Abstract
Many insects show polyphenisms, or alternative morphologies, which are based on differential gene expression rather than genetic polymorphism. Queens and workers are alternative forms of the adult female honey bee and represent one of the best known examples of insect polyphenism. Hormonal regulation of caste determination in honey bees has been studied in detail, but little is known about the proximate molecular mechanisms underlying this process, or any other such polyphenism. We report the success of a molecular-genetic approach for studying queen- and worker-specific gene expression in the development of the honey bee (Apis mellifera). Numerous genes appear to be differentially expressed between the two castes. Seven differentially expressed loci described here belong to at least five distinctly different evolutionary and functional groups. Two are particularly promising as potential regulators of caste differentiation. One is homologous to a widespread class of proteins that bind lipids and other hydrophobic ligands, including retinoic acid. The second locus shows sequence similarity to a DNA-binding domain in the Ets family of transcription factors. The remaining loci appear to be involved with downstream changes inherent to queen- or worker-specific developmental pathways. Caste determination in honey bees is typically thought of as primarily queen determination; our results make it clear that the process involves specific activation of genes in workers as well as in queens.
A key feature of insects is their ability to evolve polyphenisms, expressions of two or more morphologies with accompanying physiological and behavioral differences. The alternative forms expressed by polyphenic species can enable them to match form and physiology to environmental conditions (1). Especially intriguing are the cases in which alternative forms complement each other, performing different and mutually beneficial roles. A widespread form of such “cooperative polyphenism” is found in the highly eusocial insects. Highly eusocial insects are defined by the existence of distinct morphological phenotypes that enhance division of labor. More specifically, some colony members (i.e., workers) are largely or completely sterile, lacking fundamental reproductive equipment, whereas other colony members (queens) possess hypertrophied reproductive systems and large spermathecae and reproduce at disproportionately high rates (2, 3). Further, queens often have secondary physical characteristics such as wings, in ants and termites, that are absent in the worker caste. Workers, by contrast, can show physical traits not found in the queens of their species, including enhanced defensive and sensory organs. In highly eusocial insect species, as in all insects, adult morphology is the result of events that occur during larval development. Progress in understanding the developmental mechanisms that regulate caste determination in most eusocial insects has been frustratingly slow due in part to the small size of these organisms and the complex nature of their societies.
One of the best understood insect polyphenisms at the physiological level is the queen–worker dimorphism in honey bees (Apis mellifera) (3–6). The honey bee queen-rearing response is remarkable in terms of both speed and accuracy. New queens are reared with great reliability within 2 weeks after the disappearance of the existing queen, or in anticipation of colony splitting, yet queens are rarely produced as long as the active queen is sufficiently fertile for the colony’s needs (7). Cross-fostering experiments provided a conclusive demonstration (4) that caste determination in honey bees is decisively influenced by the environment experienced by larvae rather than by a genetic predisposition of some larvae to become queens or workers. Most importantly, queen-destined larvae receive a rich mixture of food throughout development that includes secretions from the mandibular gland of provisioning workers. Worker-destined larvae, in contrast, receive food containing this mandibular gland component for only the first 2 days of larval development (7). Honey bee queen larvae show much faster weight gains than worker larvae by the third instar of development, due primarily to enhanced feeding rates. By the end of the third instar, the corpora allata of queen-destined larvae become much enlarged relative to those of workers (8), and queen larvae begin to show substantially higher levels of the terpenoid juvenile hormone (9). The fourth larval instar (96 hours post-hatching) marks the divergence of two developmental pathways that were termed, classically, the worker and queen developmental caste programs (10).
Differentiation between the reproductive organs of developing workers and queens is well established during the fifth larval instar (6). From the early fifth instar onward, queens develop more rapidly than workers and ultimately eclose as adults several days earlier [summarized by Nijhout (5)]. The substantial physiological and morphological differences between queen and worker honey bees must reflect the differential expression of genes present in both castes. Here, we identify and describe seven genes that are differentially expressed during the bifurcation of the queen and worker developmental pathways. These genes provide a tool for examining the architecture of the developmental mechanisms that lead to insect polyphenisms.
MATERIALS AND METHODS
Rearing of Larvae.
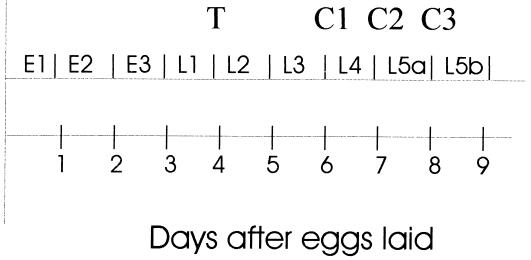
Worker- and queen-destined larvae used for preliminary screening were reared from colonies of A. mellifera in early spring, 1998, at the Carl Hayden Bee Research Laboratory (U.S. Department of Agriculture, Tucson, AZ). In each of these colonies, the queen was allowed to lay eggs over a section of empty brood cells for a period of 4 hours, after which she was excluded. After 96 hours, a subset of her offspring was transferred to standard queen-rearing cells (Fig. 1). These larvae, which had the potential to develop into new queens, were harvested after an additional 48, 72, or 96 hours concomitant with the removal of larvae of the same age that had been reared in worker cells. These time points coincide with early fourth, early fifth, and later fifth instars, respectively. Larvae were preserved at −80°C and then were weighed and placed on wet ice for ≈10 minutes before RNA extraction.
Figure 1.
Timing of Apis mellifera larval development and of the sampling regime for this project. T, larval transfer to artificial queen cells. C1, C2, and C3, collection of larvae at early fourth, early fifth, and later fifth larval instars, respectively.
Selection of Differentially Expressed Genes.
Isolation of mRNA and cDNA synthesis. To isolate messenger RNA (mRNA), queen and worker larvae were homogenized individually in a β-mercaptoethanol, guanidine thiocyanate buffer. A dT-cellulose matrix was added to this homogenate to preferentially bind mRNAs by their polyadenylated 3′ ends. After successive ethanol-salt washes [using the micro-polyA kit (Ambion, Austin, TX) or Message-Clean filters (Gene Mate, Kaysville, UT)], mRNA was eluted in 50 μl of RNase-free water. mRNA extracts from queen and worker larvae (≈1 μg) were used to generate cDNA by using reverse transcriptase (avian myeloblastosis virus; 20 units) and a poly-T synthesis primer (1uM; buffer conditions as prescribed by the PCR-Select Subtractive library kit, CLONTECH). A complementary strand for this cDNA was synthesized by using DNA polymerase I and DNA ligase, followed by treatment with RNase to remove remaining RNA and extension by T4 DNA polymerase (conditions described in ref. 11).
Suppressive-subtraction protocol.
We selected differentially expressed genes by using the suppressive-subtraction technique (11) as invoked by the PCR-Select kit (CLONTECH). In brief, cDNAs from queens and workers of the same developmental stage were subjected to blunt-end digestion by the restriction enzyme RsaI. Digests from each caste then were used as either tester cDNAs (i.e., pools of cDNA from which caste-specific genes were to be identified) or as driver cDNAs (cDNA pools used to selectively remove nondifferentially expressed genes from the tester DNAs). Tester cDNAs were split into two equimolar solutions, to which different PCR-primer adapter sequences were ligated [adapter “A” for one pool of cDNA, and adapter “B” from the other; primer sequences are described in PCR-Select kit manual (CLONTECH)]. No such adapters were ligated to the driver cDNA. After ligation, tester cDNAs were mixed with an excess of driver DNA isolated from larvae of the other caste (i.e., driver cDNA from queens was added to tester cDNAs of workers, and vice versa). Tester sequences having adapter A that remained single-stranded (did not bind to driver cDNA nor to other tester cDNAs) after the first hybridization were then allowed to hybridize with cDNAs having adapter B, again in the presence of driver cDNA. Adaptor “A-B” hybrid DNAs thereafter were amplified selectively by the PCR, leading to two pools, putatively biased toward caste-specific transcripts from genes (i) with higher expression in queens or (ii) with higher expression in workers.
Screening for Differential Expression.
PCR products from the subtracted libraries were screened for caste-specific differences after insertion into a phagemid vector by using the T/A cloning technique (Invitrogen). Plasmids were inserted into competent cells of Escherichia coli by transformation and then were grown on LB-agar media under ampicillin selection. Individual colonies that showed the presence of inserted DNA (by β-galactosidase expression) were picked, were grown in LB-ampicillin solution, were plated onto nylon membranes resting on LB-agar (Zeta-Probe, Bio-Rad Laboratories), and then were cultured overnight at 37°C. Cloned DNA then was denatured, neutralized, and affixed to the membranes by using standard methods (12).
Probes for hybridization were derived from the forward- and reverse-subtractive libraries as described in the PCR Select Differential Screening Kit (CLONTECH). Probes were radiolabeled through the incorporation of (α-32P)-dATP by random-primer labeling. Labeled probes then were purified by spin filtration (CLONTECH Chroma-spin 100 columns). Hybridization was carried out by using a rotisserie-style incubator (Gene-Roller GRH10, Savant). Membranes were prehybridized for 30 minutes at 72°C in a solution of 1 mM EDTA, 0.25 M Na2HPO4 (pH 7.2), and 7% SDS. Denatured probes (≈106 cpm signal intensity) were added and were hybridized to the cloned DNA’s overnight at 72°C. After hybridization, membranes were washed three times for 20 minutes in a low-stringency wash buffer [2× standard saline citrate (SSC)/0.5% SDS] and twice for 20 minutes in a high-stringency wash buffer (0.2× SSC/0.5% SDS). Membranes were wrapped in plastic and were exposed to film (Fuji RX) for 18–72 hours to visualize the hybridization signals.
Northern Blotting to Verify Differential Expression.
Clones representing candidate loci (i.e., those that appeared to be expressed differently based on the screening technique described above) were screened further by Northern blot analyses. These analyses were carried out with queen and worker larvae from the early fourth and early fifth larval instars. Total RNA was extracted from larvae by using the RNeasy extraction technique (Qiagen, Chatsworth, CA). For each sample, 2.5 μg of total RNA were denatured in a formamide buffer at 98°C for 5 minutes and then were loaded onto a formaldehyde-agarose gel for electrophoresis (12). RNA extracts were separated by electrophoresis for 3 hours at 10 V/cm and then were visualized by ethidium bromide staining to ensure equal concentrations of the respective RNAs (by quantifying ribosomal RNA bands). RNAs were denatured, neutralized, and transferred to nylon membranes by vacuum blotting (50 millibars of pressure for 3 hours). RNAs then were bound to the membranes by UV crosslinking.
Probes first were amplified from individual clones via PCR by using M13-Forward and M13-Reverse oligonucleotide primers and then were radiolabeled by random-prime labeling and incorporation of (α-32P)-dATP. Hybridization and membrane-washing conditions were as described above. Membranes were exposed to film (Fuji x-ray film) for between 8 hours and 12 days.
DNA Sequencing and Sequence Comparisons.
Candidate clones were amplified by using the PCR and standard cloning primers (M13-forward and M13-reverse). Clones were sequenced in both directions by using these primers and an ABI Prism 373 automated sequencing machine (Laboratory of Molecular Systematics and Evolution, University of Arizona, Tucson). DNA sequences were compared to those in the GenBank DNA and protein databases by using the blast-n and blast-x algorithms, at the DNA analysis web site maintained by the National Center for Biotechnology Information. The authors, on request, will provide sequence alignments used for inferring homology with known genes.
RESULTS
Numerous Candidate Clones Arose from the Initial Screening.
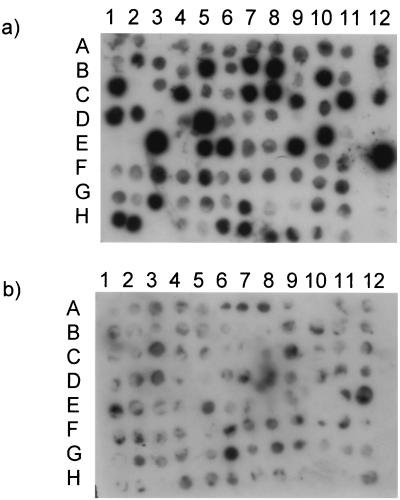
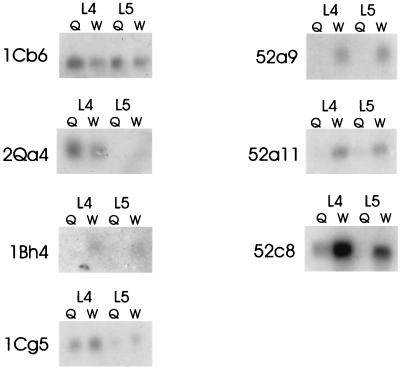
In total, we produced eight paired subtractive libraries for queens and workers, five for early fourth-instar larvae, two for early fifth-instar larvae, and one for later fifth-instar larvae. We screened 1,152 clones from these libraries, using the differential-screening protocol, generating 288 candidate differentially expressed clones (Fig. 2). The majority of the clones identified by this first screening failed to show significant differences in expression when assayed by Northern blotting. Furthermore, a substantial fraction of the 288 clones probably represented multiple captures of the same gene. Nevertheless, of 37 putatively unique clones (selected on the basis of insert size) screened by Northern-blot analyses, 7 different clones showed consistent differences in worker and queen expression (Fig. 3 and Table 1). Six of the seven clones came from subtractive libraries derived from larvae collected 48 hours after grafting whereas the last came from a library produced from larvae harvested after 72 hours. When these seven sequences were compared with those present in the GenBank nucleotide database [by using the blast-n algorithm (National Center for Biotechnology Information)], they did not match, significantly, with any existing sequences. Thus, all seven appear to be unique sequences at the DNA level. In contrast, six of the seven showed amino acid matches with proteins found in the GenBank protein databases when translated and matched by using the blast-x algorithm.
Figure 2.
Array of 96 putatively worker-expressed clones, shown after hybridization to probes derived from extracts of larval worker (a) or queen (b) mRNA. Dark signals in the top autoradiograph indicate cloned loci that were expressed at high levels by worker larvae.
Figure 3.
Autoradiographs showing Northern blot results for seven differentially expressed loci. Loci 1CB6 and 2QA4 were expressed more strongly in queen larvae than in worker larvae. Locus 1CG5 was expressed at quantitatively higher levels in workers. The remaining four loci were expressed exclusively by workers.
Table 1.
Differentially expressed loci, including estimated length of protein, in amino acids, and match probabilities resulting from a blast search of the GenBank database
| Clone | Sequence Match | Probability | Length | Expression |
|---|---|---|---|---|
| 1CB6 | Hexamerin | 0.7 | 2,400 | HQ |
| 2QA4 | Unknown | 1,450 | HQ | |
| 1BH4 | Fatty-acid Binding Protein | 3 × 10−6 | 1,400 | OW |
| 1CG5 | λ crystallin | 2 × 10−22 | 1,800 | HW |
| 52A9 | Oxidoreductase | 1 × 10−32 | 1,550 | OW |
| 52A11 | ELK-3 (Ets family) | 1.3 | 2,300 | OW |
| 52C8 | Hexamerin | 2 × 10−52 | 2,300 | OW |
The probabilities indicate the predicted number of GenBank entries that would have a similar match value by chance, HQ, higher expression in queens; HW, higher expression in workers; OW, only expressed by workers.
Queen-Biased Expression.
Locus 1CB6, whose product showed similarity to a family of insect storage proteins (hexamerins and arylphorins), was expressed at quantitatively higher levels in queens than in workers. The region encoded by this particular clone was aligned with amino acids 760–800 from larval storage protein 1 of Drosophila melanogaster (Table 1). A second cloned sequence fragment (2QA4) also was expressed at higher levels in queens than in workers. This clone was expressed by fourth-instar queen and worker larvae and appeared to be silent in fifth-instar larvae. The sequence from this clone failed to return significant matches from either the GenBank protein or DNA databases.
Worker-Biased Expression.
The locus represented by clone 1CG5 was expressed more strongly in workers than in queens (Fig. 3). Expression levels were particularly high in fourth-instar larvae and were decreased in both worker and queen fifth-instar larvae. The amino acid sequence for this locus closely matched that of λ crystallin, an apparent structural protein first described from lens tissues in mammals (13). These sequences are most similar in the region defined by amino acids 188–253 of the λ crystallin locus, where they show 66% sequence identity and 82% functional sequence similarity. The sequence from clone 1CG5 shows lower, but also significant, sequence similarity to the hydroxylacyl-coenzymeA dehydrogenases, most notably with the hydroxylacyl coA dehydrogenases from Streptomyces coelicolor and Mycobacterium tuberculosis.
Four additional clones represented loci that were turned on exclusively in workers. One of these, clone 1BH4, was closely related to cellular retinoic acid binding proteins widespread in vertebrates and to fatty-acid binding proteins more generally. This clone can be aligned with 80% of the fatty-acid binding protein described by Smith et al. (14) in the tobacco hornworm, Manduca sexta. A second worker-expressed clone, 52C8, encoded a hexamerin storage protein. The sequence from this clone was aligned with amino acids 460–660 of hexamerin larval storage protein 1 from D. melanogaster. Worker-specific clone 52A9 showed close similarity, over a 200-amino acid region, to members of the oxidoreductase enzyme family. Clone 52A11 was weakly similar to the mammalian Ets-domain transcription factor ELK-3. The sequence similarity between 52A11 and ELK-3 was highest between amino acids 210 and 300 of the ELK-3 locus (27% amino acid sequence identity, 42% functional similarity).
DISCUSSION
The evolution of castes in insect societies represents a major evolutionary transition from one level of organization to another (15). A defining feature of eusocial insects is the division of labor between colony members that reproduce and those that are largely or completely sterile. In the social Hymenoptera (ants, bees, and wasps), female larvae are channeled, often irreversibly, into either a queen or worker developmental pathway. In this study, we explored the molecular–genetic basis behind such switches. We studied a highly eusocial species, the honey bee, for which the worker–queen switch is both extreme and definitively controlled during development. In characterizing genes important for the determination of honey bee reproductive castes, we hope to provide material that can be used to study the origin and maintenance of eusociality more generally.
We identified and characterized transcripts from seven genes that are expressed differently by worker- and queen-destined larvae at a critical point in honey bee development. These genes are expressed differentially before or concurrently with physiological changes [e.g., in juvenile hormone production (6, 16)] currently used as key indicators of the start of caste differentiation. Differential expression during this time is hardly surprising, given general principles of development. Severson et al. (17) previously showed general differences in gene expression between queens and workers during the process of caste determination and differentiation. They used in vitro translation to detect the presence of differentially expressed genes in worker and queen larvae at 3-day intervals during development. Hartfelder et al. (18), examining differentiating ovaries during the late fifth larval instar, found caste-specific differences in the expression of two proteins. The molecular tools now available have allowed us to target times in early development in a way that should make a major contribution to understanding the architecture and ultimately the evolution of caste determination.
General Patterns of Expression.
Five of the seven gene products described here were expressed exclusively or primarily by worker larvae. Strikingly, four of these were completely silent in queens at the larval ages we assayed, although we found no queen-specific genes that were silent in workers. Data presented by Severson et al. (17) also suggest an early bias toward worker-transcribed genes followed by a general bias toward queen-specific gene expression in pre-pupae and pupae, periods during which much of the reproductive differentiation between queens and workers take place. This apparent worker bias in regulated genes, although somewhat counterintuitive, would fit well with known features of development and differentiation.
Immediately before the worker–queen developmental split, potential worker-destined larvae begin receiving a modified food source, worker jelly (7). Perhaps this shift in diet triggers determinative molecular events for expression of the worker pathway while queens continue to grow rapidly. Two of the four worker-exclusive genes from this life stage do appear to be related to metabolism. Assays of larval gene expression associated with diet change, along with experimental manipulations of diet, will help determine the extent to which larval gene expression is nutritionally driven.
More broadly, caste determination in social Hymenoptera is thought to involve two or more phases: an early switchpoint at which most larvae are fated to become workers and much later branches at which bipotential larvae become queens or workers (19). It is possible that early bursts of transcription in most female larvae seal their fates as workers while the remaining larvae, through the inhibition of worker-specific gene expression, remain bipotent. The generality of early worker-expressed genes that confine larvae to a worker pathway is best tested across species that show distinctly different timepoints for the commitment of larvae to a particular reproductive caste.
Characterizing the Differentially Expressed Loci.
Six of the seven differentially expressed clones showed significant matches to proteins in the GenBank database, based on their inferred amino acid sequences. Similarity to known sequences can only suggest possible functions because homologous proteins can diverge into both new structures and new functions (20). Nevertheless, the similarities shown by the six identified sequences indicate sufficiently plausible functions to warrant some discussion.
One worker-exclusive clone (52A11) showed a weak match to an important family of transcription factors (the Ets-domain family). These transcription factors appear to be involved in multiple signaling pathways important in developmental and metabolic processes throughout the eukaryotes (21). Clone 52A11 matched most closely to 90 amino acids in the mammalian Ets-family member ELK-3 (22). This particular protein is involved with transcriptional down-regulation during mouse embryogenesis.
A second worker-exclusive clone, 52A9, matched closely to members of the oxidoreductase enzyme family. Oxidoreductases form a diverse functional group. Many of the major metabolic enzymes in eukaryotes, including glucose dehydrogenase, fall into the oxidoreductase group (23), as do members of the cytochrome p450 gene family. Cytochrome p450 genes are major players in the detoxification by insects of secondary plant compounds and pesticides (24). Clone 52A9 is not a particularly close match to cytochrome p450 genes isolated in D. melanogaster (25). The worker-expressed oxidoreductase described here was expressed at high levels (Fig. 3), suggesting that it could play a major role in the metabolism of developing larvae. It is possible that this enzyme is involved in a metabolic pathway associated with the change in larval diet from royal to worker jelly. To this end, it will be interesting to determine whether the gene represented by clone 52A9 is first transcribed immediately after worker larvae are presented with worker jelly.
Storage Proteins.
Two of the seven messenger RNAs sequenced match closely with hexameric storage proteins. The hexamerins are a family of proteins found in the hemolymph and fat bodies of insects. They share the same evolutionary origin as hemocyanin proteins used for oxygen transport in Crustacea and other arthropods (26). Hexamerins store amino acids accumulated during larval development until they are used during metamorphosis or by the adult insects (27). Recently, Danty et al. (28) used antibodies to Drosophila larval storage proteins to reveal several hexamerins present in the hemolymph and antennal tissue of honey bees. Two of these hexamerins persisted longer in adult queens than in workers, suggesting caste differences in synthesis and utilization. These proteins were not sequenced fully and, consequently, it is not possible to determine whether they match our queen-biased hexamerin clone (clone 1CB6). Transcription of hexamerin is known to be sensitive to incoming nutrients; in two species of Lepidoptera, starvation inhibits hexamerin mRNA production (29, 30).
λ Crystallin and Other Crystallins.
Clone 1CG5 showed extremely high sequence similarity with a 60-amino acid section of the λ crystallin gene derived from rabbits (Oryctolagus cuniculus). λ crystallin, which makes up 7% of the protein biomass in developing rabbit eye lenses, is closely related to the hydroxylacyl coA dehydrogenase enzyme family (13). Along with several other lens crystallins, this locus provides evidence for the evolution of novel functions from loci that originally encoded basic metabolic enzymes (31). Crystallin proteins often still show some enzymatic activity, yet they occur at levels that far exceed those required for catalytic function. This overabundance of crystallins suggests that they perform additional structural or mechanical roles in the eyes (32) of both vertebrates and invertebrates (31). The fact that the honey bee sequence we present here (1CG5) and λ crystallin are more closely related to each other than either is to the hydroxyacyl enzyme family from which they were apparently derived suggests that 1CG5 performs a similarly derived, noncatalytic, function in developing larvae.
Fatty-Acid Binding Proteins.
Clone 1BH4, expressed only in worker larvae at the stages we examined, showed substantial similarity at the inferred amino acid level to a suite of fatty-acid binding proteins. These proteins, all ≈130 amino acids in length, are universally present in vertebrates, where they appear to bind retinoic acid among other molecules. The function of these proteins in insects is less well understood. Smith et al. (14) described two fatty-acid binding proteins from larvae of the tobacco hornworm, M. sexta. Conserved amino acids between these proteins and their vertebrate homologues suggested both a similar protein folding structure and similar sites for binding ligands. Mansfield et al. (33) recently described a similar fatty-acid binding protein, also in M. sexta, that closely matches the retinoic-acid binding proteins in vertebrates. This finding is particularly intriguing, in that retinoids bear structural similarity to juvenile hormone and steroid hormones and they are all synthesized from a common isoprenoid precursor in the mevalonate biosynthetic pathway (34). Juvenile hormone and ecdysteroids are implicated in both insect caste determination (35) and insect development more generally (5).
Implications for Honey Bee Caste Determination.
Differential gene expression in honey bee larvae undoubtedly plays a role in guiding the divergence in developmental pathways as well as generating at least some of the morphological and anatomical differences between the castes. We are optimistic that genes identified by the described strategy and method of reciprocal subtractive hybridization will speed the identification of processes that transduce nutritional signals to hormonal signals or will translate these hormonal signals into developmental responses. Our identification of loci that show similarity to the fatty-acid/retinoic-acid binding proteins and to Ets-family transcription factors is of interest because of the importance of these proteins in other insects. Studying the structure and transcriptional regulation of these loci should provide a better understanding of the process of caste determination in honey bees. Additional screenings of PCR products from our subtracted libraries certainly should yield additional candidate loci. As more loci accumulate, it should be possible to determine the covariance in expression between loci with respect to developmental stage and caste. Such analyses should be useful for uniting loci into similar developmental pathways.
Relevance to Other Social Insects and Insect Polyphenisms Generally.
Comparative gene-expression studies can be used to address questions of the origin of physical caste and the nature of caste specialization. Differentially expressed genes, such as those identified in the present study, can be used to screen other social insects for homologous genes that are expressed differentially. Thanks to the well described caste switchpoint in honey bees, this species can be used to generate numerous candidate genes that can then be tested for differential expression in less tractable species. Such comparative studies can be used to infer whether some caste-determining processes are homologous across social insects, despite the fact that higher eusociality clearly has had multiple origins (2, 36, 37).
One fundamental division in social insects is that between the highly eusocial species with morphologically distinct castes and the “primitively” eusocial species that lack them. In primitively eusocial insects, behavioral dominance hierarchies and physiological differences between queens and subordinates might reflect, or drive, differences in gene expression between these colony members. Comparative gene-expression studies can help determine whether processes used in regulating reproduction in adult nest mates are homologous to mechanisms used in reproductive caste determination.
Comparative gene-expression techniques also might be used to answer persistent questions related to the origin and affinities of specialized workers and queens in highly eusocial insects. The fact that the ancestors of modern-day workers must have been fertile females has been used to argue that the sterile worker is the most divergent caste (38). In fact, as pointed out by Michener (3), most of the queen’s specializations are “structural and behavioral losses compared with workers and with females of solitary bees” (ref. 3, p. 92). The solitary bees ancestral to honey bees would have had a more generalized range of behaviors than that shown by honey bee queens. Although honey bee queens have both a greatly hypertrophied reproductive system and specialized glands involved in communication with their worker offspring, they do not build or maintain the nest, care for brood, or forage. It is plausible that the evolution of honey bee queens, and of queens in the other highly eusocial insect species, has involved more often the suppression of particular genetic pathways than the induction of new ones. Tests for the derivation of particular worker and queen traits should be carried out for species showing a wide range of social behaviors and caste dichotomies. It will be particularly interesting to compare patterns in highly eusocial taxa with those found in relatives that show lesser degrees of differentiation [e.g., some wasps (37)] and social species that lack true polyphenisms [e.g., some halictid bees (39)].
Social insects, by virtue of the comparative material available, can serve as a model group for exploring how gene expression changes during the evolution of a major innovation and transition in life history (15). More generally, a greater understanding of the molecular genetics of caste determination in honey bees will provide broad insight into the mechanisms regulating insect polyphenisms. Phenotype-specific gene expression is almost certainly involved in trophic polyphenisms found in aphids (1) and caterpillars (40) and in the production of defender larval forms in parasitic wasps (41). Comparisons across insects of the molecular–genetic bases of polyphenisms should enhance our understanding of the developmental and evolutionary basis of alternative forms. Juvenile hormone, which plays an important role in honey bee caste determination, seems to be a common, though certainly not universal, mediator of a wide variety of polyphenisms in insects (35). Studies of gene expression should help clarify the mechanisms by which juvenile hormone and similar agents affect different developmental programs.
Evolutionary explanations for the diversification of polyphenisms (1, 42) depend on the assumption that polyphenisms reflect differential gene expression, yet this assumption has been supported primarily by indirect evidence (e.g., 43). Direct evidence of morph-specific gene expression, such as that described here, is a fundamental step in elucidating both the mechanisms and evolution of phenotypic diversity producible by a single genome.
Acknowledgments
We thank G. DeGrandi-Hoffman and J. Martin (U.S. Department of Agriculture–Carl Hayden Bee Research Laboratory, Tucson) for their careful rearing of queen and worker larvae and for their advice regarding the sampling of larvae. B. Dunkov, H. Hagedorn, J. Isoe, R. Meisfeld, F. W. Plapp, J. Seger, B. Tabashnik, J. Tu, and two anonymous reviewers offered valuable advice. M. J. West-Eberhard offered particularly insightful comments on the submitted manuscript. A National Institutes of Health postdoctoral training grant (5T32-AI-07475) administered through the Center for Insect Science, University of Arizona, supported J.D.E. Further research support was provided by a grant from the University of Arizona Foundation to D.E.W.
Footnotes
References
- 1.Moran N A. Am Nat. 1992;139:971–989. [Google Scholar]
- 2.Wilson E O. The Insect Societies. Cambridge, MA: Belknap; 1971. [Google Scholar]
- 3.Michener C D. The Social Behavior of the Bees. Cambridge, MA: Harvard Univ. Press; 1974. [Google Scholar]
- 4.Weaver N. Ann Entomol Soc Am. 1957;50:283–294. [Google Scholar]
- 5.Nijhout H F. Insect Hormones. Princeton, N. J.: Princeton Univ. Press; 1994. [Google Scholar]
- 6.Hartfelder K, Engels W. Curr Topics Dev Biol. 1998;40:45–77. doi: 10.1016/s0070-2153(08)60364-6. [DOI] [PubMed] [Google Scholar]
- 7.Laidlaw H H. In: The Hive and the Honey Bee. Graham J M, editor. Hamilton, IL: Dadant; 1992. pp. 989–1042. [Google Scholar]
- 8.Dogra G S, Ulrich G M, Rembold H. Z Naturforsch. 1977;32:637–642. [Google Scholar]
- 9.Rachinsky A, Strambi C, Strambi A, Hartfelder K. Gen Comp Endocrinol. 1990;79:31–38. doi: 10.1016/0016-6480(90)90085-z. [DOI] [PubMed] [Google Scholar]
- 10.Goewie E A. Med Landbouwhogeschool (Wageningin) 1978;75:1–75. [Google Scholar]
- 11.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, et al. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Mulders J W M, Hendriks W, Blankesteijn W M, Bloemendal H, de Jong W W. J Biol Chem. 1988;263:15462–15466. [PubMed] [Google Scholar]
- 14.Smith A F, Tsuchida K, Hanneman E, Suzuki T C, Wells M A. J Biol Chem. 1992;267:380–384. [PubMed] [Google Scholar]
- 15.Maynard Smith J, Szathmary L. The Major Transitions in Evolution. New York: Freeman; 1995. [Google Scholar]
- 16.Rachinsky A, Hartfelder K. Naturwissenschaften. 1991;78:270–272. [Google Scholar]
- 17.Severson D W, Williamson J L, Aiken J M. Insect Biochem. 1989;19:215–220. [Google Scholar]
- 18.Hartfelder K, Kostlin K, Hepperle C. Roux’s Arch Dev Biol. 1995;205:73–80. doi: 10.1007/BF00188845. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler D E. Am Nat. 1986;128:13–34. [Google Scholar]
- 20.Gerhardt J, Kirschner M. Cells, Embryos and Evolution. Malden, MA: Blackwell; 1997. [Google Scholar]
- 21.Sharrocks A D, Brown A L, Ling Y, Yates P R. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 22.Shore P, Bisset L, Lakey J, Waltho J P, Virden R, Sharrocks A D. J Biol Chem. 1995;270:5805–5811. doi: 10.1074/jbc.270.11.5805. [DOI] [PubMed] [Google Scholar]
- 23.Cavener D R. J Mol Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 24.Taylor M, Feyereisen R. Mol Biol Evol. 1996;13:719–734. doi: 10.1093/oxfordjournals.molbev.a025633. [DOI] [PubMed] [Google Scholar]
- 25.Dunkov B C, Rodriguez-Arnaiz R, Pittendrigh B, Ffrench-Constant R H, Feyereisen R. Mol Gen Genet. 1996;251:290–297. doi: 10.1007/BF02172519. [DOI] [PubMed] [Google Scholar]
- 26.Burmester T, Scheller K. J Mol Evol. 1996;42:713–728. doi: 10.1007/BF02338804. [DOI] [PubMed] [Google Scholar]
- 27.Telfer W H, Kunkel J G. Annu Rev Entomol. 1991;36:205–228. doi: 10.1146/annurev.en.36.010191.001225. [DOI] [PubMed] [Google Scholar]
- 28.Danty E, Arnold G, Burmester T, Huet J-C, Huet D, Pernollet J-C, Masson C. Insect Biochem Mol Biol. 1998;28:387–397. doi: 10.1016/s0965-1748(98)00011-3. [DOI] [PubMed] [Google Scholar]
- 29.Memmel N A, Ray A, Kumaran A K. Roux’s Arch Dev Biol. 1988;197:496–502. doi: 10.1007/BF00385683. [DOI] [PubMed] [Google Scholar]
- 30.Webb B, Riddiford L. Dev Biol. 1988;130:671–681. doi: 10.1016/0012-1606(88)90359-4. [DOI] [PubMed] [Google Scholar]
- 31.Tomarev S I, Piatigorsky J. Eur J Biochem. 1996;235:449–465. doi: 10.1111/j.1432-1033.1996.00449.x. [DOI] [PubMed] [Google Scholar]
- 32.Piatigorsky J Biol Chem. 1992;267:4277–4280. [PubMed] [Google Scholar]
- 33.Mansfield S G, Cammer S, Alexander S C, Muehleisen D P, Gary R S, Tropsha A, Bollenbacher W E. Proc Natl Acad Sci USA. 1998;95:6825–6830. doi: 10.1073/pnas.95.12.6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon M A, Boehm M F, Heyman R A, Mangelsdorf D J. Proc Natl Acad Sci USA. 1995;92:6157–6160. doi: 10.1073/pnas.92.13.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nijhout H F, Wheeler D E. Q Rev Biol. 1982;57:109–133. [Google Scholar]
- 36.Cameron S A. Proc Natl Acad Sci USA. 1993;90:8687–8691. doi: 10.1073/pnas.90.18.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Donnell S. Annu Rev Entomol. 1998;43:323–346. doi: 10.1146/annurev.ento.43.1.323. [DOI] [PubMed] [Google Scholar]
- 38.de Wilde J. In: Phase and Caste Determination in Insects: Endocrine Aspects. Luscher M, editor. Oxford: Pergamon; 1976. pp. 5–20. [Google Scholar]
- 39.Mueller U G. J Kans Entomol Soc. 1997;69:116–138. [Google Scholar]
- 40.Greene E. Science. 1989;243:643–646. doi: 10.1126/science.243.4891.643. [DOI] [PubMed] [Google Scholar]
- 41.Grbic M, Strand M R. Proc Nat Acad Sci USA. 1998;95:1097–1101. doi: 10.1073/pnas.95.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West-Eberhard M J. In: Molds, Molecules, and Metazoa: Growing Points in Evolutionary Biology. Grant P R, Horn H, editors. Princeton, N. J.: Princeton Univ. Press; 1992. pp. 57–75. [Google Scholar]
- 43.West-Eberhard M J. In: Natural History and Evolution of Paper Wasps. Turillazzi S, West-Eberhard M J, editors. Oxford: Oxford Univ. Press; 1996. pp. 290–317. [Google Scholar]