Abstract
Purpose:
To evaluate the feasibility of obtaining dried blood spots (DBS) from newborn screening archives for subjects in epidemiologic studies and using these specimens for genotyping, and to evaluate the potential for bias in their use.
Methods:
We attempted to locate DBS at Washington State's archives for 230 participants in a previous case-control study of childhood cancer, who were born 1978-1990. We compared characteristics of children for whom we did and did not locate specimens, and attempted genetic polymorphism analyses (11 polymorphisms, 82-480 bp amplicons).
Results:
We retrieved specimens for 203 (88%) children, including 199 (94%) born in months when a DBS catalog was available. Among the latter, the proportion with specimens located varied by birth place (e.g., hospital, home), maternal education and prenatal smoking, but did not vary significantly by race/ethnicity. All genotyping assays were completed for all specimens, and among controls genotype distributions were in Hardy-Weinberg equilibrium and similar to previous reports.
Conclusions:
Newborn screening archives have potential to provide specimens for epidemiologic studies conducting genotyping and perhaps other assays, but the possibility that reliance on these resources could bias risk estimates must be considered.
Keywords: Epidemiologic Methods, Bias (Epidemiology), Selection Bias, Biological Specimen Banks
Introduction
Many countries universally screen infants for selected in-born disorders using Guthrie cards, specially formulated filter paper used to absorb approximately 13 mm spots of blood collected shortly after birth. In many regions these dried blood spots (DBS) are stored, and epidemiologists have recognized the potential value of these population-based archives [1-3]. Most commonly, anonymous random specimens have been used, for example to estimate the population prevalence of HTLV-I antibodies [4], or to provide a control group in a study of childhood cancer and glutathione-S transferase (GST) polymorphisms [5]. Occasionally, researchers have used such archives to collect specimens for specific individuals participating in new or existing epidemiologic studies, the potential benefits including cost-efficiency, reduced subject burden, inclusion of deceased subjects, and use of pre-diagnosis specimens. Examples include a large prospective cohort study evaluating the maternal-fetal transmission of toxoplasmosis [6], case-control studies of childhood cancer and chemical metabolism genes [7, 8], and case series demonstrating genetic alterations associated with childhood leukemia [9]. DBS have also been used to measure possible indicators of infection [10-12], inflammation [13, 14], nutritional status [15-17], and environmental exposure [18, 19].
Previously we demonstrated the feasibility of collecting DBS archived for up to 25 years for child participants from an existing case-control study (brain tumors), and using these specimens for genotyping [7]. Here we detail our methods and assess the potential for bias in risk estimates introduced by reliance on archived DBS. This has not been examined previously, and future studies using these specimens likely will focus on childhood diseases, especially often fatal ones, as did we.
Methods
Subject identification
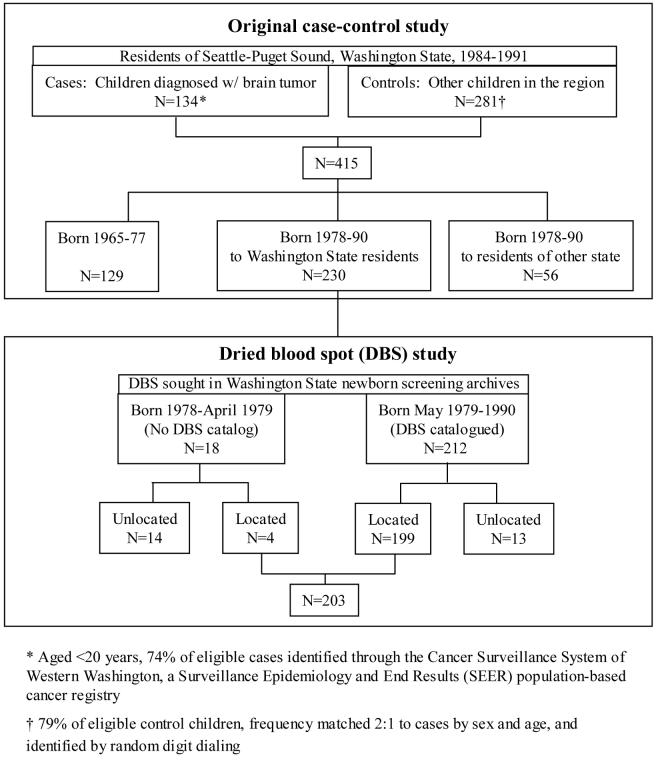
Institutional Review Board approvals were received from the Fred Hutchinson Cancer Research Center and the Washington State Department of Health (WDOH) prior to the conduct of this study. We included children from an earlier case-control study of brain tumors [20]. Briefly, cases were diagnosed <20 years of age, during 1984-1991, while living in the Seattle-Puget Sound region of Washington State covered by a population-based cancer registry. Controls in the same region were identified by random digit dialing [21], frequency matched to cases 2:1 by sex and age. Of eligible children, 415 participated (134 (74%) cases and 281 (79%) controls, Figure 1). Mothers of all subjects and 83% of fathers were interviewed following informed consent obtained in writing or via telephone.
Figure 1.
Children for whom an archived dried blood spot (DBS) was sought, and number for whom a DBS was located, overall and by presence of specimen cataloguing
WDOH screens >99% of Washington newborns, and archives all DBS for 21.5 years without special climate controls. Using the date, hospital and city of residence at birth for children in the original case-control study, we identified those born after 1977 (1978-90) to Washington state residents — children for whom a DBS card might have remained at the WDOH Newborn Screening Program archives. Birth in a hospital in Washington state was not required because WDOH recommends a second card be collected in the physician's office a week or two after birth (about 80-90% newborns participated during the study years). Of the 415 children in the original study, 230 (55%) were born after 1977 to Washington state residents (Figure 1).
Specimen collection
We abstracted data on these 230 eligible children's sex, birth date, and hospital/location of birth; and the first and last name (at the time of the interview) of the child, mother, and father. Blind to case status, WDOH staff matched the list with the catalog of DBS cards (N=212), or for births before May 1979 searched uncatalogued cards manually (N=18, Figure 1). When the card was located, staff double-checked the match, and verified at least one complete (fully saturated without punches) DBS would remain in the archives. A complete (when available) or partial DBS was clipped from the card (or two cards, when a unique match could not be determined). For quality assurance, 21 duplicate or quadruplicate specimens for a convenience sample of 6% cases and 6% controls were collected.
Maintenance of confidentiality
Specimens were labeled only with a new randomly assigned identification number. Prior to release from the archives, specimens were “anonymized” [22-24]. We created a file with non-identifying interview data linked to the new identification number. We then destroyed the link with the identifiers, and the list used for matching.
Genotyping
The University of Washington Center for Ecogenetics and Environmental Health Functional Genomics Core Laboratory used approximately one third of each DBS (six 3mm punches) for genotyping. DNA was extracted using a commercial kit [7]; GSTM1 and GSTT1 null status were determined using a single assay (215, 268 and 480 bp amplicons) with co-amplification of a region of the β-globin gene to verify double-nulls [25]; and nine single nucleotide polymorphisms (GSTP1I105V, CYP2D6*3 (2549delA), CYP2E1*5 (RsaI, C-1053T), EPHX1Y113H, EPHX1H139R, PON1C-108T, PON1Q192R, PON1L55M and PON2S311C) were determined using separate TaqMan™ assays (82-227 bp amplicons) [7, 25, 26]. These polymorphisms affect xenobiotic metabolism.
Statistical analysis
With Fisher's two-sided exact chi-square test in Stata [27] we: compared characteristics of children with and without specimens located (to examine the potential for a biased sample), checked Hardy-Weinberg equilibrium of genotypes (to examine assay accuracy with these specimens/potential for environmental contamination), and among controls compared genotypes with selected demographic characteristics (to examine whether sample composition might influence risk estimates).
Results
At least one specimen was located for 203 (88%) subjects for whom we attempted to locate a DBS — 199 (94%) born in catalogued months, and four (22%) born in earlier, uncatalogued months (Figure 1).
Specimen and match quality
Of the 203 children with a DBS, we included 13 (6% eligible subjects) by accepting a partial specimen, and three (1% eligible subjects, one singleton and two with a same-sex twin) by collecting specimens for two equally well matching children (Table 1). For 172 (85%), the birth date, hospital, sex, mother's first name, and a family last name on the DBS card(s) agreed with the list provided to WDOH. An additional 11 (5%) fully matched except that the card was collected at a doctor's office in the birth city, instead of the hospital. For the remaining 20 matches, one part of the birth date (N=6), sex (N=4), last name (N=4), mother's first name (N=2), or multiple variables (N=4) did not match or were missing, but for 14 (70%), additional information supported the match (e.g., child's first name on the card).
Table 1.
Specimen and match quality for subjects for whom a dried blood spot was located
| N=203 n (%) |
|
|---|---|
| Quality of primary* dried blood spot | |
| Complete (fully saturated and without previous punches from screening) | 190 (94) |
| Partial (punched or not fully saturated) | 13 (6) |
| Number of children in the catalog who matched with the subject | |
| One child | 200 (99) |
| Two children† | 3 (1) |
| Matching variables known and in agreement | |
| All (birth date, hospital, sex, mother's first name, family last name) | 172 (85) |
| All but hospital (specimen collected at doctor's office in city of birth) | 11 (5) |
| All but one part of the birth date | 6 (3) |
| All but sex | 4 (2) |
| All but family last name | 4 (2) |
| All but mother's first name | 2 (1) |
| Multiple variables did not agree/were missing | 4 (2) |
Excludes 21 secondary specimens obtained for quality assurance
Equally well matching, includes two subjects with same-sex twins and one singleton
Factors associated with failure to locate a DBS
Just over half of children for whom a specimen could not be located (14 children, 6% eligible subjects) were born during uncatalogued months. Twelve of the other 13 unlocated children had one of the following characteristics: five (2% eligible subjects) were born in the earliest three-year period when a catalogue existed but was indexed by the child's last name only (not computerized); five (2% eligible subjects) were born in a hospital that sent DBS to an out-of-state laboratory; one (0.4% eligible subjects) was born in a neighboring state; and one (0.4% eligible subjects) was born at home. Consistent with this, among the 212 (92%) children born in catalogued months, the proportion with DBS located was markedly greater for children born in an in-state non-federal hospital (97%) than elsewhere (30%, p<0.0001, Table 2; includes 2/2 children born in a birth center, 1/2 born at home, and none in federal/out-of-state hospitals). It was somewhat better among children with interviewed fathers (95% vs. 80% without paternal interviews, p=0.02) or mothers who did not smoke during pregnancy (96% vs. 85% with mothers who smoked, p=0.02). It appeared slightly better among cases (98% vs. 92% controls, p=0.07), children whose mothers had a college degree (100% vs. 92% without a degree, p=0.04), those living at the same residence at birth and interview (98% vs. 92% who moved, p=0.14), and Hispanic or non-white children (100% vs. 93% of non-Hispanic white children), although the latter comparison was based on particularly small numbers (p=1.0). Multivariate analyses were not possible, but among the 13 children without DBS, all four whose fathers were not interviewed were born in an in-state non-federal hospital, while prenatal smoking was prevalent regardless of birth location (3/6 born in an in-state non-federal hospital, 3/7 born elsewhere, data not shown).
Table 2.
Proportion dried blood spots (DBS) collected among subjects born in a catalogued month, by selected characteristics of subjects
| DBS Located N=199 |
DBS Unlocated N=13 |
% Collected | p-value* | |
|---|---|---|---|---|
| Case status | ||||
| Case | 65 | 1 | 98 | |
| Control | 134 | 12 | 92 | p=0.07 |
| Birth year | ||||
| 1978-1983 | 98 | 8 | 92 | |
| 1984-1990 | 101 | 5 | 95 | p=0.57 |
| Born in an in-state non-federal hospital | ||||
| Yes | 196 | 6 | 97 | |
| No† | 3 | 7 | 30 | p<0.0001 |
| Sex | ||||
| Male | 126 | 7 | 95 | |
| Female | 73 | 6 | 92 | p=0.56 |
| Mother's race/ethnicity | ||||
| Non-Hispanic white | 185 | 13 | 93 | |
| Hispanic or non-white | 14 | 0 | 100 | p=1.0 |
| Mother's education (college degree) | ||||
| Yes | 52 | 0 | 100 | |
| No | 147 | 13 | 92 | p=0.04 |
| Mother smoked during pregnancy | ||||
| Yes | 33 | 6 | 85 | |
| No | 166 | 7 | 96 | p=0.02 |
| Interview and birth residence same | ||||
| Yes | 76 | 2 | 98 | |
| No | 123 | 11 | 92 | p=0.14 |
| Father completed interview | ||||
| Yes | 183 | 9 | 95 | |
| No | 16 | 4 | 80 | p=0.02 |
Fisher's exact, two-sided
Birth center, home, federal hospital or out-of-state location
Genotypes
We completed all assays for all children. Genotyping confirmed that the two possible matches for one singleton control were from two individuals with different genotypes for some polymorphisms, leaving 136 (99.3%) controls and all 66 cases. Agreement between the primary and replicate specimens was 99.1%.
Among the 136 controls, 52.2% were GSTM1 null, 15.4% were GSTT1 null, and 10.3% were double-null (Table 3). Variant allele frequencies for the other polymorphisms were 0.018-0.44, and all genotype frequencies were in Hardy-Weinberg equilibrium. Results for all polymorphisms were similar to those reported previously for healthy Caucasians. Eight (6%) controls had Hispanic or non-white parent(s), and compared to controls with two non-Hispanic white parents, they more often carried PON1192R (p<0.01) and CYP2E1*5 (p=0.06) alleles (data not shown), consistent with previous observations [28-31].
Table 3.
Genotyping results using archived dried blood spots for control children
| Polymorphism (variant genotype or allele) |
Hardy- Weinberg Equilibrium* |
Frequency† Among Controls |
Previously Reported Frequency† Among controls‡ |
|---|---|---|---|
| GSTM1/GSTT1 | |||
| GSTM1 (Null) | n/a | 52.2% | 53.1-53.5%[25, 32, 34] |
| GSTT1 (Null) | n/a | 15.4% | 15.0-19.7% [25, 32, 34] |
| Both Null | n/a | 10.3% | 10.4% [32] |
| Other polymorphisms§ | |||
| GSTP1I105V (V) | p=0.86 | 0.42 | 0.37[25] |
| CYP2D6 (*3) | p=1.0 | 0.018 | 0.018 [33] |
| CYP2E1 (*5) | p=1.0 | 0.029 | 0.038 [32] |
| EPHX1Y113H (H) | p=0.84 | 0.29 | 0.32 [32] |
| EPHX1H139R (R) | p=0.81 | 0.23 | 0.22 [32] |
| PON1C-108T (T) | p=0.38 | 0.44 | 0.44-0.62 [28, 29] |
| PON1Q192R (R) | p=0.56 | 0.33 | 0.24-0.38 [28, 29] |
| PON1L55M (M) | p=0.25 | 0.34 | 0.28-0.46 [28, 29] |
| PON2S311C (C) | p=0.42 | 0.31 | 0.20-0.25 [28, 29] |
Significant disequilibrium indicated by p<0.05, exact chi square
Genotype frequency (GSTM1 and GSTT1 null) or variant allele frequency (all other polymorphisms)
Healthy white individuals/control subjects; when available [32], includes genotype/allele frequencies from International Project on Genetic Susceptibility to Environmental Carcinogens (GSEC) database containing information on 12,525 white individuals without cancer
Single nucleotide polymorphisms determined by individual TaqMan assays
Abbreviations: n/a=not applicable; GST=glutathione S-transferase; CYP=cytochrome P450; EPHX=microsomal epoxide hydrolase; PON=paraoxonase
Among non-Hispanic white controls, children of mothers with some college education were more likely than other children to carry the CYP2E1*5 allele (p=0.04) and the PON1-108C allele (p=0.07); and children of mothers who smoked during pregnancy were more likely than other children to carry the EPHX1139H allele (p=0.12). Genotyping data were only available for three children born outside a hospital, precluding consideration of this factor.
Discussion
We collected DBS from newborn screening archives for 94% of children born in months when DBS cards were catalogued, similar to a study in another state [8]. All our genotyping assays were completed for all children with DBS, with genotype/allele frequencies similar to studies using DNA from fresh blood [25, 28, 29, 32-34]. These results are encouraging, given that many states archive DBS for several years [35] and might allow them to be used for research [2]. It is important, however, to consider how bias might be introduced.
Screening participation
In Washington state, newborn screening is nearly universal [35], but here and elsewhere non-participation is unlikely to be random. Unscreened children are perhaps more likely to have been born at home, died shortly after birth, or have parents who object to standard medical care. The one child without a specimen and born at home perhaps was not screened.
Archive coverage
We sought a DBS for all children born to all state residents, even those born out of state or in hospitals that sent screening cards to another state, because these subjects could have participated in the recommended secondary screening in-state. Excluding them a priori would have been inappropriate because they likely differed from others. We located none, affecting our collection effort somewhat. In regions where residents more commonly cross state lines for childbirth, attempting to access the neighboring state's archives may minimize bias that could otherwise be introduced by incomplete archive coverage.
Matching information
The mother's first name and child's date and hospital of birth were often adequate for matching. Important exceptions were the short periods when the DBS catalog was unavailable or searchable only by last name. Some children's last names may have changed between birth and interview. Children without a specimen located were more likely to have changed residence since birth (likely associated with a name change) and their fathers were less likely to have been interviewed (providing additional matching information).
In rare instances we were unable to narrow matches to a single screened child. For each we collected a DBS from each of the two matching children, and retained their assay results when concordant, because such children may differ from others (e.g. common name, incomplete matching information, large birth hospital, twin).
With such archives one cannot rule out collecting a DBS for the wrong subject. However, our match quality was generally excellent, suggesting this problem may be minimal.
These potential sources of bias related to matching are probably less common in studies accessing archives with computerized catalogues searchable by several variables, and that can determine the child's birth name prior to matching via interview or linkage to other data (e.g. birth certificates).
Specimen availability
A DBS was available from all located cards, but had we not accepted a partial DBS, we would have excluded several children, and potentially introduced bias. Specimens may be less available for subjects with only one card, fewer spots collected per card (because of birth year or other factors), ‘positive’ newborn screening program results, or with parents requesting the specimen not be archived/used for research.
Specimen usability
Despite storage for 25 years, at times without air conditioning, all specimens yielded genotyping results. However, the climate in western Washington is moderate, and our assays mainly required short amplicons. In a separate study in another state using similar methods on DBS of similar age, GST genotyping was completed for 97% of children born in the later years, but only 3% born in earlier years, when specimens reached high temperatures during storage [8]. Nevertheless, DNA in DBS stored in tropical conditions for at least ten years was stable enough for genotyping of all of specimens with assays requiring 255, 674 and 1039 bp amplicons [36]. However, only 60% of slightly older specimens in that study were usable for the assay with the longest amplicon, consistent with another report [3] that genotype might not be determinable for all subjects for assays requiring long amplicons. Specimen age and storage conditions might also affect the ability to obtain results for assays assessing something other than genotype.
Assay accuracy
All the genotype/allele frequencies observed among controls were consistent with previous reports and in Hardy-Weinberg equilibrium, and there was no evidence of under-representation of GSTT1 expressers (480 bp, the longest amplicon) or over-representation of GSTM1 expressers or double-null children (215-268 bp, shorter amplicons). It appears that any DNA fragmentation or environmental contamination (e.g. from existing punches [37] or contact with adjacent cards [12]) were not sufficient to alter results, even though the DBS cards were stored in direct contact with one another. However, the TaqMan assays we used each required amplicons of a single, modest length, and considered the relative amount of each allele, not just their presence. In addition, we flame-sterilized the punch between specimens.
Assays measuring characteristics other than genotype might be less accurate with dried than with fresh blood, and accuracy might also depend on storage time and conditions. Some assays might also be less resilient to possible contamination of the DBS, although potential counter strategies exist (e.g. comparing amounts of viral DNA on index and adjacent DBS cards to determine which is truly positive [12], and comparing amounts of environmental chemicals in blood-saturated vs. blood-free portions of the card).
Potential effect on risk estimates
Some potential sources of bias could affect risk estimates by altering the composition of the study groups. Our results indicate that potentially important differences (e.g., maternal smoking and education) may exist in children for whom archived DBS cannot be located. Other studies have observed non-random differences between children for whom a located specimen can and cannot be assayed [8, 36]. The magnitude and direction of the effect, if any, on risk estimates might be difficult to predict. We located DBS for a somewhat greater percentage of cases than controls, possibly because proportionally more cases had fathers interviewed, and none had out-of-state specimens. Further, some characteristics associated with locating a specimen, maternal education and possibly smoking, were also associated with genotype. If these associations applied equally for cases and controls for whom we failed to locate a DBS (largely born to mothers without a college degree), we might expect a slight under-representation of, for example, the PON1−108T allele among both cases and controls in a study focused on this polymorphism [7]. And, because proportionally more control DBS specimens were not located, an odds ratio of 1.7 for case status in relation to the PON1−108T allele might be biased slightly upward. In another study using DBS [8], in which birth year was strongly associated with ability to obtain genotyping results, cases and controls were matched on birth date, so even if there were racial/ethnic changes in the population over time, presumably risk estimates were not materially affected.
Bias in the form of measurement error would arise if results from dried blood, or that stored for a long time in non-optimal conditions, are not as accurate as those from fresh blood. This did not appear to occur with our genotyping, but may with other assays. If the resulting measurement error does not differ between comparison groups, risk estimates may be biased toward null.
The effect of other potential sources of bias could be less straightforward. For example, it seems unlikely that environmental contamination could cause true heterozygotes (TaqMan assays), double expressers (GST assays), and viral-DNA-positive subjects to be misclassified. For the remaining subjects (those truly homozygous, GST null, and viral-DNA-negative, respectively), it would be more difficult to rule out the possibility that environmental contamination could alter assay results (e.g. [12]). If such misclassification occurred and was non-differential, one would generally expect risk estimates to be biased toward null (e.g. if based on inherently dichotomous categories such as viral DNA positive/negative), but exceptions exist (e.g. odds ratios based only on the two homozygous categories would not be biased). Similarly, bias may or may not occur if the wrong specimen is obtained. For example, in our previous study using population-based controls, unconfounded odds ratios might be similar to those observed even had we obtained the wrong specimen for all controls; our risk estimates were very similar when we used randomly selected archive controls [7]. Only in the absence of a true association might cases' specimens be substituted in this manner without bias. However, studies that track match quality can assess whether questionable matches affect risk estimates.
Most potential sources of bias related to reliance on archived DBS are likely minimal, or can be reduced. In some situations reliance on archived DBS might reduce bias in risk estimates, compared to approaches in which specimens are obtained after diagnosis and only from surviving individuals. The potential for bias does not preclude the use of these specimens for genotyping and perhaps other assays, but careful consideration of how bias might enter into results is warranted.
Acknowledgements
This work supported by NIEHS T32ES07262, NIEHS P30ES07033 and 1 R03 CA106011 from the National Institutes of Health; and contract N01-CN-05230 from the National Cancer Institute. We thank the Washington State Department of Health Newborn Screening Program for their generous participation in this project, especially Mr. Michael Glass and Mr. Michael Ginder; the University of Washington Center for Ecogenetics and Environmental Health Functional Genomics Core Laboratory, especially Dr. Federico M. Farin and Ms. Hannah-Malia A. Viernes; and Ms. Randi Niemer.
Abbreviations and Acronyms
- bp
Base pairs
- CYP
Cytochrome P450
- DBS
Dried blood spot
- EPHX1
Microsomal epoxide hydrolase
- GST
Glutathione S-transferase
- GSTM1
Glutathione S-transferase mu 1
- GSTP1
Glutathione S-transferase pi 1
- GSTT1
Glutathione S-transferase theta 1
- HTLV-I
Human T-cell leukemia virus type 1
- PON
Paraoxonase
- WDOH
Washington State Department of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parker SP, Cubitt WD. The use of the dried blood spot sample in epidemiological studies. J Clin Pathol. 1999;52:633–9. doi: 10.1136/jcp.52.9.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McEwen JE, Reilly PR. Stored Guthrie cards as DNA “banks”. Am J Hum Genet. 1994;55:196–200. [PMC free article] [PubMed] [Google Scholar]
- 3.Hannelius U, Lindgren CM, Melen E, Malmberg A, von Dobeln U, Kere J. Phenylketonuria screening registry as a resource for population genetic studies. J Med Genet. 2005;42:e60. doi: 10.1136/jmg.2005.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker SP, Taylor MB, Ades AE, Cubitt WD, Peckham C. Use of dried blood spots for the detection and confirmation of HTLV-I specific antibodies for epidemiological purposes. J Clin Pathol. 1995;48:904–7. doi: 10.1136/jcp.48.10.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnette P, Scholl R, Blandford M, Ballard L, Tsodikov A, Magee J, et al. High-throughput detection of glutathione s-transferase polymorphic alleles in a pediatric cancer population. Cancer Epidemiol Biomarkers Prev. 2004;13:304–13. doi: 10.1158/1055-9965.epi-03-0178. [DOI] [PubMed] [Google Scholar]
- 6.Lebech M, Petersen E. Neonatal screening for congenital toxoplasmosis in Denmark: presentation of the design of a prospective study. Scand J Infect Dis Suppl. 1992;84:75–9. [PubMed] [Google Scholar]
- 7.Searles Nielsen S, Mueller BA, De Roos AJ, Viernes HM, Farin FM, Checkoway H. Risk of brain tumors in children and susceptibility to organophosphorus insecticides: the potential role of paraoxonase (PON1) Environ Health Perspect. 2005;113:909–13. doi: 10.1289/ehp.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klotz J, Bryant P, Wilcox HB, Dillon M, Wolf B, Fagliano J. Population-based retrieval of newborn dried blood spots for researching paediatric cancer susceptibility genes. Paediatr Perinat Epidemiol. 2006;20:449–52. doi: 10.1111/j.1365-3016.2006.00749.x. [DOI] [PubMed] [Google Scholar]
- 9.Taub JW, Ge Y. The prenatal origin of childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2004;45:19–25. doi: 10.1080/1042819031000149403. [DOI] [PubMed] [Google Scholar]
- 10.Jardi R, Rodriguez-Frias F, Buti M, Schaper M, Valdes A, Martinez M, et al. Usefulness of dried blood samples for quantification and molecular characterization of HBV-DNA. Hepatology. 2004;40:133–9. doi: 10.1002/hep.20275. [DOI] [PubMed] [Google Scholar]
- 11.McDade TW, Stallings JF, Angold A, Costello EJ, Burleson M, Cacioppo JT, et al. Epstein-Barr virus antibodies in whole blood spots: a minimally invasive method for assessing an aspect of cell-mediated immunity. Psychosom Med. 2000;62:560–7. doi: 10.1097/00006842-200007000-00015. [DOI] [PubMed] [Google Scholar]
- 12.Lewensohn-Fuchs I, Osterwall P, Forsgren M, Malm G. Detection of herpes simplex virus DNA in dried blood spots making a retrospective diagnosis possible. J Clin Virol. 2003;26:39–48. doi: 10.1016/s1386-6532(02)00019-7. [DOI] [PubMed] [Google Scholar]
- 13.McDade TW, Burhop J, Dohnal J. High-sensitivity enzyme immunoassay for C-reactive protein in dried blood spots. Clin Chem. 2004;50:652–4. doi: 10.1373/clinchem.2003.029488. [DOI] [PubMed] [Google Scholar]
- 14.Skogstrand K, Thorsen P, Norgaard-Pedersen B, Schendel DE, Sorensen LC, Hougaard DM. Simultaneous Measurement of 25 Inflammatory Markers and Neurotrophins in Neonatal Dried Blood Spots by Immunoassay with xMAP Technology. Clin Chem. 2005;51:1854–66. doi: 10.1373/clinchem.2005.052241. [DOI] [PubMed] [Google Scholar]
- 15.Erhardt JG, Craft NE, Heinrich F, Biesalski HK. Rapid and simple measurement of retinol in human dried whole blood spots. J Nutr. 2002;132:318–21. doi: 10.1093/jn/132.2.318. [DOI] [PubMed] [Google Scholar]
- 16.O'Broin SD, Gunter EW. Screening of folate status with use of dried blood spots on filter paper. Am J Clin Nutr. 1999;70:359–67. doi: 10.1093/ajcn/70.3.359. [DOI] [PubMed] [Google Scholar]
- 17.McDade TW, Shell-Duncan B. Whole blood collected on filter paper provides a minimally invasive method for assessing human transferrin receptor level. J Nutr. 2002;132:3760–3. doi: 10.1093/jn/132.12.3760. [DOI] [PubMed] [Google Scholar]
- 18.Di Martino MT, Michniewicz A, Martucci M, Parlato G. EDTA is essential to recover lead from dried blood spots on filter paper. Clin Chim Acta. 2004;350:143–150. doi: 10.1016/j.cccn.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Burse VW, DeGuzman MR, Korver MP, Najam AR, Williams CC, Hannon WH, et al. Preliminary investigation of the use of dried-blood spots for the assessment of in utero exposure to environmental pollutants. Biochem Mol Med. 1997;61:236–9. doi: 10.1006/bmme.1997.2603. [DOI] [PubMed] [Google Scholar]
- 20.Gurney JG, Mueller BA, Davis S, Schwartz SM, Stevens RG, Kopecky KJ. Childhood brain tumor occurrence in relation to residential power line configurations, electric heating sources, and electric appliance use. Am J Epidemiol. 1996;143:120–8. doi: 10.1093/oxfordjournals.aje.a008718. [DOI] [PubMed] [Google Scholar]
- 21.Hartge P, Brinton LA, Rosenthal JF, Cahill JI, Hoover RN, Waksberg J. Random digit dialing in selecting a population-based control group. Am J Epidemiol. 1984;120:825–33. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]
- 22.The American Society of Human Genetics (ASHG) ASHG report. Statement on informed consent for genetic research. Am J Hum Genet. 1996;59:471–4. [PMC free article] [PubMed] [Google Scholar]
- 23.Therrell BL, Hannon WH, Pass KA, Lorey F, Brokopp C, Eckman J, et al. Guidelines for the retention, storage, and use of residual dried blood spot samples after newborn screening analysis: statement of the Council of Regional Networks for Genetic Services. Biochem Mol Med. 1996;57:116–24. doi: 10.1006/bmme.1996.0017. [DOI] [PubMed] [Google Scholar]
- 24.Newborn screening: A blueprint for the future executive summary: newborn screening task force report. Pediatrics. 2000;106:386–8. [PubMed] [Google Scholar]
- 25.Kelada SN, Stapleton PL, Farin FM, Bammler TK, Eaton DL, Smith-Weller T, et al. Glutathione S-transferase M1, T1, and P1 polymorphisms and Parkinson's disease. Neurosci Lett. 2003;337:5–8. doi: 10.1016/s0304-3940(02)01286-7. [DOI] [PubMed] [Google Scholar]
- 26.Farin FM, Janssen P, Quigley S, Abbott D, Hassett C, Smith-Weller T, et al. Genetic polymorphisms of microsomal and soluble epoxide hydrolase and the risk of Parkinson's disease. Pharmacogenetics. 2001;11:703–8. doi: 10.1097/00008571-200111000-00009. [DOI] [PubMed] [Google Scholar]
- 27.StataCorp . Stata statistical software: release 8.0. Stata Corporation; College Station, Texas: 2003. [Google Scholar]
- 28.Brophy VH, Jarvik GP, Furlong CE. PON1 Polymorphisms. In: Costa LG, Furlong CE, editors. Paraoxonase (PON1) in Health and Disease: Basic and Clinical Aspects. Kluwer Academic Publishers; Boston: 2002. [Google Scholar]
- 29.Chen J, Kumar M, Chan W, Berkowitz G, Wetmur JG. Increased influence of genetic variation on PON1 activity in neonates. Environ Health Perspect. 2003;111:1403–9. doi: 10.1289/ehp.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland N, Furlong C, Bastaki M, Richter R, Bradman A, Huen K, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ Health Perspect. 2006;114:985–91. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens EA, Taylor JA, Kaplan N, Yang CH, Hsieh LL, Lucier GW, et al. Ethnic variation in the CYP2E1 gene: polymorphism analysis of 695 African-Americans, European-Americans and Taiwanese. Pharmacogenetics. 1994;4:185–92. doi: 10.1097/00008571-199408000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–48. [PubMed] [Google Scholar]
- 33.Tamminga WJ, Wemer J, Oosterhuis B, de Zeeuw RA, de Leij LF, Jonkman JH. The prevalence of CYP2D6 and CYP2C19 genotypes in a population of healthy Dutch volunteers. Eur J Clin Pharmacol. 2001;57:717–22. doi: 10.1007/s002280100359. [DOI] [PubMed] [Google Scholar]
- 34.Chen CL, Liu Q, Relling MV. Simultaneous characterization of glutathione S-transferase M1 and T1 polymorphisms by polymerase chain reaction in American whites and blacks. Pharmacogenetics. 1996;6:187–91. doi: 10.1097/00008571-199604000-00005. [DOI] [PubMed] [Google Scholar]
- 35.National Newborn Screening and Genetics Resource Center (NNSGRC) National Newborn Screening Program - 2000. Austin, TX: 2002. [Google Scholar]
- 36.Chaisomchit S, Wichajarn R, Janejai N, Chareonsiriwatana W. Stability of genomic DNA in dried blood spots stored on filter paper. Southeast Asian J Trop Med Public Health. 2005;36:270–3. [PubMed] [Google Scholar]
- 37.Caggana M, Conroy JM, Pass KA. Rapid, efficient method for multiplex amplification from filter paper. Hum Mutat. 1998;11:404–9. doi: 10.1002/(SICI)1098-1004(1998)11:5<404::AID-HUMU8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]