3-L1 cells were very low under the conditions used. However, in a separate experiment with different homogenates using a lower (0.5 μM) AEA concentration, an assay pH of 8 and incubation time up to 120 min, hydrolysis of AEA by 3T3-L1 homogenates was clearly seen (inset to Figure 4).
Figure 4.
Hydrolysis of 2 μM [3H-Et]AEA by either homogenates of RBL2H3, R3327 AT-1, PC-3 and 3T3-L1 cells. The homogenates and assays were conducted at pH 9. Data are means±s.e.m., n=3. RBL2H3 cells (20 μg protein per assay) were also assayed at the 40 and 60 min time points, in these experiments. Rates of hydrolysis (as % of added AEA) of 15±0.9 and 18±2 were found for the 40 and 60 min incubations, respectively. In the inset, 3T3-L1 cells (23±1 μg protein per assay) were assayed over a longer time frame using homogenates made and assayed at a pH value of 8 and an [AEA] of 0.5 μM. (means±s.e.m., n=4) AEA, anandamide (arachidonoylethanolamide).
Discussion
The aim of the study was to determine whether the ability of genistein to inhibit the uptake of AEA into cells could be ascribed to its ability to interfere with caveolae-dependent endocytotic processes or to its ability to block FAAH. Our data would suggest that the latter is the primary determinant of genistein action, since the ability of the compound to reduce AEA uptake in the different cell lines correlated very well with that seen for the selective FAAH inhibitor, URB597. Furthermore, daidzein, which reportedly lacks the tyrosine kinase inhibitory effects of genistein (Akiyama et al., 1987) but which shared with genistein its FAAH inhibitory actions, affected AEA uptake in the same manner as genistein. Given that the other compounds or conditions used to inhibit caveolae-dependent endocytosis in the study of McFarland et al. (2004) either also inhibit FAAH (500 μM N-ethylmaleimide), reduce the concentration of AEA available for uptake (reduction of assay temperature to 18°C), or are just as effective at reducing the degree of AEA retained by wells alone as they are at inhibiting the cellular uptake (combination of 25 μg ml−1 nystatin and 10 μg ml−1 progesterone) (see Thors et al., 2007), there is at present no hard evidence that AEA is taken into cells by an endocytotic process. The association of the fluorescent AEA derivative SKM 4-45-1 (3′,6′-bis(acetyloxy)-3-oxospiro[isobenzofuran-1 (3H),9′-[9H]xanthen]-5-yl]-2-[[1-oxo-5Z,8Z,11Z,14Z-eicosatetraenyl]amino]ethyl ester carbamic acid) with markers for caveolae (McFarland et al., 2004) may reflect an association with cholesterol in such regions of the plasma membrane before membrane translocation and delivery to intracellular FAAH, given that changes in the cellular cholesterol content can influence AEA uptake (Bari et al., 2005) and that the AEA homologue N-myristoylethanolamine can form complexes with cholesterol (Ramakrishnan et al., 2002).
The above discussion concerns endocytosis as a postulated route of AEA uptake. A separate issue concerns the manner in which FAAH contributes to AEA uptake. URB597 has been used in the present study as a selective inhibitor of FAAH, the assumption being made that effects of this compound upon AEA uptake are secondary to inhibition of this enzyme. Certainly, the rate of hydrolysis of 100 nM AEA (added concentration) by intact RBL2H3 cells (4.7 pmol per well over a 20 min incubation period, corresponding to ∼13% of the added AEA) would mean that a considerable proportion of the AEA accumulated by these cells in the uptake experiments would be metabolized. There are reports in the literature that URB597 can affect targets in addition to FAAH, the most notable being its ability to block the binding of [3H]LY2183240 (5-biphenyl-4-ylmethyl-tetrazole-1-carboxylic acid dimethylamide) to cell membranes (Ki value ∼12 nM) (Dickason-Chesterfield et al., 2006). However, the relevance of this binding to the uptake of AEA is uncertain (see Alexander and Cravatt, 2006, for data demonstrating the multiplicity of pharmacological actions of LY218240; and Dickason-Chesterfield et al., 2006 for data showing that the uptake inhibitors (S)- and (R)-N-(1-(4-hydroxyphenyl)-2-hydroxyethyl)oleamide (OMDM)-1 and OMDM-2 are very poor inhibitors of this binding site), and while the long-term uptake of AEA into RBL2H3 cells was highly sensitive to URB597 (IC50 value 1.5 nM), it was not at all sensitive to this compound in HeLa cells (IC50 value 65 μM), which do not express FAAH (Dickason-Chesterfield et al., 2006). Furthermore, the ability of URB597 to inhibit uptake is shared by other structurally different FAAH inhibitors, such as oleoyl oxazolopyridine (see Kaczocha et al., 2006 and references therein). Thus, we feel it justified in ascribing the URB597-sensitive component of the uptake to the component dependent upon FAAH.
The variation of sensitivity of the cells to URB597 is a notable property of the uptake, given that all the cells used in this study hydrolyse AEA (Bisogno et al., 1997; Ruiz-Llorente et al., 2004; Gasperi et al., 2007; present study). However, there were considerable differences in the rates of AEA hydrolysis by homogenates of the cells, with RBL2H3>R3327 AT-1>PC-3≈3T3-L1. If it is assumed that the rates of hydrolysis seen in the cell homogenates reflect the activity of FAAH in the vicinity of AEA once it has been taken up by the intact cells (rather than, say, different sensitivities of the enzyme to proteolytic digestion during the homgenate preparation), then it would be expected that the URB597-sensitive component of the uptake would be much lower for the PC-3 and 3T3-L1 cells than for the RBL3H3 cells, with the R3327 AT-1 cells being in between these extremes. This interpretation is entirely consistent with the data of Deutsch et al. (2001), who showed that Hep2 cells, which had a very low AEA hydrolysing capacity, accumulated about half the AEA of that seen for N18 neuroblastoma cells (which hydrolysed AEA at the rate of 0.87 nmol h−1 106 cells−1), and that transfection of Hep2 cells with FAAH approximately doubled their ability to accumulate AEA.
While the present study supports the role of FAAH in regulating AEA uptake in cells with significant expression of the enzyme, it does not shed light upon the precise mechanisms as to how this is brought about. FAAH is usually considered to be located within the cell, although there is evidence that the enzyme is also associated with the inner surface of the cell membrane (Tsou et al., 1998; Morozov et al., 2004; Oddi et al., 2005). The simplest suggestion is that FAAH acts to ‘drive' the uptake by maintaining the extracellular/intracellular AEA gradient, although this may be an indirect result of the removal of AEA from intracellular carriers that have a limited capacity to bind this compound and therefore become saturated (McFarland and Barker, 2004).
A final observation is that the degree of uptake remaining after inhibition of FAAH was much the same for all the four cell types. This would suggest that either the processes involved in the FAAH-independent cellular accumulation of AEA are rather universal, or alternatively that several different proteins can act as carriers, so that differences in individual expression patterns between cells are minimized. Given that AEA crosses biological membranes very rapidly (Bojesen and Hansen, 2005), the most likely site for such processes are intracellular, although binding to cell surface receptors that utilize AEA (such as the CB1 receptor, Ortega-Gutiérrez et al., 2004; see also Ruiz-Llorente et al., 2004, although these latter authors used high (50 μM) concentrations of antagonists) may contribute in some cells to the apparent accumulation of AEA. Hillard and Jarrahian (2000) suggested that intracellular sequestration of the AEA would provide an explanation for the ability of AEA to be concentrated within cells. Certainly, such a process would be likely to be common to all cells. Whether this process requires a facilitated transport mechanism awaits elucidation.
Acknowledgments
We are grateful to Ingrid Persson and Britt Jacobsson for expert technical assistance. The research was supported by grants from the Swedish Research Council (Grant no. 12158, medicine) and the Research Funds of the Medical Faculty, Umeå University.
Abbreviations
- AEA
anandamide (arachidonoylethanolamide)
- BSA
bovine serum albumin
- FAAH
fatty acid amide hydrolase
- KRH buffer
Krebs–Ringer HEPES buffer
- LY2183240
5-biphenyl-4-ylmethyl-tetrazole-1-carboxylic acid dimethylamide
- OA
oleic acid
- OMDM-1 and OMDM-2
(S)- and (R)-N-(1-(4-hydroxyphenyl)-2-hydroxyethyl)oleamide
- URB597
(3′-(aminocarbamoyl)[1,1′-biphenyl]-3-yl)-cyclohexylcarbamate
Conflict of interest
The authors state no conflict of interest.
References
- Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S-i, Itoh N, et al. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- Alexander JP, Cravatt BF. The putative endocannabinoid transport blocker LY2183240 is a potent inhibitor of FAAH and several other brain serine hydrolases. J Am Chem Soc. 2006;128:9699–9704. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]
- Bari M, Paradisi A, Pasquariello N, Maccarrone M. Cholesterol-dependent modulation of type 1 cannabinoid receptors in nerve cells. J Neurosci Res. 2005;81:275–283. doi: 10.1002/jnr.20546. [DOI] [PubMed] [Google Scholar]
- Bisogno T, Maurelli S, Melck D, De Petrocellis L, Di Marzo V. Biosynthesis, uptake, and degradation of anandamide and palmitoylethanolamide in leukocytes. J Biol Chem. 1997;272:3315–3323. doi: 10.1074/jbc.272.6.3315. [DOI] [PubMed] [Google Scholar]
- Bojesen IN, Hansen HS. Membrane transport of anandamide through resealed human red blood cell membranes. J Lipid Res. 2005;46:1652–1659. doi: 10.1194/jlr.M400498-JLR200. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Wilson SJ, Barbier AJ, Fowler CJ. A simple stopped assay for fatty acid amide hydrolase avoiding the use of a chloroform extraction phase. J Biochem Biophys Methods. 2004;60:171–177. doi: 10.1016/j.jbbm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Cayabyab FS, Schlichter LC. Regulation of an ERG K+ current by Src tyrosine kinase. J Biol Chem. 2002;277:13673–13681. doi: 10.1074/jbc.M108211200. [DOI] [PubMed] [Google Scholar]
- Couet J, Li S, Okamoto T, Scherer PE, Lisanti MP. Molecular and cellular biology of caveolae. Paradoxes and plasticities. Trends Cardiovasc Med. 1997;7:103–110. doi: 10.1016/S1050-1738(97)00001-7. [DOI] [PubMed] [Google Scholar]
- Deutsch DG, Chin SA. Enzymatic synthesis and degradation of anandamide, a cannabinoid receptor agonist. Biochem Pharmacol. 1993;46:791–796. doi: 10.1016/0006-2952(93)90486-g. [DOI] [PubMed] [Google Scholar]
- Deutsch DG, Glaser ST, Howell JM, Kunz JS, Puffenbarger RA, Hillard CJ, et al. The cellular uptake of anandamide is coupled to its breakdown by fatty-acid amide hydrolase. J Biol Chem. 2001;276:6967–6973. doi: 10.1074/jbc.M003161200. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz J-C, et al. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- Dickason-Chesterfield AK, Kidd SR, Moore SA, Schaus JM, Liu B, Nomikos GG, et al. Pharmacological characterization of endocannabinoid transport and fatty acid amide hydrolase inhibitors. Cell Mol Neurobiol. 2006;26:407–423. doi: 10.1007/s10571-006-9072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal R, Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974;139:715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperi V, Fezza F, Pasquariello N, Bari M, Oddi S, Finazzi Agrò A, et al. Endocannabinoids in adipocytes during differentation and their role in glucose uptake. Cell Mol Life Sci. 2007;64:219–229. doi: 10.1007/s00018-006-6445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser ST, Kaczocha M, Deutsch DG. Anandamide transport: a critical review. Life Sci. 2005;77:1584–1604. doi: 10.1016/j.lfs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Harmon AW, Patel YM, Harp JB. Genistein inhibits CCAAT/enhancer-binding protein ß /C/EBPß) activity and 3T3-L1 adipogenesis by increasing C/EBP homologous protein expression. Biochem J. 2002;367:203–208. doi: 10.1042/BJ20020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillard CJ, Jarrahian A. The movement of N-arachidonoylethanolamine (anandamide) across cellular membranes. Chem Phys Lipids. 2000;108:123–134. doi: 10.1016/s0009-3084(00)00191-2. [DOI] [PubMed] [Google Scholar]
- Jeong HY, Son SM, Kim YK, Yun MR, Lee SM, Kim CD. Tyrosine kinase-mediated activation of NAD(P)H oxidase enhances proliferative capacity of diabetic vascular smooth muscle cells. Life Sci. 2005;76:1747–1757. doi: 10.1016/j.lfs.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Kaczocha M, Hermann A, Glaser ST, Bojesen IN, Deutsch DG. Anandamide uptake is consistent with rate-limited diffusion and is regulated by the degree of its hydrolysis by fatty acid amide hydrolase. J Biol Chem. 2006;281:9066–9075. doi: 10.1074/jbc.M509721200. [DOI] [PubMed] [Google Scholar]
- Karlsson M, Påhlsson C, Fowler CJ. Reversible, temperature-dependent, and AM404-inhibitable adsorption of anandamide to cell culture wells as a confounding factor in release experiments. Eur J Pharm Sci. 2004;22:181–189. doi: 10.1016/j.ejps.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Kathuria S, Gaetani S, Fegley D, Valiño F, Duranti A, Tontini A, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- McFarland MJ, Barker EL. Anandamide transport. Pharmacol Ther. 2004;104:117–135. doi: 10.1016/j.pharmthera.2004.07.008. [DOI] [PubMed] [Google Scholar]
- McFarland MJ, Porter AC, Rakhshan FR, Rawat DS, Gibbs RA, Barker EL. A role for caveolae/lipid rafts in the uptake and recycling of the endogenous cannabinoid anandamide. J Biol Chem. 2004;279:41991–41997. doi: 10.1074/jbc.M407250200. [DOI] [PubMed] [Google Scholar]
- Morozov YM, Ben-Ari Y, Freund TF. The spatial and temporal pattern of fatty acid amide hydrolase expression in rat hippocampus during postnatal development. Eur J Neurosci. 2004;20:459–466. doi: 10.1111/j.1460-9568.2004.03507.x. [DOI] [PubMed] [Google Scholar]
- Nichols B. Caveosomes and endocytosis of lipid rafts. J Cell Sci. 2003;116:4707–4714. doi: 10.1242/jcs.00840. [DOI] [PubMed] [Google Scholar]
- Oddi S, Bari M, Battista N, Barsacchi D, Cozzani I, Maccarrone M. Confocal microscopy and biochemical analysis reveals spatial and functional separation between anandamide uptake and hydrolysis in human keratinocytes. Cell Mol Life Sci. 2005;62:386–395. doi: 10.1007/s00018-004-4446-8. [DOI] [PubMed] [Google Scholar]
- Ortega-Gutiérrez S, Hawkins EG, Viso A, López-Rodríguez ML, Cravatt BF. Comparison of anandamide transport in FAAH wild-type and knockout neurons: evidence for contributions by both FAAH and the CB1 receptor to anandamide uptake. Biochemistry. 2004;43:8184–8190. doi: 10.1021/bi049395f. [DOI] [PubMed] [Google Scholar]
- Paylor B, Holt S, Fowler CJ.The potency of the fatty acid amide hydrolase inhibitor URB597 is dependent upon the assay pH Pharmacol Res 200654481–485.Erratum published in Pharmacol Res 55:80 (2007) [DOI] [PubMed] [Google Scholar]
- Rakhshan F, Day TA, Blakely RD, Barker EL. Carrier-mediated uptake of the endogenous cannabinoid anandamide in RBL2H3 cells. J Pharmacol Exp Ther. 2000;292:960–967. [PubMed] [Google Scholar]
- Ramakrishnan M, Kenoth R, Kamlekar RK, Chandra MS, Radhakrishnan TP, Swamy MJ. N-Myristoylethanolamine-cholesterol (1:1) complex: first evidence from differential scanning calorimetry, fast-atom-bombardment mass spectrometry and computational modelling. FEBS Letts. 2002;531:343–347. doi: 10.1016/s0014-5793(02)03553-6. [DOI] [PubMed] [Google Scholar]
- Roig J, Traugh JA. p21-activated protein kinase γ-PAK is activated by ionizing radiation and other DNA-damaging agents. Similarities and differences to α-PAK. J Biol Chem. 1999;274:31119–31122. doi: 10.1074/jbc.274.44.31119. [DOI] [PubMed] [Google Scholar]
- Ruiz-Llorente L, Ortega-Gutiérrez S, Viso A, Sánchez MG, Sánchez AM, Fernández C, et al. Characterization of an anandamide degradation system in prostate epithelial PC-3 cells: synthesis of new transporter inhibitors as tools for this study. Br J Pharmacol. 2004;141:457–467. doi: 10.1038/sj.bjp.0705628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Fowler CJ. Measurement of saturable and non-saturable components of anandamide uptake into P19 embryonic carcinoma cells in the presence of fatty acid-free bovine serum albumin. Chem Phys Lipids. 2005;134:131–139. doi: 10.1016/j.chemphyslip.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Schmid PC, Zuzarte-Augustin ML, Schmid HHO. Properties of rat liver N-acylethanolamine amidohydrolase. J Biol Chem. 1985;260:14145–14149. [PubMed] [Google Scholar]
- Skogseth H, Larsson E, Halgunset J. Inhibitors of tyrosine kinase inhibit the production of urokinase plasminogen activator in human prostatic cancer cells. APMIS. 2005;113:332–339. doi: 10.1111/j.1600-0463.2005.apm_113504.x. [DOI] [PubMed] [Google Scholar]
- Thors L, Alajakku K, Fowler CJ. The ‘specific' tyrosine kinase inhibitor genistein inhibits the enzymic hydrolysis of anandamide: implications for anandamide uptake. Br J Pharmacol. 2007;150:951–960. doi: 10.1038/sj.bjp.0707172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thors L, Fowler CJ. Is there a temperature-dependent uptake of anandamide into cells. Br J Pharmacol. 2006;149:173–181. doi: 10.1038/sj.bjp.0706831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou K, Nogueron MI, Muthian S, Sañudo-Peña MC, Hillard CJ, Deutsch DG, et al. Fatty acid amide hydrolase is located preferentially in large neurons in the rat central nervous system as revealed by immunochemistry. Neurosci Letts. 1998;254:137–140. doi: 10.1016/s0304-3940(98)00700-9. [DOI] [PubMed] [Google Scholar]
- Westermann M, Steiniger F, Richter W. Belt-like localisation of caveolin in deep caveolae and its re-distribution after cholesterol depletion. Histochem Cell Biol. 2005;123:613–620. doi: 10.1007/s00418-004-0750-5. [DOI] [PubMed] [Google Scholar]