Abstract
The DNA sequence of the second hypervariable region of the mitochondrial control region of the Neandertal type specimen, found in 1856 in central Europe, has been determined from 92 clones derived from eight overlapping amplifications performed from four independent extracts. When the reconstructed sequence is analyzed together with the previously determined DNA sequence from the first hypervariable region, the Neandertal mtDNA is found to fall outside a phylogenetic tree relating the mtDNAs of contemporary humans. The date of divergence between the mtDNAs of the Neandertal and contemporary humans is estimated to 465,000 years before the present, with confidence limits of 317,000 and 741,000 years. Taken together, the results support the concept that the Neandertal mtDNA evolved separately from that of modern humans for a substantial amount of time and lends no support to the idea that they contributed mtDNA to contemporary modern humans.
The role of Neandertals with respect to the evolution of anatomically modern humans is controversial. Although some paleontologists view Neandertals as a distinct branch in hominid evolution that became extinct without any direct genetic contribution to present-day humans (1), others consider the Neandertals to be among the direct ancestors of modern Europeans (2). Recently, as a part of an interdisciplinary project of the Rheinisches Landesmuseum Bonn (3, 4), the DNA sequence of the first hypervariable region (HVRI) of the mtDNA from the Neandertal type specimen was determined (5). When compared with HVRI sequences of contemporary humans, the Neandertal mtDNA tended to fall outside the variation of modern humans. Furthermore, phylogenetic analyses suggested that the Neandertal mtDNA was an outgroup to the mtDNAs of modern humans, and the age of the most recent common ancestor (MRCA) of the mtDNAs of the Neandertal and modern humans was estimated to be about four times older than the age of the MRCA of modern human mtDNAs. These results indicate that the Neandertal mtDNA gene pool evolved for a substantial time period as an entity distinct from modern humans and give no indication that Neandertals contributed mtDNA to modern humans (5, 6).
However, because these analyses were based on a DNA sequence of only 333 bp, the results are less than conclusive. For example, the support for the placement of the Neandertal mtDNA outside the variation of modern human mtDNA in the phylogenetic tree was merely 89% (5). To better estimate the relationship of the Neandertal mtDNA to the current mtDNA gene pool, we have determined 340 bp of the second mtDNA HVR (HVRII) from the Neandertal type specimen and analyzed the relationship of the combined sequences to the contemporary human mtDNA gene pool.
MATERIALS AND METHODS
Experimental Procedures.
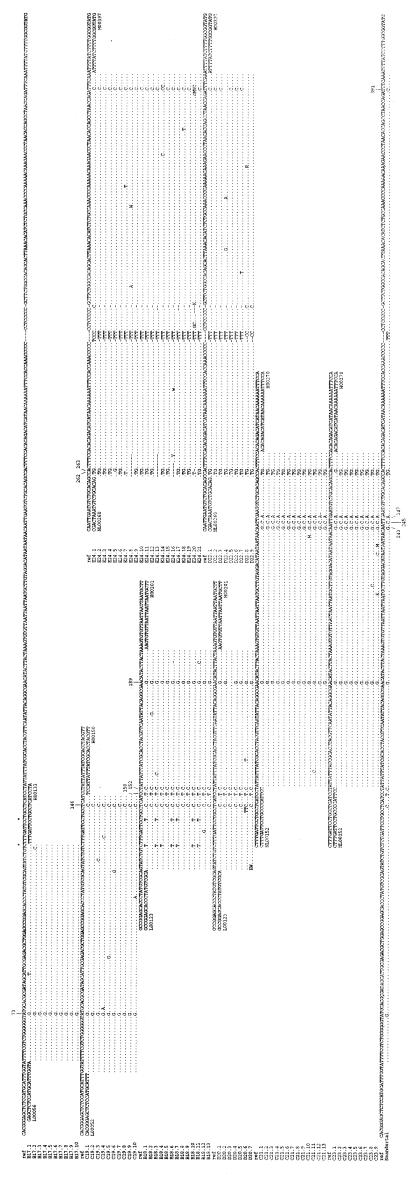
Sampling, precautions against contamination, DNA extraction, PCR amplifications, cloning of PCR products, and sequencing of clones were performed as described (5). Extracts A, B, and C were prepared previously (5), whereas extracts D and E were prepared for this work from 0.4 g of bone each. Extracts A, B, and C were known to yield PCR products that contained various proportions of modern human mtDNA sequences in addition to the Neandertal sequence (5). To test the degree of contamination of extracts D and E, PCRs were performed for a part of HVRI, which had been determined previously (primers L16209 and H16271; ref. 5). The PCR products were cloned, and 10 clones each were sequenced. In both extracts, 8 of 10 clones carried 7 substitutions and an adenosine insertion was determined for the Neandertal in this region (5), whereas two clones were identical to the reference sequence (data not shown). Thus, as in the case of extracts A and B, a small proportion of contaminating sequences is present in extracts D and E. Extract C, which was prepared in another laboratory, contains a majority of contaminating modern human DNA (5). Thus, whenever this extract was used, primers specific for putative Neandertal sequences determined from adjacent segments were used (cf. Fig. 1). There were 6, 2, and 19 clones derived from amplifications C21, C23, and E24 (Fig. 1), respectively, that contained sequences with only one or no difference from the reference sequence (7). These clones (not shown) were considered contaminants and were not included in the reconstruction of the Neandertal sequence. For the other amplifications, all clones sequenced and the primers used are shown in Fig. 1.
Figure 1.
HVRII mtDNA sequences of clones derived from PCR products from Neandertal bone extracts. Dots indicate base identity to the human reference sequence (7). Clone designations are made up of a letter (B, C, D, E) that refers to DNA extracts followed by the number of the amplification performed and, after the dot, the number of the individual clones. Primers used are given for the first clone of each PCR. Numbers in primer designations indicate the number of the 3′ base according to ref. 7, and L and H refer to the light and heavy strands of mtDNA. The sequences of the H primers are reverse and complemented. Standard designations are used for unidentified bases. The numbers of the positions at which substitutions in the Neandertal were found are indicated.
Alignments and Sequence Analyses.
For the analysis of the HVRI and HVRII sequences, the Neandertal sequences were aligned to a data set of 663 contemporary mtDNA lineages, i.e., distinct mtDNA sequences found among 682 contemporary humans (8). All human sequences with ambiguities in the reported sequences were excluded before the analysis. In addition, nine mtDNA lineages from seven common chimpanzees and two bonobos (“pygmy chimpanzees”) were used (9–14). At positions where insertions/deletions occurred between the sequences of the apes, humans and the Neandertal were excluded from the alignment. Sequence comparisons thus were based on a total of 600 nucleotide positions, encompassing positions 16024–16365 and 73–340, but excluding positions 16078, 16166, 252, 291, 299, and 317–321 (numbering according to ref. 7). Pairwise sequence differences were calculated by using unpublished software by A. von Haeseler (Max Planck Institute for Evolutionary Anthropology Leipzig, Germany).
The transition/transversion ratio was estimated by using the 663 human lineages and the program puzzle 4.0 (15), and genetic distances between pairs of sequences were calculated according to the F84 model using the “ML” option of the program “dnadist” from the phylip package (16). A neighbor-joining tree (17) was produced by using the program “neighbor” from phylip, and the support for branches in this tree was calculated with the likelihood mapping option in puzzle 4.0 (18). For the estimation of the age of the MRCAs, pairwise genetic distances were calculated with the Tamura—Nei algorithm (19) as implemented in puzzle 4.0. The parameters κ (transition/transversion ratio), τ (purine/pyrimidine transition ratio), and α, which describes the distribution of the evolutionary rates of individual nucleotide positions (20, 21), were estimated from the human sequences by using puzzle 4.0. Means and SDs of the genetic distances within and between species were calculated by using the program excel 4.0.
The rate of nucleotide substitution per position per year per lineage was calculated by the method of Tamura and Nei (19), using as a calibration point the divergence of humans and chimpanzees 4–5 million years ago (22, 23). The upper and lower confidence limits of this rate were estimated by using, respectively, the upper 95% confidence limit of the mean genetic distance and the younger date for the human—chimpanzee split, and the lower confidence limit of the genetic distance and the older date for the human—chimpanzee split. These two rates, in turn, were used to calculate the lower and upper limits of the age of the MRCA of the human and chimpanzee mtDNA gene pools.
RESULTS AND DISCUSSION
Retrieval of the Neandertal HVRII.
Primers designed to amplify human and chimpanzee mtDNA sequences were used to amplify a 122-bp segment (including primers) of HVRII from extract B, prepared from the 0.4 g of the right humerus of the Neandertal type specimen (24). A weak amplification product could be visualized on an agarose gel. This product was reamplified and cloned in a plasmid vector, and the inserts of 13 clones were sequenced (Fig. 1, B18). All clones carried four identical substitutions from the contemporary human reference sequence (7). Furthermore, two other positions differed from the reference sequence in six and seven clones, respectively, and four more positions showed substitutions in single clones. The same primers were used to amplify the same DNA segment from a different extract (Fig. 1, D20). Among the seven clones sequenced, the four substitutions found in all clones of the first amplification were found in all of these clones, with the sole exception of a G at position 189 in one of the clones. Neither the four singleton substitutions nor the two substitutions observed in several clones from the first amplification were seen among the clones from the second amplification. However, three more singleton substitutions were observed at positions that showed no variation among the first amplification. Substitutions that are not reproducible between amplifications are likely to be due to nucleotide misincorporations during the PCR, which may be induced by damage present in ancient DNA (25). That two nonreproducible substitutions occur in a large proportion of clones from one of the amplifications indicates that the amplifications start from very few template molecules, a supposition that agrees with the quantitation performed previously (5). When the sequence with the four substitutions seen in both amplifications was compared with a collection of 951 contemporary human mtDNA control region lineages (8), this combination of substitutions was not found, although all four positions are variable among humans.
To determine the rest of the HVRII sequence, additional amplifications partly overlapping with the above segment were performed from three different extracts, one of which had been prepared in another laboratory (extract C, ref. 5). All sequence positions from position 57 to 396 were scored by using 9–21 clones from at least two independent amplifications. Nucleotide states reproducible between amplifications were inferred to exist in the original template molecules.
Authenticity.
The sequence determined was considered to be derived from the mtDNA of the Neandertal individual for the following reasons. (i) An analysis of the state of preservation of amino acids in the bone (5) has shown that the conditions under which the bone has been preserved are compatible with macromolecular preservation. (ii) The DNA sequence could be amplified reproducibly from different extracts. Because the amount of bone available for extractions is limited, the HVRII sequence was not reproduced from an independent extract in another laboratory. However, this was done for the HVRI sequence determined previously (5). For the HVRII sequence, the extract prepared at Penn State University (extract C) yielded the same sequence as extracts prepared in Munich. (iii) The DNA sequence falls as an outgroup to modern human sequences in phylogenetic analyses (see below), an observation that may be taken to support that it is derived from the bone. However, in some cases, divergent mtDNA sequences derived from amplifications of contemporary DNA containing nuclear insertions of mtDNA segments have been misidentified as ancient sequences (26). Therefore, we designed a primer pair (NL152, cf. Fig. 1 and NH243 5′-TGG CTG TGC A A CAT TTA GTC-3′) that matches the sequence from the Neandertal type specimen and not contemporary human mtDNA sequences. Under amplification conditions that allow less than one copy of the Neandertal sequence per genome of human genomic DNA to be amplified, these primers failed to produce products in amplifications attempted from nine Africans, six Europeans, eight Asians, and three Australians/Oceanians. This makes a nuclear insertion an unlikely source of the sequence. (iv) If some form of miscoding DNA damage that was highly sequence-specific were prevalent in the Neandertal DNA molecules, this would result in nucleotide substitutions that would be reproduced between independent amplifications and thus would be mistaken for substitutions in the authentic Neandertal DNA sequence. We consider this unlikely because 37 of 38 positions in which a substitutional difference between the Neandertal and reference sequence are observed in HVRI and HVRII also show differences among modern humans, chimpanzees, and bonobos. Because 295 of 600 positions studied are variable in this data set, this would be an extremely unlikely result (P < 1.13 × 10−11) if the substitutions in the Neandertal were generated by a process different from the process generating the differences in the contemporary species, for example, some form of postmortem chemical damage.
Sequence Comparisons.
Among the 340 positions determined for HVRII, 11 transitional differences from the reference sequence (7) were identified. In addition, an insertion of three thymine residues occurs in a C-rich region after position 307 that shows length variation in humans (8).
To estimate the relationship of the Neandertal mtDNA to that of contemporary humans, positions 73–340 of HVRII were joined with positions 16024–16365 of HVRI and aligned to the homologous sequences from 151 Africans, 472 Europeans, 41 Asians, 10 Native Americans, and 15 Australian/Oceanians as well as 7 chimpanzees and 2 bonobos. Positions at which insertions/deletions occurred were excluded. Some humans shared the same sequences such that the data set could be collapsed to 663 different mitochondrial lineages.
The contemporary human mtDNA lineages differ at an average of 10.9 positions from one another and at 35.3 positions from the Neandertal (Table 1). Thus, on average, the Neandertal mtDNA has more than three times as many differences from modern human sequences as the latter have between them. In addition to the substitutions, the Neandertal sequences carry an insertion of an A after position 16263 in HVRI as well as the insertion of three T residues in HVRII. It may be noted that a small fraction (0.037%) of the interhuman comparisons are larger than the smallest distance (29 substitutions) between the Neandertal and humans.
Table 1.
Pairwise sequence differences between the Neandertal, humans, and some apes
| Humans (n = 663) | Neandertal (n = 1) | Chimpanzees/ bonobos (n = 9) | |
|---|---|---|---|
| Humans | 10.9 ± 5.1 | 35.3 ± 2.3 | 93.4 ± 7.1 |
| (1–35) | (29–43) | (78–113) | |
| Neandertal | 94.1 ± 5.7 | ||
| (84–103) | |||
| Chimpanzees/ | 54.8 ± 24 | ||
| bonobos | (1–81) |
Shown are means ± SD and ranges (in parentheses) of pairwise sequence differences. The chimpanzee and bonobo DNA sequences used come from two P. troglodytes troglodytes, one P. troglodytes verus, four P. troglodytes spec., and two bonobos (Pan paniscus).
It has been suggested that Neandertals were among the direct ancestors of modern Europeans (2, 27). European mtDNAs therefore might be expected to have fewer nucleotide differences from the Neandertal mtDNA than mtDNAs from Africa or Asia. The modern human sequences from the different continents therefore were compared separately with the Neandertal sequence. The mtDNAs from Africa, Europe, and Asia were found to carry 34.4 ± 2.7, 35.8 ± 2.1, and 33.8 ± 2.0 differences from the Neandertal sequence, respectively. The modern human lineages displaying the fewest differences (29 substitutions) to the Neandertal mtDNA were found in Africa, but the closest lineages in Asia and Europe were almost as similar to the Neandertal (30 and 31 differences, respectively). Thus, the mtDNA gene pools of modern humans on these continents are equidistant from the Neandertal mtDNA.
Because of the stochastic nature of the accumulation of mutations and the heterogeneity of substitution rates among nucleotide positions, it is not surprising that some contemporary mtDNAs have fewer differences from the Neandertal mtDNA than the maximum number of differences seen among contemporary mtDNAs. This cannot be taken as an indication that these contemporary mtDNAs are more closely related in a phylogenetic sense to the Neandertal mtDNA than they are to other contemporary mtDNAs, as has been implied recently (28). Rather, phylogenetic analyses are needed to elucidate this question.
Phylogenetic Analysis.
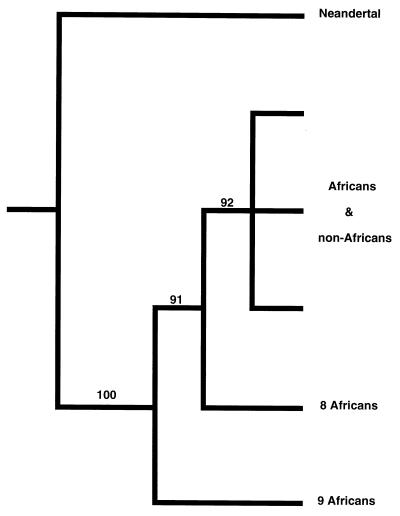
The observed nucleotide differences among all pairs of sequences of the Neandertal, the 663 modern humans, and the 7 chimpanzees and 2 bonobos were corrected for multiple substitutions, and a phylogenetic tree was constructed by using the neighbor-joining algorithm (17). In this tree the Neandertal forms the outgroup to the modern human mtDNAs (Fig. 2). The reliability of the branch connecting the Neandertal mtDNA with that of the modern humans was tested by the likelihood-mapping approach (18), where quartets of sequences involving the Neandertal, a chimpanzee or bonobo, and two humans were analyzed such that the probability of sampling each human sequence at least once was 0.96. For each quartet, the likelihood of each of the three possible tree topologies was calculated. In 100% of the 1,898,505 quartets tested the most likely topology had the Neandertal mtDNA as an outgroup to that of the humans. Furthermore, among the modern human mtDNAs, nine and eight African sequences were found to branch off on the first and the second branch after the Neandertal, respectively. These branches were supported by 91% and 92% of quartets, respectively.
Figure 2.
A schematic phylogenetic tree of the Neandertal mtDNA and 663 modern human mtDNA lineages. Only the relevant branches are shown. The tree was rooted with seven chimpanzees and two bonobos. Numbers on internal branches indicate quartet puzzling support values (18).
Thus, the phylogenetic analysis shows that the line leading to the Neandertal mtDNA diverged before the most recent common ancestor of the modern human mtDNA gene pool existed. Furthermore, as has been pointed out earlier (5, 29–31), the branching pattern among human mtDNAs suggests an African origin of the modern human mtDNA gene pool.
Dates of Divergences.
For the estimation of the ages of MRCAs of different groups of mtDNAs, the observed nucleotide differences were corrected for multiple substitutions by using the Tamura–Nei algorithm (17). The resulting genetic distances and the estimated age of the modern human–chimpanzee split of 4–5 million years (22, 23) were used to calculate the substitution rate of 0.94 × 10−7 substitutions per site per year per lineage with 5.92 × 10−8 and 1.38 × 10−7 as the lower and upper confidence limits. These estimates are in reasonable agreement with previous rate estimations for the mtDNA control region (32, 33). Using these rates, the age of the MRCA of the Neandertal and modern human mtDNAs was estimated to be 465,000 years, with confidence limits of 317,000 and 741,000 years. This age is significantly older than that of the MRCA of modern human mtDNAs, which, by the same procedure, was determined to be 163,000 years, with 111,000 and 260,000 years as confidence limits. Finally, the age of the MRCA of the mtDNAs of the seven chimpanzees and the two bonobos was calculated as 2,844,000 years (confidence limits: 1,940,000 and 4,534,000 years).
Relative Divergence Between Neandertals and Humans.
In western Europe, Neandertals and modern humans coexisted from approximately 40,000 years ago to less than 30,000 years ago (34). The implications of that coexistence in terms of culture and genetic relationships are a matter of debate. The results presented here indicate that the mtDNA gene pools of these two hominid forms had diverged for a substantial time before they came into contact. To put the extent of genetic differentiation that had resulted into perspective, a useful comparison may be the differentiation found today among chimpanzees and bonobos. The number of differences between the Neandertal and modern humans is 35.5 ± 2.3, about half that between chimpanzees and bonobos (75.7 ± 4.6). Unfortunately, HVRII sequences are not available for different subspecies of chimpanzees. However, if the analysis is confined to 312 bp of HVRI, the average difference between modern humans and the Neandertal is 25.6 ± 2.2, whereas that among 19 bonobos is 17.7 ± 8.5, among 10 central chimpanzees (Pan troglodytes troglodytes) is 14.6 ± 8.1, among 25 western chimpanzees (P. troglodytes verus) is 21.8 ± 9.7, and among 108 eastern chimpanzees (P. troglodytes schweinfurthii) is 7.9 ± 3.0. The observed differences between the subspecies varies from 19.7 ± 2.9 between central and eastern chimpanzees and 36.2 ± 6.1 and 33.0 ± 4.5 between western and central, and western and eastern chimpanzees, respectively. Thus, the average observed difference between the Neandertal mtDNA and the mtDNA of modern humans exceeds that occurring within chimpanzee subspecies and within bonobos, but is less than what is found between two of three pairwise comparisons between currently recognized subspecies of chimpanzees. When the differences are corrected for multiple substitutions, this general situation remains unchanged. However, mtDNA sequences from more Neandertal individuals are needed to obtain a better understanding of the extent of separation between the mtDNA gene pools of Neandertals and modern humans.
CONCLUSIONS
The divergence of the Neandertal mtDNA from the line leading to the contemporary human mtDNA gene pool is almost 3-fold older than the deepest divergence among contemporary human mtDNAs. The extent of sequence divergence exceeds that found within current chimpanzee subspecies. This shows that the Neandertal mtDNA and the human ancestral mtDNA gene pool have evolved as separate entities for a substantial period of time and gives no support to the notion that Neandertals should have contributed mtDNA to the modern human gene pool.
Acknowledgments
We are indebted to F. G. Zehnder and H.-E. Joachim (Rheinisches Landesmuseum Bonn) for permission to remove samples, to I. Boschi and V. Pascali for unpublished human mtDNA sequences, to P. Gagneux and U. Gerloff for unpublished chimpanzee and bonobo mtDNA sequences, to C. Capelli, S. Meyer, K. Strimmer, L. Vigilant, A. von Haeseler, and W. Schartau for discussion and technical help, and to the Boehringer–Ingelheim Fonds and the Deutsche Forschungsgemeinschaft for financial support.
ABBREVIATIONS
- HVRI and HVRII
first and second hypervariable region, respectively
- MRCA
most recent common ancestor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF142095).
References
- 1.Stringer C, McKie R. African Exodus. London: Random House; 1996. [Google Scholar]
- 2.Wolpoff M, Caspari R. Race and Human Evolution. New York: Simon & Schuster; 1997. [Google Scholar]
- 3.Schmitz R W, Pieper P, Bonte W, Krainitzki H. In: Advances in Forensic Sciences, Forensic Odontology. Jacob B, Bonte W, editors. Vol. 7. Berlin: Köster; 1995. pp. 42–44. [Google Scholar]
- 4.Schmitz R W. Doctoral thesis. Germany: University of Cologne; 1996. [Google Scholar]
- 5.Krings M, Stone A, Schmitz R-W, Krainitzki H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 6.Nordborg M. Am J Hum Genet. 1998;63:1237–1240. doi: 10.1086/302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–474. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 8.Handt O, Meyer S, von Haeseler A. Nucleic Acids Res. 1998;26:126–129. doi: 10.1093/nar/26.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnason U, Xu X, Gullberg A. J Mol Evol. 1996;42:145–152. doi: 10.1007/BF02198840. [DOI] [PubMed] [Google Scholar]
- 10.Foran D R, Hixson J E, Brown W M. Nucleic Acids Res. 1988;16:5841–5861. doi: 10.1093/nar/16.13.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg T L, Ruvolo M. Mol Biol Evol. 1997;14:976–984. doi: 10.1093/oxfordjournals.molbev.a025841. [DOI] [PubMed] [Google Scholar]
- 12.Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocher T D, Wilson A C. In: Evolution of Life: Fossils, Molecules and Culture. Osawa S, Honjo T, editors. Tokyo: Springer; 1991. pp. 391–413. [Google Scholar]
- 14.Morin P A, Moore J J, Chakraborty R, Jin L, Goodall J, Woodruff D S. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 15.Strimmer K, von Haeseler A. J Mol Evol. 1996;13:964–969. [Google Scholar]
- 16.Felsenstein J. phylip. Seattle: University of Washington; 1994. , Version 3.5. [Google Scholar]
- 17.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Strimmer K, von Haeseler A. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Nei M. J Mol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 20.Wakeley J. J Mol Evol. 1993;37:613–623. doi: 10.1007/BF00182747. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 22.Adachi J, Hasegawa M. J Mol Evol. 1995;40:622–628. doi: 10.1007/BF00160510. [DOI] [PubMed] [Google Scholar]
- 23.Takahata N, Satta Y, Klein J. Theor Popul Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- 24.King W. Q J Sci. 1864;1:88–97. [Google Scholar]
- 25.Höss M, Jaruga P, Zastawny T H, Dizdaroglu M, Pääbo S. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zischler H, Höss M, Handt O, von Haeseler A, van der Kuyl A C, Goudsmit J, Pääbo S. Science. 1995;268:1193. doi: 10.1126/science.7605504. [DOI] [PubMed] [Google Scholar]
- 27.Smith F H, Falsetti A B, Donelly S M. Yearbook Phys Anthropol. 1989;32:35–68. [Google Scholar]
- 28.Wolpoff M. Evol Anthropol. 1998;7:1–3. [Google Scholar]
- 29.Cann R L, Stoneking M, Wilson A C. Nature (London) 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 30.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson A C. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 31.Zischler H, Geisert H, von Haeseler A, Pääbo S. Nature (London) 1995;378:489–492. doi: 10.1038/378489a0. [DOI] [PubMed] [Google Scholar]
- 32.Jazin E, Soodyall H, Jalonen P, Lindholm E, Stoneking M, Gyllensten U. Nat Genet. 1998;18:109–110. doi: 10.1038/ng0298-109. [DOI] [PubMed] [Google Scholar]
- 33.Ward R H, Frazier B L, Dew-Jager K, Pääbo S. Proc Natl Acad Sci USA. 1991;88:8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hublin J-J, Barosso Ruiz C, Medina Lara P, Fontugne M, Reyss J-L. C R Acad Sci Paris. 1995;321:931–937. [Google Scholar]