Abstract
A meta-analysis, unlike a literature review, synthesizes previous studies into new results. Pooled data from 211 studies measured ligand binding affinities at human (Hs) or rat (Rn) cannabinoid receptors CB1 and CB2. Cochrane methods were modified for this non-clinical analysis. Meta-regression detected data heterogeneity arising from methodological factors: use of sectioned tissues, lack of PMSF and choice of radioligand. Native brain tissues exhibited greater affinity (lower nM) than transfected cells, but the trend fell short of significance, as did the trend between centrifugation and filtration methods. Correcting for heterogeneity, mean Ki values for Δ9-tetrahydrocannabinol differed significantly between HsCB1 and RnCB1 (25.1 and 42.6 nM, respectively) but not between HsCB1 and HsCB2 (25.1 and 35.2). Mean Kd values for HsCB1, RnCB1 and HsCB2 of CP55,940 (2.5, 0.98, 0.92) and WIN55,212-2 (16.7, 2.4, 3.7) differed between HsCB1 and RnCB1 and between HsCB1 and HsCB2. SR141716A differed between HsCB1 and RnCB1 (2.9 and 1.0 nM). Anandamide at HsCB1, RnCB1 and HsCB2 (239.2, 87.7, 439.5) fell short of statistical differences due to heterogeneity. We consider these Kd and Ki values to be the most valid estimates in the literature. Sensitivity analyses did not support the numerical validity of cannabidiol, cannabinol, 2-arachidonoyl glycerol and all ligands at RnCB2. Aggregate rank order analysis of CB1 distribution in the brain (pooled from 119 autoradiographic, immunohistochemical and in situ hybridization studies) showed denser HsCB1 expression in cognitive regions (cerebral cortex) compared to RnCB1, which was relatively richer in movement-associated areas (cerebellum, caudate-putamen). Implications of interspecies differences are discussed.
Keywords: anandamide, cannabinoid, cannabis, directed molecular evolution, endocannabinoid, meta-analysis, pharmacogenetics, species specificity, structure-activity relationship, tetrahydrocannabinol
Introduction
Δ9-Tetrahydrocannabinol (THC) primarily affects animals via cannabinoid receptors type 1 and 2 (CB1 and CB2). CB1 and CB2 activate intracellular G-proteins, which transduce signal to a variety of effectors, such as ion channels, adenylyl cyclase, phospholipase C and the mitogen-activated protein kinase cascade (Howlett, 2002). The first step in this signal transduction involves the binding of a ligand to a receptor. Receptor–ligand binding is measured as affinity, expressed in molar units, usually in a nanomolar range (Pertwee, 2005). Researchers studying cannabinoid often perform experiments with an underlying null hypothesis (implicit, if unstated) that receptors are functionally equivalent between species. Method sections in published papers often make statements such as ‘Chinese Hamster Ovary (CHO) cells transfected with CB1 receptors,' without identifying the CB1 species. Publications regarding rodent receptors are frequently generalized to human orthologues. A few studies have compared human and rodent receptors, but the studies lacked statistical power. Given the molecular divergences between human and rodent orthologues (McPartland et al., 2006), the null hypothesis seems flawed.
In the absence of an adequate cross-species comparison, we conducted a meta-analysis. Broadly defined, meta-analysis includes any methodology that combines data from independent studies. A valid meta-analysis identifies sources of heterogeneity between studies and manages heterogeneity by placing defined limits upon data selection (Glass et al., 1981). Heterogeneity in cannabinoid receptor–ligand binding studies may arise from methodological factors, such as the use of native tissues versus transfected cells, use of enzyme inhibitors and choice of radioligand. These factors can be explored in a subgroup meta-regression analysis. Meta-regression may determine why affinity values for 2-AG (sn-2-arachidonoyl glycerol), for example, ‘look like a random compilation of numbers' (R Mechoulam, personal communication).
The Cochrane Collaboration (www.cochrane.org) leads meta-analysis research worldwide, although they focus upon healthcare interventions (Higgins and Green, 2005). The Cochrane ‘effect size' or ‘standardized mean difference' pertains to randomized, controlled trials (for instance to the size of a treatment effect between experiment and control) and is not applicable to our literature, where direct interspecies comparisons were rarely measured and controls or placebos were not tested. Non-medical researchers have emphasized the metric for meta-analysis should not be limited to effect sizes (Boyce et al., 2005), which agrees with the theory set out by Cochrane himself (Cochran, 1954). We adopted all relevant aspects of the Cochrane protocol, because of its proven validity and robustness.
This meta-analysis focused on in vitro studies of radioligand binding affinities, as well as the distribution of CB1 receptors in the brain as measured by radioligand, immunohistochemistry and in situ hybridization methods. We have described the studies that met inclusion criteria, explored methodological heterogeneities among the studies, pooled data from appropriate studies and concluded with post hoc sensitivity analyses of our models and methods. From these results, we can draw conclusions regarding phenotypic divergences between orthologues.
Methods
Search strategy and study selection
We briefly describe the analysis here. For details regarding inclusion criteria, heterogeneity tests, subgroup analysis, quality assessment and publication bias, see Supplementary file, Extend Methods at the British Journal of Pharmacology website (http://www.nature.com/bjp). MEDLINE was searched for articles published in any language through December 2006, using the following keywords: affinity, anandamide (AEA), cannabinoid, endocannabinoid, tetrahydrocannabinol, tritiated and 2-arachidonoyl glycerol. Retrieved articles were screened for supporting citations and antecedent sources were retrieved. Investigators of original studies were contacted to obtain clarifications and missing data. Three groups of reviewers independently considered studies for inclusion, and resolved disagreements by consensus. To be accepted for analysis, an article had to meet the following criteria: (1) the study examined CB1 or CB2 of human (Homo sapiens, Hs) or rat (Rattus norvegicus, Rn) origin. For sensitivity analyses, we also included studies of rhesus macaque (Macaca fascicularis, Mf) and mouse (Mus musculus, Mm). Data were limited to normal wild types. Studies of chimeric cell lines were excluded (for example, F-11 and NG108-15 cells), as were studies of tissues with unidentified receptors (for example, foetal lung fibroblasts), (2) studies were limited to nine frequently tested cannabinoids: N-arachidonoyl ethanolamine (AEA), cannabidiol (CBD), cannabinol (CBN), CP55,940, HU210, SR141716A, THC, WIN55,212-2 and 2-AG, (3) studies were limited to two aspects of cannabinoid ligand binding:
CB1 and CB2 affinity studies that reported ligand Kd or Ki, measured in molar units. Fifteen IC50 studies were also included after investigators provided information that enabled conversion of IC50 to Ki using the Cheng–Prusoff equation.
CB1 receptor distribution studies, limited to: (i) CB1 studies of the brain, excluding spinal cord, retina and the peripheral nervous system; (ii) studies of adult animals; (iii) studies that used radioligand binding, immunohistochemistry or in situ hybridization methods.
Data extraction
We conducted a prospective meta-analysis, where studies were identified and determined to be eligible before the results were synthesized. Reviewers used piloted, standardized data extraction sheets. The following data were extracted from affinity studies: the species of CB1 or CB2 receptor, each ligand and its mean Kd and/or Ki, sample size, sample variance and methodological covariates. Five methodological covariates were selected for data extraction, chosen by a priori hypotheses: (1) use of brain homogenates versus brain sections; (2) use of phenylmethylsulphonyl fluoride (PMSF) to prevent the breakdown of AEA by catabolic enzymes; (3) use of centrifugation versus rapid filtration for the separation of free and bound radioligand; (4) differences between Kd and Ki values, and Ki variance, due to the use of dissimilar radioligands; (5) use of native tissues versus heterologously expressed systems (transfected cells).
For the meta-analysis of CB1 distribution studies, extracted data included the following: the species of CB1, the mean receptor density per brain region, study size and several methodological covariates: (1) whether the study used radioligand binding, immunohistochemistry or in situ hybridization; (2) which probe was used (that is, which radioligand, tagged antibody or labelled oligonucleotide); (3) whether the study measured receptor density from autoradiographs or from dissected brain regions. Decisions regarding data extraction met consensus before data were extracted for synthesis. When more than one publication described the results of a single experiment, we extracted data from the publication with the most complete information regarding that experiment.
Data synthesis
Data were synthesized quantitatively or qualitatively. Quantitatively, cannabinoid affinity at four receptors (HsCB1, RnCB1, HsCB2, RnCB2) was synthesized twice, as a pooled mean and as a pooled weighted mean. The weighted mean adjusted each study's mean by its inverse variance, because larger studies with less variance should carry more ‘weight' in a meta-analysis. To determine whether pooling was statistically appropriate, the coefficient of variation (CV) was determined for each pooled mean. CV measures data dispersion of a probability distribution, defined as the ratio of the standard deviation to the mean (Reed et al., 2002). We applied the Cochrane ‘skew test' to the CV (Higgins and Green, 2005): a skewed mean with excessive heterogeneity was defined by a CV⩾1 (that is, standard deviation⩾mean). For means with excessive heterogeneity, we applied a random effects model (Higgins and Green, 2005), and performed meta-regressions upon subgroups, based on methodological covariates described above. Sources of heterogeneity identified via meta-regressions were removed from synthesis. To these adjusted means, we reapplied the CV-skew test. Persistently skewed means were submitted to Grubb's test, using an outlier calculator (GraphPad, www.graphpad.com/quickcalcs/Grubbs1.cfm). Studies with data outliers were inspected for methodological irregularities, and in some cases were also removed from synthesis, as noted in Supplementary Table 1.
The distribution data for CB1 receptors in brain were synthesized as scalar transformations (ranked orders) or as narrative comparisons. Scalar transformations were performed upon every study that reported CB1 density in ⩾2 brain regions. The relative density of each brain region was ranked (arrayed) from highest value to lowest value. Rank orders from individual studies were then aggregated using a bubble-sort algorithm (Sese and Morishita, 2001). Narrative comparisons were used to describe aspects of CB1 distribution that were not quantified in original studies.
Sensitivity analysis
A series of analyses were performed to judge a priori hypotheses and the robustness of the results. We began by identifying additional methodological covariates based upon post hoc observations. The validity and numerical precision of pooled Ki and Kd values were tested, using three approaches: (1) Ki and Kd values in original studies were transformed into aggregate rank orders. For every study that examined ⩾2 ligands per receptor, ligands were ranked from highest to lowest affinity; individual studies were then aggregated using a bubble-sort algorithm, (2) pooled Ki and Kd values for Hs and Rn were compared to studies in the literature that made direct interspecies comparisons, (3) affinity values for Rn were compared to affinity values from Mm studies. Rn and Mm cross-species differences undoubtedly exist, but the rodents share similarity on a molecular level—RnCB1 and MmCB1 are 99.8% identical (diverging at one residue out of 473 amino acids), compared with RnCB1 and HsCB1 differing at 12 residues plus a codon deletion.
Sensitivity analyses for CB1 brain distribution data also utilized cross-species comparisons. Regional rank orders for RnCB1 were examined for consistency with MmCB1 rank orders, and HsCB1 rank orders were examined for consistency with two other primates, rhesus macaque (Macaca fascicularis, Mf) and baboon (Papio hamadryas, Ph).
Results of data extraction and synthesis
Receptor–ligand affinity
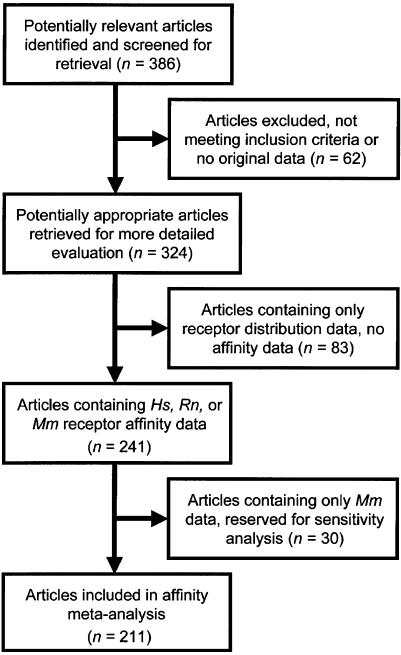
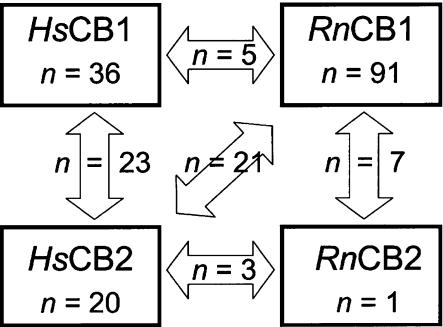
A flow diagram of article selection for receptor–ligand affinity meta-analysis is shown in Figure 1. A total of 211 affinity studies concerning Hs and Rn orthologues met the inclusion criteria, and collectively reported 676 affinity values, presented in Supplementary Table 1. Most studies measured affinity at one receptor, but some studies compared ligand affinities between orthologues or paralogues (Figure 2). Thirty studies of Mm orthologues met the inclusion criteria, presented in Supplementary Table 2. No affinity studies of Mf were identified that met inclusion criteria.
Figure 1.
Flow diagram of article selection for ligand affinity meta-analysis.
Figure 2.
Diagram showing all the 211 articles included in the receptor–ligand affinity meta-analysis. The numbers of articles that tested ligand affinity at one receptor are indicated in boxes. The numbers of articles that compared ligand affinity at two receptors are indicated in arrows. Additionally, four articles compared all four receptors (not illustrated).
Weighted means were compromised by missing data. Many studies omitted information regarding sample size or sample variance, or the studies did not state whether their variances were s.d.s or s.e.s (see notes for each study in Supplementary Tables 1 and 2). Therefore, we conducted heterogeneity (CV-skew) tests on unweighted means. Results from the CV-skew tests (Supplementary Table 3) indicated a significant degree of heterogeneity among studies, making them inappropriate to pool in a fixed-effects model. This supported our decision to employ subgroup meta-regression.
Meta-regression demonstrated statistically significant differences between studies that used brain sections versus studies that used brain homogenates (Table 1). Native tissues produced lower Ki values than transfected cells, but differences fell short of significance. Centrifugation assays produced lower Ki values than filtration assays, but differences were not significant (except for AEA, see Discussion). Kd values were significantly different from Ki values, and the difference was primarily due to Ki studies that used [3H]SR141716A. The use of PMSF significantly lowered the Ki of AEA in brain tissues, but PMSF did not change Ki values in transfected cells.
Table 1.
Subgroup meta-regression: effects of methodological factors upon mean affinity values
| Methodological factora | Affinity mean (nM), standard error, sample size | Difference between means |
|---|---|---|
| Brain homogenates versus | 3.4±1.1 (n=77) | Statistical difference, P=0.0001 |
| Brain sections | 20.8±3.1 (n=10) | |
| Native tissues versus | 1.6±0.76 (n=13) | No difference, P=0.12 |
| Transfected cells | 3.2±0.45 (n=38) | |
| Filtration of radioligand versus | 1.0±0.10 (n=56) | No difference, P=0.28 |
| Centrifugation of radioligand | 0.87±0.10 (n=20) | |
| Kd versus | 0.98±0.12 (n=51) | Statistical difference, P=0.0001 |
| Ki | 8.2±1.5 (n=26) | |
|
Ki values of CP55,940 obtained using: [3H]CP55,940 [3H]SR141716A [3H]WIN55212-2 [3H]HU243 |
1.1±4.55 (n=14) 19.4±3.41 (n=25) (a) 0.89±4.55 (n=14) 1.3±7.62 (n=5) |
(a)=statistical difference to all other ligands, P=0.002 |
| Brain tissue with PMSF versus | 205.8±301.9 (n=30) | Statistical difference, P=0.0001 |
| Brain tissue without PMSF | 2500.9±413.3 (n=16) | |
| CB1-transfected cells with PMSF versus | 196.5±123.1 (n=6) | No difference, P=0.29 |
| CB1-transfected cells without PMSF | 396.4±100.5 (n=9) | |
| CB2-transfected cells with PMSF versus | 477.9±385.5 (n=9) | No difference, P=0.45 |
| CB2-transfected cells without PMSF | 916.1±348.7 (n=11) |
Abbreviation: PMSF, phenylmethylsulphonyl fluoride.
All meta-regressions were performed on the single ligand and receptor that provided the greatest amount of extractable data; CP55,940 at RnCB1 data were used for all methodological factors, except the three PMSF versus no PMSF meta-regressions. For the PMSF versus no PMSF analyses, the brain tissue comparison used AEA at RnCB1 data, the CB1-transfected cells used AEA at HsCB1 data and the CB2-transfected cells used AEA at HsCB2 data. Statistical significances (P-values) were calculated with Mann–Whitney U-test (Systat; Evanston, IL, USA).
Based on meta-regression results, we withdrew heterogeneous data derived from studies that used sectioned brain tissue, studies of AEA in brain homogenates that lacked PMSF and Ki studies that used [3H]SR141716A. Adjusted means were submitted to Grubb's test and several outliers were removed from synthesis (Supplementary Table 1). The remaining data were utilized for calculating pooled means. Mean Kd values for [3H]CP55,940, [3H]WIN55212-2 and [3H]SR141716A are presented in Table 2. Mean Kd values presented in bold typeface had numerical validities confirmed by sensitivity analyses (described below). Differences between validated means were tested for statistical significance, accompanied by Kd ratios. A Kd ratio <1 indicated that ligand affinity was greater for the Hs orthologue (a small Kd value indicating high affinity). Conversely, a Kd ratio >1 indicated that ligand affinity was greater for the Rn orthologue. Several interspecies differences were statistically significant. Mean Ki values for THC, CBD, CBN, AEA, 2-AG and HU210 are presented in Table 3. Robust data at HsCB1 and RnCB1 receptors demonstrated interspecies differences for THC, whereas AEA was not significantly different between HsCB1 and RnCB1 (despite a relatively large Kd ratio of 2.7), due to remaining heterogeneity among studies. Data regarding CBD, CBN, 2-AG, and all the comparisons at RnCB2 were less robust, hampered by heterogeneity and small sample sizes. We also compared paralogues HsCB1 and HsCB2 and recovered significant differences (Table 3); comparisons between paralogues RnCB1 and RnCB2 were hampered again by small samples sizes at RnCB2.
Table 2.
Meta-analytic Kd values for radioligands at human (Hs) and rat (Rn) cannabinoid receptors
| Radioligand | HsCB1 | RnCB1 | HsCB2 | RnCB2 | HsCB1 | HsCB2 | RnCB1 | RnCB2 |
|---|---|---|---|---|---|---|---|---|
| Kd±s.e. | Kd±s.e. | Kd±s.e. | Kd±s.e. | Kd±s.e. | Kd±s.e. | Kd±s.e. | Kd±s.e. | |
| [3H]CP55,940 | 2.5±0.39 (n=28) | 0.98±0.12 (n=51) | 0.92±0.16 (n=18) | 0.84±0.30 (n=8) | 2.5±0.39 (n=28) | 0.92±0.16 (n=18) | 0.98±0.12 (n=51) | 0.84±0.30 (n=8) |
| Difference between species, P=0.001; Kd ratio=2.6 | No difference, P=0.32; Kd ratio=1.1 | Difference between CB1 and CB2, P=0.001; Kd ratio=2.7 | No difference, P=0.19; Kd ratio=1.2 | |||||
| [3H]WIN55212 | 16.7±1.71 (n=6) | 2.4±0.348 (n=13) | 3.7±1.19 (n=7) | 35.6 (n=2) | 16.7±1.71 (n=6) | 3.7±1.19 (n=7) | 2.4±0.348 (n=13) | ND |
| Difference between species, P=0.001; Kd ratio=7.0 | Difference between CB1 and CB2, P=0.003; Kd ratio=4.5 | |||||||
| [3H]SR141716A | 2.9±0.54 (n=10) | 1.0±0.22 (n=19) | ND | ND | 2.9±0.54 (n=10) | ND | 1.0±0.22 (n=19) | ND |
| Difference between species, P=0.002; Kd ratio=2.9 | ||||||||
Abbreviations: Hs, Homo sapiens; Rn, Rattus norvegicus.
The results in the table are expressed as Kd±s.e. (nM) and the number of studies extracted for determination of pooled means. Means in bold typeface had their numerical validity confirmed by sensitivity analyses. Differences between validated means were tested for statistical significance with Mann–Whitney U-test (Systat; Evanston, IL, USA). ND=no Kd data in Supplementary Table 1.
Table 3.
Meta-analytic Ki values for ligands at human (Hs) and rat (Rn) cannabinoid receptors
| Ligand | HsCB1 | RnCB1 | HsCB2 | RnCB2 | HsCB1 | HsCB2 | RnCB1 | RnCB2 |
|---|---|---|---|---|---|---|---|---|
| Ki±s.e. | Ki±s.e. | Ki±s.e. | Ki±s.e. | Ki±s.e. | Ki±s.e. | Ki±s.e. | Kd±s.e. | |
| THC | 25.1±5.54 (n=16) | 42.6±5.01 (n=18) | 35.2±5.86 (n=16) | 13.0±7.70 (n=3) | 25.1±5.54 (n=16) | 35.2±5.86 (n=16) | 42.6±5.01 (n=18) | 13.0±7.70 (n=3) |
| Difference between species, P=0.010; Ki ratio=0.59 | No difference between CB1 and CB2, P=0.12; Ki ratio=0.71 | |||||||
| CBD | ND | 2210.5±558.08 (n=6) | 2860 (n=1) | 1000 (n=1) | ND | 2860 (n=1) | 2210.5±558.08 (n=6) | 1000 (n=1) |
| CBN | 525.3±308.1 (n=3) | 368.0±121.1 (n=8) | 168.2±32.16 (n=7) | ND | 525.3±308.1 (n=3) | 168.2±32.16 (n=7) | 368.0±121.1 (n=8) | ND |
| AEA | 239.2±61.77 (n=16) | 87.7±11.32 (n=26) | 439.5±95.89 (n=22) | 267.8±67.94 (n=5) | 239.2±61.77 (n=16) | 439.5±95.89 (n=22) | 87.7±11.32 (n=26) | 267.8±67.94 (n=5) |
| No difference between species, P=0.23; Ki ratio=2.7 | No difference between CB1 and CB2, P=0.06; Ki ratio=0.54 | |||||||
| 2-AG | 3423.6±3288.24 (n=3) | 1180.5±538.59 (n=4) | 1193.8±327.71 (n=9) | 1900.0±1800.0 (n=2) | 3423.6±3288.24 (n=3) | 1193.8±327.71 (n=9) | 1180.5±538.59 (n=4) | 1900.0±1800.0 (n=2) |
| HU210 | 0.25±0.069 (n=9) | 0.34±0.102 (n=7) | 0.40±0.095 (n=15) | ND | 0.25±0.069 (n=9) | 0.40±0.095 (n=15) | 0.34±0.102 (n=7) | ND |
| No difference between species, P=0.49; Ki ratio=0.74 | No difference between CB1 and CB2, P=0.34; Ki ratio=0.63 | |||||||
Abbreviations: AEA, anandamide; CB1, cannabinoid receptor subtype 1; CB2, cannabinoid receptor subtype 2; CBD, cannabidiol; cannabinol; Hs, Homo sapiens; Rn, Rattus norvegicus; 2-AG, sn-2 arachidonoyl glycerol.
The results in the table are expressed as Ki±s.e. (nM) and the number of studies extracted for determination of pooled means. Means in bold typeface had their numerical validity confirmed by sensitivity analyses. Differences between validated means were tested for statistical significance with Mann–Whitney U-test (Systat; Evanston, IL, USA). ND=no Ki data in Supplementary Table 1.
CB1 receptor distribution
A total of 119 CB1 receptor distribution studies met the inclusion criteria (87 studies reported only CB1 distribution data, the remaining 32 studies also contained affinity data), cited and extracted in Supplementary Table 4. A subset of publications compared CB1 density in ⩾2 brain regions, enabling us to transform relative densities into rank orders, arrayed from the highest density to the lowest density. Aggregate rank orders derived from this subset of publications are presented in Supplementary Table 5. Heterogeneity tests could not be calculated for receptor distribution data, because the results were qualitative (rank orders). Subgroup analysis was based on the methodology presented in Supplementary Table 5. Autoradiography, immunohistochemistry and in situ hybridization methods varied in their sensitivity and level of resolution. Results of autoradiographic studies broadly agreed with each other, irrespective of the radioligand used. Autoradiographic study results generally agreed with studies that used regional dissection and studies that used immunohistochemical methods, allowing for the high degree of variability in brain tissue and the low sample sizes. Many immunohistochemical studies reported subcellular or ultrastructural findings, but did not provide regional comparisons for our meta-analysis. Rank orders from in situ hybridization studies differed from rank orders reported in autoradiographic and immunohistochemical studies (see Discussion). Therefore, in situ hybridization studies were excluded and the remaining studies (autoradiography, regional dissection and immunocytochemistry) were assembled in the following rank orders:
HsCB1
Substantia nigra>globus pallidus internus>globus pallidus externus>dentate gyrus>hippocampus>cerebral cortex>putamen>caudate>cerebellum>amygdala>thalamus=hypothalamus.
RnCB1
Substantia nigra>globus pallidus externus>entopeduncular nucleus (homologue of GPi)>cerebellum>hippocampus=caudate-putamen (striatum)>dentate gyrus>cerebral cortex>amygdala>hypothalamus>thalamus.
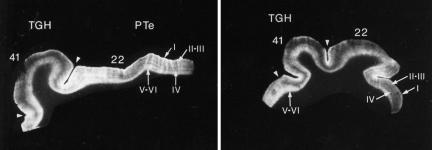
Narrative comparisons were employed by some researchers to describe aspects of CB1 distribution that were not quantified: Glass et al. (1997) reported that CB1 expression in human forebrain and thalamus showed greater complexities than CB1 expression patterns seen in rat brain. The human cortex in Brodmann's area 22 (the planum temporale, part of Wernicke's area, the temporal lobe) showed greater CB1 density in the left hemisphere than the right hemisphere (Figure 3), whereas rat studies showed no lateralization. Harkany et al. (2005) reported significant area-specific differences in expression of CB1 in primate (lemur, Microcebus murinus) compared with a largely homogenous distribution of CB1 afferents in the rat. Eggan and Lewis (2006) reported similar CB1 distribution within the neocortex of the human and the long-tailed macaque (Macaca fascicularis).
Figure 3.
Autoradiograms of human cortex with [3H]CP55,940, showing greater CB1 density (brightness) in the left hemisphere (left panel) compared to the right hemisphere (right panel). In both panels, Brodmann's area 22, labelled ‘22,' lies to the right of the transverse gyrus of Heschl (TGH), marked by arrow heads. Reproduced from Glass (1995), with permission. CB1, cannabinoid receptor subtype 1; TGH, transverse gyrus of Heschl.
Results of sensitivity testing
Additional methodological covariates
For affinity studies, post hoc observations identified eight additional methodological covariates:
Ligand affinity at CB1 may vary among specific brain regions, although regional differences have not been significant in studies reporting small sample sizes (Rodriguez de Fonseca et al., 1994; Breivogel et al., 1997).
In native brain tissues, ligand affinity may vary depending on the usage of synaptosome fractions (primarily presynaptic nerve terminals) versus whole-membrane homogenates, which contain pre- and postsynaptic nerve terminals (Steffens et al., 2004).
Rodent studies used a variety of inbred lines; ligand affinity may vary among mouse strains (Hungund and Basavarajappa, 2000) and rat strains (Arnold et al., 2001; Hoffman et al., 2005).
Affinities for AEA and WIN55212-2 may have been affected by additional binding targets in mouse brain (Breivogel et al., 2001) and presumably human brain.
Affinity may vary among transfected cell types. CB1 receptors are expressed at the cell surface in some recombinant systems, such as AtT-20 cells, HEK 293 cells and Xenopus oocytes, yet are not well expressed in CHO and COS-7 cells (Andersson et al., 2003). However, Felder et al. (1995) reported non-significant differences between AtT-20 and CHO cells.
Studies were included whether or not they used methylarachidonoyl fluorophosphonate, which may be ≈70 000-fold more potent than PMSF at preventing 2-AG degradation (Savinainen et al., 2003).
Various autoradiographic studies used different ligand concentrations. Lower concentrations of tritiated ligand (that is, closer to ligand Kd) may be sensitive to the coupling state of the receptor and therefore be measuring differences in coupling rather than receptor Bmax.
Studies used a variety of vehicles (for example, ethanol, dimethylsulphoxide, bovine serum albumin or none), detergents (for example, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)), buffer concentrations (particularly Na+ and Mg2+ with agonists) and other reagents, which may have affected affinity data. Studies rarely described methods that prevented photo-oxidation of cannabinoids, which are sensitive to light.
Testing the numerical validity of pooled means
Numerical validity was tested in three ways. First, the pooled means in Tables 2 and 3 were scalar-transposed into rank orders, ranked from the highest affinity (HU210) to the lowest affinity (CBD). These rank orders were compared to rank orders derived from studies that directly examined ⩾2 ligands per receptor. Studies that examined ⩾2 ligands per receptor are presented in Supplementary Table 6. These studies were aggregated using a bubble-sort algorithm, and the aggregate rank orders are presented in Supplementary Table 7. Rank orders from pooled means generally agreed with rank orders from the bubble-sort at HsCB1, RnCB1 and HsCB2, but not at RnCB2 receptors (see comparisons in Supplementary Table 7).
Second, we compared pooled means in Tables 2 and 3 to studies that made direct comparisons between receptors, which were listed in Supplementary Table 1, column seven. Means derived from direct comparisons were presented in Supplementary Table 8. For example, the pooled affinity of THC at HsCB1 versus HsCB2 (Table 3), when expressed as a ratio, was (25.1/35.2 nM=) 0.71. This was consistent with eight studies that directly compared THC at HsCB1 versus HsCB2; the ratios in the eight studies ranged from 0.1 to 1.6, with a mean ratio of 0.75 (Supplementary Table 8). Affinity ratios from pooled means generally agreed with affinity ratios derived from studies that made direct comparisons at HsCB1, RnCB1 and HsCB2, excepting a dearth of data for CBD, CBN and 2-AG. Lack of data at RnCB2 again prevented the validation of pooled means at those receptors, except for CP55,940 and AEA. Direct comparisons of WIN55212-2 diverged widely in the literature; HsCB1 versus HsCB2 ratios varied from 1.3 to 28.5, and HsCB1 versus RnCB1 ratios varied from 0.45 to 70.5 (Supplementary Table 1, column seven).
Third, we tested the precision of pooled means at RnCB1 and RnCB2 by comparing them to MmCB1 and MmCB2 affinity data (Supplementary Table 2). Pooled means of MmCB1 and MmCB2 data are presented in Supplementary Table 9 and compared to RnCB1 and RnCB2 data. This sensitivity analysis agreed with the previous two sensitivity analyses, except for two surprises: the pooled mean for THC at RnCB2 (13.0 nM; Table 3) was supported by a similar pooled mean at MmCB2 (13.4 nM; Supplementary Table 9). The pooled mean for 2-AG at RnCB1 (1180.5 nM; Table 3) was supported by a similar pooled mean at MmCB2 (1626.6 nM; Supplementary Table 9).
Testing the validity of CB1 distribution rank orders
The CB1 distribution rank order for RnCB1 was broadly consistent with the MmCB1 rank order (Supplementary Table 5). The most prominent exception was seen in cerebellum, which ranked fourth in rat and second in mouse. The CB1 distribution rank order for HsCB1 deviated somewhat from that seen in the rhesus macaque, especially regarding the relative rank orders of cerebellum and cerebral cortex (Supplementary Table 5). This sensitivity analysis was hampered, however, by a low sample size in the other primates (n=3), and possible MfCB1 staining artefacts (K Mackie, personal communication).
Discussion
Critics may refute meta-analyses as mixtures of apples and oranges. We feel, however, that it can be a powerful tool to deal with questions not addressed by individual studies. We have collected individual studies and compared them, as a group, to a null hypothesis: orthologues of cannabinoid receptors do not differ between species. Data heterogeneity was obviated by subgroup meta-regression and by putting limits on data selection. Meta-analysis of individual studies provided an interpretive context not available in any single study, and enabled the adjudication of inconsistencies between studies. Meta-analysis also uncovered data lacunae. For example, the affinity of CBD at HsCB1 receptors has never been reported in the peer-reviewed literature. This is a notable oversight, given the recent evaluation of CBD in several human clinical trials (Guy et al., 2004).
Meta-analysis is particularly appropriate when combining studies that have small sample sizes; the median sample size for affinity studies was n=3. Meta-analyses in the medical literature commonly ‘weight' larger studies that have less variance. Our weighted means lacked validity because many affinity studies did not provide the information required for weighting: sample size and sample variance. For this meta-analysis, weighting may not have been particularly advantageous, because 80% of affinity studies used identical sample sizes (n=3) and studies that employed larger samples also reported larger variances.
Receptor–ligand affinity
Despite strict exclusion criteria, our search strategy identified 211 receptor–ligand affinity studies for meta-analysis. In comparison, recent literature reviews have cited an average of 28 studies (Diaz-Laviada and Ruiz-Llorente, 2005; Pertwee, 2005; Demuth and Molleman, 2006). The seeming wealth of information in 211 studies, however, was winnowed by subgroup analysis, then partitioned among nine cannabinoid ligands, and further shared by four receptors (HsCB1, HsCB2, RnCB1 and RnCB2).
Meta-regression identified several methodological covariates that generated heterogeneity in pooled means (Table 1). Studies that used brain sections differed significantly from the majority of studies that used brain homogenates. Use of PMSF significantly lowered the Ki of AEA in brain tissues, but not in transfected cells. Use of PMSF upon spleen tissues could not be tested, because of a paucity of data. For that same reason, we did not test the effects of PMSF upon 2-AG affinity. Use of centrifugation versus filtration did not generate significantly different Ki values in any ligands except AEA in native brain tissues. In that assay, centrifugation produced Ki values that did not vary in the presence or absence of PMSF (originally demonstrated by Sheskin et al., 1997). Mean Kd (saturation binding) values differed from mean Ki (competition binding) values and the difference was primarily due to Ki studies that used [3H]SR141716A (Table 1), as originally demonstrated by Thomas et al. (1998). When unlabelled agonists were assayed with [3H]SR141716A, they produced biphasic competition curves (Houston and Howlett, 1998). Kearn et al. (1999) noted that [3H]SR141716A labelled two populations of receptors. In our meta-regression, [3H]WIN55212-2 produced mean Ki values that differed from mean Ki values of [3H]CP55,940, [3H]HU243 and [3H]BAY38-7271, but this trend fell short of statistical significance. Petitet et al. (1996) first noted this phenomenon; Thomas et al. (2005) attributed differences to WIN55212-2's divergent binding domain.
Meta-regression of native tissues versus heterologous expression systems produced controversial results. At RnCB1, CP55,940 exhibited greater affinity in native versus transfected receptors, although the difference fell short of significance (Table 1). This is surprising, considering the differences in receptor stoichiometry among heterologous expression systems. For example, CB1 receptors are expressed at the cell surface in AtT20 cells, HEK 293 cells and Xenopus oocytes, but not well expressed at the cell surface in CHO cells and COS-7 cells (Andersson et al., 2003). Six studies in the literature made direct comparisons between native versus transfected: five reported slightly greater affinity (but not statistically different) in native tissues (Felder et al., 1992, 1995; Mauler et al., 2002; Thomas et al., 2005; Paugh et al., 2006), whereas Rhee et al. (1997) reported greater affinity in transfected cells (see data in Supplementary Table 1). If native tissue does indeed produce higher affinities than transfected cells, we may be underreporting interspecies differences for ligands that demonstrate greater affinity for Hs than Rn (for example, THC), because RnCB1 studies are primarily assayed in native tissues, whereas HsCB1 studies primarily use transfected cells. Contrarily, ligands with greater affinity at Rn than Hs (for example, WIN55212, CP55,940 and SR141716A) may actually exert less interspecies differences than estimated by our meta-analysis. The interspecies differences reported in our meta-analysis should be confirmed by the gold standard: a large, adequately powered, in vitro comparative study (LeLorier et al., 1997). Interestingly, the Ki of THC at HsCB1 places it around the median affinity for current small-molecule drugs, which is 20 nM (Overington et al., 2006).
Receptor distribution
Autoradiography studies that used different radioligands were broadly consistent with each other. This agreed with Breivogel et al. (1997), who made a direct comparison between [3H]WIN55212-2 and [3H]SR141716A and reported a good correlation (r=0.89) in brain region Bmax values. Studies that used regional dissection showed greater variability in their results, perhaps due to the difficulty of obtaining clean regional dissections. For example, dissected thalamus, a region with low Bmax, may be contaminated by fragments from the entopeduncular nucleus, a region with high Bmax (Breivogel et al., 1997). Immunohistochemistry studies used a variety of tagged antibodies raised against N-terminal or C-terminal regions, but their results were broadly consistent (Supplementary Table 5). In agreement, Harkany et al. (2005) directly compared a variety of N-terminus and C-terminus epitopes and reported nearly identical staining patterns, after correcting for methodological considerations that produced staining artefacts. Conversely, Egertova and Elphick (2000) suggested that antibodies directed against the C-terminus were rendered non-immunoreactive by phosphorylation of amino-acid residues in the C-terminus, and it remains untested whether the transient binding of interacting proteins such as GASP1 (Martini et al., 2007) can alter antibody recognition. Furthermore, antibodies directed against the N-terminus cannot label either splice variant of the CB1 receptor. Thus immunohistochemical approaches risk labelling only a subset of total receptors.
Immunohistochemistry studies broadly correlated with radioligand results, given that antibodies specifically label CB1 receptors, whereas [3H]CP55,940 binds to CB1 as well as CB2 receptors and CB2 receptors may be present in the brain (Van Sickle et al., 2005; Gong et al., 2006). Furthermore, cannabinoid ligands may bind to targets other than CB1 receptors (Breivogel et al., 2001; Pertwee et al., 2005; Baker et al., 2006), although the extent of the contribution of these ‘new' receptors in tissue preparations has yet to be elucidated (Petitet et al., 2006). In situ hybridization studies yielded different rank orders than immunohistochemistry and radioligand studies, due to the fact that labelled oligonucleotides: (1) identified mRNA in brain regions that may not have translated into the corresponding protein product; (2) identified mRNA in cell bodies (perikarya), whereas the corresponding protein product (CB1) translocated to distal sites before inserting into cell membranes. For example, many neurons with cell bodies in the striatum project axons to the globus pallidus and substantia nigra. Thus, high levels of CB1 mRNA were expressed in the striatum and negligible levels were found in the globus pallidus and substantia nigra. And the reverse was true regarding the corresponding protein product (CB1), with high densities in the globus pallidus, entopenducular nucleus and substantia nigra, but only moderate densities in the striatum. In situ hybridization studies used a variety of probes, but their results were broadly consistent with each other (Supplementary Table 5).
Interspecies differences could be discerned in CB1 receptor distribution rank orders: HsCB1 densities were relatively greater in brain regions associated with cognitive function (frontal cortex) and memory (hippocampus), whereas RnCB1 densities were relatively greater in movement-associated areas (cerebellum) and the caudate-putamen. These differences were first described by Herkenham et al. (1990). They noted that human brain, compared with rat (and rhesus, surprisingly), expressed greater CB1 densities in the cortex and amygdala, and lesser densities in the cerebellum. Romero et al. (1998) were the first to show that mouse brain, compared to rat brain, exhibited even greater densities in the cerebellum compared to cortex.
Narrative comparisons in the literature described more subtle interspecies differences. According to Glass et al. (1997), the human cortex showed CB1 enrichment in the left (not the right) planum temporale, which is part of Wernicke's area in the temporal lobe. This region is associated with verbal language development, human communication and musical talent. CB1 in the human forebrain expressed a greater density and complexity than in rat forebrain; this difference likely mediates a THC ‘high' in humans that may be absent in rats. Similarly, CB1 distribution is more elaborate in the human dorsomedial nucleus, an area that mediates emotional tones from the amygdaloid complex and an area that contributes to the formation of human personality. According to Harkany et al. (2005), CB1 architecture was more complex in primate than rodent, in the cingulate and frontal cortex (regions associated with motor control), hippocampus (associated with memory and learning), amygdala (emotional behaviours) and parietotemporal areas (visual and auditory information processing). In contrast, only minor differences between the primate and rodent were seen in basal forebrain territories (associated with olfaction). Eggan and Lewis (2006) compared the human neocortex with that of the long-tailed macaque (Macaca fascicularis). Human CB1 cytoarchitecture was slightly more complex in prefrontal area 46 (associated with the ability to retain and manipulate visual spatial information) and occipital area 17 (the primary visual cortex).
Conclusion
Sensitivity analyses provided confidence in meta-analytic estimates of Kd (CP55,940, WIN55,212-2, SR141716A) and Ki (THC, AEA, HU210) distilled from studies on HsCB1, HsCB2 and RnCB1 receptors. We consider those pooled Kd and Ki values to be the most valid estimates in the literature, able to be generalized to studies that employed tissue homogenates (not tissue sections) and tritiated agonists (not tritiated inverse agonists), applied PMSF in AEA assays and used either filtration or centrifugation of radioligands. Meta-regression observed a trend towards greater affinity at native tissues compared to heterologous expression systems, but fell short of significance. In other words, heterogeneity arising between studies that used native tissues versus transfected cells is likely due to pipetting inaccuracies and other experimental errors, rather than due to the methodological dichotomy. We lack confidence in our results at RnCB2 receptors, as well as the pooled Ki values of 2-AG, CBD and CBN, due to small sample sizes and lack of validation in sensitivity analyses.
Only 0.05 mg kg−1 of THC affects a human (Isbell et al., 1967; Ohlsson et al., 1980), whereas it may take 3 mg kg−1 of THC to affect a rat (Jarbe et al., 1976; Browne and Weissman, 1981; Wiley et al., 1995). Comparing doses across in vivo animal models is controversial, because drug discrimination in the rat and the human perception of ‘high' may not involve the same populations of receptors; furthermore only limited pharmacokinetic data exists for cannabinoids across species. Nevertheless this 60-fold disparity suggests phenotypic differences exist between species. Our meta-analysis confirmed differences in ligand binding between human and rat orthologues of CB1 and CB2 receptors The fact that synthetic cannabinoids (for example, WIN55,212-2) discriminated between human and rat orthologues did not surprise us. After all, ligands have been synthesized for that very purpose (Astolfi et al., 1997), and WIN55,212-2 induces behavioural changes that are strikingly different between species (Haller et al., 2007). The R-enantiomer of AM1241 acts as an agonist at HsCB2, but an inverse agonist at RnCB2, and MmCB2 (Bingham et al., 2007). Species differences in ligand affinity are the norm at vanilloid receptors (Szallasi, 1994; Huang et al., 2002) and melanocortin receptors (Schioth et al., 2005). Phytocannabinoids that discriminate between human and rat orthologues are more interesting from an evolutionary viewpoint, although the concept is not without precedents; Russo et al. (2005) showed that CBD discriminated between human and rat orthologues of the 5-HT1A receptor.
This study did not evaluate functional (efficacy) assays, such as cannabinoid inhibition of cAMP accumulation by forskolin-stimulated adenylyl cyclase, or cannabinoid stimulation of radiolabelled non-hydrolysable guanosine 5′-O-(3-thio)-triphosphate ([35S]GTPγS). Given the disparate affinities for THC and AEA at human versus rat receptors, we hypothesize that efficacies also vary between species. We plan to explore this possibility by conducting our next meta-analysis with efficacy data, to better assess how cannabinoid ligands interact with cannabinoid receptors to evoke responses.
External data objects
Acknowledgments
This work was partially supported by an unrestricted grant from GW Pharmaceuticals, Salisbury, UK. We thank all the scientists, especially Ken Mackie, Lumir Hanuš and Raphael Mechoulam, who responded to our queries regarding their published research.
Abbreviations
- AEA
anandamide
- CB1
cannabinoid receptor subtype 1
- CB2
cannabinoid receptor subtype 2
- CBD
cannabidiol
- CBN
cannabinol
- CV
coefficient of variation
- Hs
human
- Mf
rhesus macaque
- Mm
mouse
- PMSF
phenylmethylsulphonyl fluoride
- Rn
rat
- THC
Δ9-tetrahydrocannabinol
- 2-AG
sn-2 arachidonoyl glycerol
Conflict of interest
The authors state no conflict of interest.
Supplementary Information accompanies the paper on British Journal of Pharmacology website (http://www.nature.com/bjp)
References
- Andersson H, D'Antona AM, Kendall DA, Von Heijne G, Chin CN. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Mol Pharmacol. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- Arnold JC, Topple AN, Mallet PE, Hunt GE, McGregor IS. The distribution of cannabinoid-induced Fos expression in rat brain: differences between the Lewis and Wistar strain. Brain Res. 2001;921:240–255. doi: 10.1016/s0006-8993(01)03127-4. [DOI] [PubMed] [Google Scholar]
- Astolfi M, Patacchini R, Maggi M, Manzini S. Improved discriminatory properties between human and murine tachykinin NK1 receptors of MEN 10930: a new potent and competitive antagonist. Neuropeptides. 1997;31:373–379. doi: 10.1016/s0143-4179(97)90074-3. [DOI] [PubMed] [Google Scholar]
- Baker D, Pryce G, Davies WL, Hiley CR. In silico patent searching reveals a new cannabinoid receptor. Trends Pharmacol Sci. 2006;27:1–4. doi: 10.1016/j.tips.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Bingham B, Jones PG, Uveges AJ, Kotnis S, Lu P, Smith VA, et al. Species-specific in vitro pharmacological effects of the cannabinoid receptor 2 (CB2) selective ligand AM1241 and its resolved enantiomers Br J Pharmacol 2007: 2007 Jun 4 [E-pub ahead of print] [DOI] [PMC free article] [PubMed]
- Boyce MS, Irwin LL, Barker R. Demographic meta-analysis: synthesizing vital rates for spotted owls. J Appl Ecol. 2005;42:38–49. [Google Scholar]
- Breivogel CS, Griffin G, Di Marzo V, Martin BR. Evidence for a new G protein-coupled cannabinoid receptor in mouse brain. Mol Pharmacol. 2001;60:155–163. [PubMed] [Google Scholar]
- Breivogel CS, Sim LJ, Childers SR. Regional differences in cannabinoid receptor/G-protein coupling in rat brain. J Pharmacol Exp Ther. 1997;282:1632–1642. [PubMed] [Google Scholar]
- Browne RG, Weissman A. Discriminative stimulus properties of delta 9-tetrahydrocannabinol: mechanistic studies. J Clin Pharmacol. 1981;21:227S–234S. doi: 10.1002/j.1552-4604.1981.tb02599.x. [DOI] [PubMed] [Google Scholar]
- Cochran W. The combination of estimates from different experiments. Biometrics. 1954;10:101–124. [Google Scholar]
- Demuth DG, Molleman A. Cannabinoid signalling. Life Sci. 2006;78:549–563. doi: 10.1016/j.lfs.2005.05.055. [DOI] [PubMed] [Google Scholar]
- Diaz-Laviada I, Ruiz-Llorente L. Signal transduction activated by cannabinoid receptors. Mini Rev Med Chem. 2005;5:619–630. doi: 10.2174/1389557054368808. [DOI] [PubMed] [Google Scholar]
- Egertova M, Elphick MR. Localisation of cannabinoid receptors in the rat brain using antibodies to the intracellular C-terminal tail of CB. J Comp Neurol. 2000;422:159–171. doi: 10.1002/(sici)1096-9861(20000626)422:2<159::aid-cne1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Eggan SM, Lewis DA. Immunocytochemical distribution of the cannabinoid CB1 receptor in the primate neocortex: a regional and laminar analysis. Cereb Cortex. 2006;17:175–191. doi: 10.1093/cercor/bhj136. [DOI] [PubMed] [Google Scholar]
- Felder CC, Joyce KE, Briley EM, Mansouri J, Mackie K, Blond O, et al. Comparison of the pharmacology and signal transduction of the human cannabinoid CB1 and CB2 receptors. Mol Pharmacol. 1995;48:443–450. [PubMed] [Google Scholar]
- Felder CC, Veluz JS, Williams HL, Briley EM, Matsuda LA. Cannabinoid agonists stimulate both receptor- and non-receptor-mediated signal transduction pathways in cells transfected with and expressing cannabinoid receptor clones. Mol Pharmacol. 1992;42:838–845. [PubMed] [Google Scholar]
- Glass GV, McGaw B, Smith ML. Meta-analysis in Social Research. SAGE Publications: Beverly Hills, CA; 1981. [Google Scholar]
- Glass M. Doctorate Dissertation, Department of Pharmacology and Clinical Pharmacology. University of Auckland: Auckland; 1995. p. 220. [Google Scholar]
- Glass M, Dragunow M, Faull RL. Cannabinoid receptors in the human brain: a detailed anatomical and quantitative autoradiographic study in the fetal, neonatal and adult human brain. Neuroscience. 1997;77:299–318. doi: 10.1016/s0306-4522(96)00428-9. [DOI] [PubMed] [Google Scholar]
- Gong JP, Onaivi ES, Ishiguro H, Liu QR, Tagliaferro PA, Brusco A, et al. Cannabinoid CB2 receptors: immunohistochemical localization in rat brain. Brain Res. 2006;1071:10–23. doi: 10.1016/j.brainres.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Guy G, Robson R, Strong K, Whittle B. The Medicinal Use of Cannabis. Royal Society of Pharmacists: London; 2004. [Google Scholar]
- Haller J, Matyas F, Soproni K, Varga B, Barsy B, Nemeth B, et al. Correlated species differences in the effects of cannabinoid ligands on anxiety and on GABAergic and glutamatergic synaptic transmission. Eur J Neurosci. 2007;25:2445–2456. doi: 10.1111/j.1460-9568.2007.05476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkany T, Dobszay MB, Cayetanot F, Hartig W, Siegemund T, Aujard F, et al. Redistribution of CB1 cannabinoid receptors during evolution of cholinergic basal forebrain territories and their cortical projection areas: a comparison between the gray mouse lemur (Microcebus murinus, primates) and rat. Neuroscience. 2005;135:595–609. doi: 10.1016/j.neuroscience.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, de Costa BR, et al. Cannabinoid receptor localization in brain. Proc Natl Acad Sci USA. 1990;87:1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JPT, Green S.Cochrane Handbook for Systematic Reviews of Interventions 4.2.5 The Cochrane Library 2005John Wiley & Sons, Ltd.: Chichester, UK; [updated May 2005]. In: Higgins JPT, Green S (eds) [Google Scholar]
- Hoffman AF, Macgill AM, Smith D, Oz M, Lupica CR. Species and strain differences in the expression of a novel glutamate-modulating cannabinoid receptor in the rodent hippocampus. Eur J Neurosci. 2005;22:2387–2391. doi: 10.1111/j.1460-9568.2005.04401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DB, Howlett AC. Differential receptor-G-protein coupling evoked by dissimilar cannabinoid receptor agonists. Cell Signal. 1998;10:667–674. doi: 10.1016/s0898-6568(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Howlett AC. The cannabinoid receptors. Prostaglandins Other Lipid Mediat. 2002;68–69:619–631. doi: 10.1016/s0090-6980(02)00060-6. [DOI] [PubMed] [Google Scholar]
- Huang SM, Bisogno T, Trevisani M, Al-Hayani A, De Petrocellis L, Fezza F, et al. An endogenous capsaicin-like substance with high potency at recombinant and native vanilloid VR1 receptors. Proc Natl Acad Sci USA. 2002;99:8400–8405. doi: 10.1073/pnas.122196999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungund BL, Basavarajappa BS. Distinct differences in the cannabinoid receptor binding in the brain of C57BL/6 and DBA/2 mice, selected for their differences in voluntary ethanol consumption. J Neurosci Res. 2000;60:122–128. doi: 10.1002/(SICI)1097-4547(20000401)60:1<122::AID-JNR13>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Isbell H, Gorodetzsky CW, Jasinski D, Claussen U, Von Spulak F, Korte F. Effects of (−)delta-9-trans-tetrahydrocannabinol in man. Psychopharmacologia. 1967;11:184–188. doi: 10.1007/BF00401256. [DOI] [PubMed] [Google Scholar]
- Jarbe TU, Johansson JO, Henriksson BG. Characteristics of tetrahydrocannabinol (THC)-produced discrimination in rats. Psychopharmacology (Berlin) 1976;48:181–187. doi: 10.1007/BF00423258. [DOI] [PubMed] [Google Scholar]
- Kearn CS, Greenberg MJ, DiCamelli R, Kurzawa K, Hillard CJ. Relationships between ligand affinities for the cerebellar cannabinoid receptor CB1 and the induction of GDP/GTP exchange. J Neurochem. 1999;72:2379–2387. doi: 10.1046/j.1471-4159.1999.0722379.x. [DOI] [PubMed] [Google Scholar]
- LeLorier J, Gregoire G, Benhaddad A, Lapierre J, Derderian F. Discrepancies between meta-analyses and subsequent large randomized, controlled trials. N Engl J Med. 1997;337:536–542. doi: 10.1056/NEJM199708213370806. [DOI] [PubMed] [Google Scholar]
- Martini L, Waldhoer M, Pusch M, Kharazia V, Fong J, Lee JH, et al. Ligand-induced down-regulation of the cannabinoid 1 receptor is mediated by the G-protein-coupled receptor-associated sorting protein GASP1. FASEB J. 2007;21:802–811. doi: 10.1096/fj.06-7132com. [DOI] [PubMed] [Google Scholar]
- Mauler F, Mittendorf J, Horvath E, De Vry J. Characterization of the diarylether sulfonylester (−)-(R)-3-(2-hydroxymethylindanyl-4-oxy)phenyl-4,4,4-trifluoro-1-sulfonate (BAY 38-7271) as a potent cannabinoid receptor agonist with neuroprotective properties. J Pharmacol Exp Ther. 2002;302:359–368. doi: 10.1124/jpet.302.1.359. [DOI] [PubMed] [Google Scholar]
- McPartland JM, Matias I, Di Marzo V, Glass M. Evolutionary origins of the endocannabinoid system. Gene. 2006;370:64–74. doi: 10.1016/j.gene.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Ohlsson A, Lindgren JE, Wahlen A, Agurell S, Hollister LE, Gillespie HK. Plasma delta-9 tetrahydrocannabinol concentrations and clinical effects after oral and intravenous administration and smoking. Clin Pharmacol Ther. 1980;28:409–416. doi: 10.1038/clpt.1980.181. [DOI] [PubMed] [Google Scholar]
- Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there. Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- Paugh SW, Cassidy MP, He H, Milstien S, Sim-Selley LJ, Spiegel S, et al. Sphingosine and its analog, the immunosuppressant 2-amino-2-(2-[4-octylphenyl]ethyl)-1,3-propanediol, interact with the CB1 cannabinoid receptor. Mol Pharmacol. 2006;70:41–50. doi: 10.1124/mol.105.020552. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Pharmacological actions of cannabinoids. Handb Exp Pharmacol. 2005;168:1–51. doi: 10.1007/3-540-26573-2_1. [DOI] [PubMed] [Google Scholar]
- Pertwee RG, Thomas A, Stevenson LA, Maor Y, Mechoulam R. Evidence that (−)-7-hydroxy-4′-dimethylheptyl-cannabidiol activates a non-CB(1), non-CB(2), non-TRPV1 target in the mouse vas deferens. Neuropharmacology. 2005;48:1139–1146. doi: 10.1016/j.neuropharm.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Petitet F, Donlan M, Michel A. GPR55 as a new cannabinoid receptor: still a long way to prove it. Chem Biol Drug Des. 2006;67:252–253. doi: 10.1111/j.1747-0285.2006.00370.x. [DOI] [PubMed] [Google Scholar]
- Petitet F, Marin L, Doble A. Biochemical and pharmacological characterization of cannabinoid binding sites using [3H]SR141716A. Neuroreport. 1996;7:789–792. doi: 10.1097/00001756-199602290-00026. [DOI] [PubMed] [Google Scholar]
- Reed GF, Lynn F, Meade BD. Use of coefficient of variation in assessing variability of quantitative assays. Clin Diagn Lab Immunol. 2002;9:1235–1239. doi: 10.1128/CDLI.9.6.1235-1239.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee MH, Vogel Z, Barg J, Bayewitch M, Levy R, Hanus L, et al. Cannabinol derivatives: binding to cannabinoid receptors and inhibition of adenylylcyclase. J Med Chem. 1997;40:3228–3233. doi: 10.1021/jm970126f. [DOI] [PubMed] [Google Scholar]
- Rodriguez de Fonseca F, Gorriti MA, Fernandez-Ruiz JJ, Palomo T, Ramos JA. Downregulation of rat brain cannabinoid binding sites after chronic delta 9-tetrahydrocannabinol treatment. Pharmacol Biochem Behav. 1994;47:33–40. doi: 10.1016/0091-3057(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Romero J, Fernandez-Ruiz JJ, Vela G, Ruiz-Gayo M, Fuentes JA, Ramos JA. Autoradiographic analysis of cannabinoid receptor binding and cannabinoid agonist-stimulated [35S]GTP gamma S binding in morphine-dependent mice. Drug Alcohol Depend. 1998;50:241–249. doi: 10.1016/s0376-8716(98)00036-2. [DOI] [PubMed] [Google Scholar]
- Russo EB, Burnett A, Hall B, Parker KK. Agonistic properties of cannabidiol at 5-HT1a receptors. Neurochem Res. 2005;30:1037–1043. doi: 10.1007/s11064-005-6978-1. [DOI] [PubMed] [Google Scholar]
- Savinainen JR, Saario SM, Niemi R, Jarvinen T, Laitinen JT. An optimized approach to study endocannabinoid signaling: evidence against constitutive activity of rat brain adenosine A1 and cannabinoid CB1 receptors. Br J Pharmacol. 2003;140:1451–1459. doi: 10.1038/sj.bjp.0705577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schioth HB, Haitina T, Ling MK, Ringholm A, Fredriksson R, Cerda-Reverter JM, et al. Evolutionary conservation of the structural, pharmacological, and genomic characteristics of the melanocortin receptor subtypes. Peptides. 2005;26:1886–1900. doi: 10.1016/j.peptides.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Sese J, Morishita S. Rank aggregation method for biological databases. Genome Inform. 2001;12:506–507. [Google Scholar]
- Sheskin T, Hanus L, Slager J, Vogel Z, Mechoulam R. Structural requirements for binding of anandamide-type compounds to the brain cannabinoid receptor. J Med Chem. 1997;40:659–667. doi: 10.1021/jm960752x. [DOI] [PubMed] [Google Scholar]
- Steffens M, Engler C, Zentner J, Feuerstein TJ. Cannabinoid CB1 receptor-mediated modulation of evoked dopamine release and of adenylyl cyclase activity in the human neocortex. Br J Pharmacol. 2004;141:1193–1203. doi: 10.1038/sj.bjp.0705706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szallasi A. The vanilloid (capsaicin) receptor: receptor types and species differences. Gen Pharmacol. 1994;25:223–243. doi: 10.1016/0306-3623(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Thomas BF, Francisco ME, Seltzman HH, Thomas JB, Fix SE, Schulz AK, et al. Synthesis of long-chain amide analogs of the cannabinoid CB1 receptor antagonist N-(piperidinyl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyra zole-3-carboxamide (SR141716) with unique binding selectivities and pharmacological activities. Bioorg Med Chem. 2005;13:5463–5474. doi: 10.1016/j.bmc.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Thomas BF, Gilliam AF, Burch DF, Roche MJ, Seltzman HH. Comparative receptor binding analyses of cannabinoid agonists and antagonists. J Pharmacol Exp Ther. 1998;285:285–292. [PubMed] [Google Scholar]
- Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K, et al. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- Wiley JL, Lowe JA, Balster RL, Martin BR. Antagonism of the discriminative stimulus effects of delta 9-tetrahydrocannabinol in rats and rhesus monkeys. J Pharmacol Exp Ther. 1995;275:1–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.