Abstract
Alfalfa (Medicago sativa) and Arabidopsis were used as model systems to examine molecular mechanisms underlying developmental effects of a microsomal UDP-glucuronosyltransferase-encoding gene from pea (Pisum sativum; PsUGT1). Alfalfa expressing PsUGT1 antisense mRNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter exhibited delayed root emergence, reduced root growth, and increased lateral root development. The timing of root emergence in wild-type and antisense plants was correlated with the transient accumulation of auxin at the site of root emergence. Cell suspension cultures derived from the antisense alfalfa plants exhibited a delay in cell cycle from 24-h in the wild-type plants to 48-h in the antisense plants. PsUGT1::uidA was introduced into Arabidopsis to demonstrate that, as in alfalfa and pea, PsUGT1 expression occurs in regions of active cell division. This includes the root cap and root apical meristems, leaf primordia, tips of older leaves, and the transition zone between the hypocotyl and the root. Expression of PsUGT1::uidA colocalized with the expression of the auxin-responding reporter DR5::uidA. Co-expression of DR5::uidA in transgenic Arabidopsis lines expressing CaMV35S::PsUGT1 revealed that ectopic expression of CaMV35S::PsUGT1 is correlated with a change in endogenous auxin gradients in roots. Roots of ecotype Columbia expressing CaMV35S::PsUGT1 exhibited distinctive responses to exogenous naphthalene acetic acid. Completion of the life cycle occurred in 4 to 6 weeks compared with 6 to 7 weeks for wild-type Columbia. Inhibition of endogenous ethylene did not correct this early senescence phenotype.
The root cap meristem of higher plants, especially pea (Pisum sativum) and corn (Zea mays), has long been a favored model system to study the plant cell cycle because of its physical accessibility and ease of manipulation (Barlow, 2003). The cap meristem gives rise to organized tiers of cells dedicated to specific functions, including cell division in the meristem, starch synthesis and gravity sensing in the central columella, and starch degradation and secretion in the cap periphery. On the basis of the structurally regulated turnover of root cap development in maize, Barlow (1975) proposed that products of genes expressed in a particular cell tier, which is dedicated to a specific process such as mitosis, have a high probability of playing a role in executing the process during the time when that is active.
The cell cycle in the cap meristem was long presumed to be a constitutively active process that operates in tandem with the cell cycle in the apical meristem (Sievers and Braun, 1996). More recently, it has become clear that in many species, the cap meristem is regulated independently of the apical meristem, in response to diverse endogenous and environmental signals (for review, see Hawes et al., 2000, 2003). One factor controlling mitosis in the cap meristem in legumes is the presence of root border cells (or “sloughed root cap” cells) on the cap surface. Border cells release an extracellular signal, Factor B, which acts to prevent cap turnover by suppressing mitosis in the cap meristem (Brigham et al., 1998). Upon removal of border cells by immersion of the root tip into water for 30 to 60 s (or simply dilution of Factor B by dipping the tip into water), induction of renewed cap turnover occurs in correlation with a nearly instantaneous global switch in gene expression throughout the cap and a 8-fold increase in mitosis in the meristem within minutes (Brigham et al., 1998; Woo et al., 1999). Root cap development in legumes and cereals thus can be induced and tightly synchronized from one plant to another by the non-destructive removal of border cells from the cap periphery (Hawes and Lin, 1990). The use of the inducible root cap system to examine the cell cycle in higher plants makes it possible to examine factors controlling mitosis in synchronized cells within organized tissue, without the need for growth factor starvation or lengthy exposure to toxic chemical agents to block the process (Magyar et al., 1997; Planchais et al., 2000). Unfortunately, the best-characterized plant model, Arabidopsis, does not produce border cells; its root caps therefore are not subject to experimental induction and synchronization (Hawes et al., 2000). However, the cell cycle in plants is highly conserved, such that functional analysis of putative cell cycle-associated genes, once identified using the inducible root cap systems in cereals or legumes, ultimately can be carried out in Arabidopsis (Doonan and Forbert, 1997; den Boer and Murray, 2000a, 2000b; Breyne and Zabeau, 2001; Potuschak and Doerner, 2001).
In previous studies, the inducible root cap system in pea was used to examine predictions of Barlow's (1975) model (Woo et al., 1994, 1995; Woo and Hawes, 1997). The following approach was used to test the prediction that the pool of genes expressed in the cap meristem during the time when cell division is active, is enriched in genes controlling cell division: mRNAs whose expression is localized to the cap meristem during the 5-min window of time when mitosis is induced following border cell removal were isolated (Woo and Hawes, 1997). One of the genes corresponding to selected mRNAs encodes a previously unknown microsomal enzyme that transfers UDP-glucuronide to an unknown substrate; preliminary analysis of this unknown substrate indicates that it is probably a flavonoid (Woo et al., 2002). Expression of this pea UDP glucuronosyltransferase (PsUGT1), which is present in the pea genome in a single copy, is temporally and spatially correlated with the induction of mitosis and is required for cell survival (Woo et al., 1999). Thus, PsUGT1 mRNA levels increase 6-fold just before increased mitosis in the cap meristem is evident and then drop to background expression levels as mitosis drops to background levels. Moreover, inhibiting the inducible, meristem-localized expression of PsUGT1 in transgenic pea hairy roots by expressing PsUGT1 antisense mRNA under the control of its own promoter is lethal.
Inhibition of a PsUGT1 homolog in alfalfa (Medicago sativa) by expression of PsUGT1 antisense mRNA under the control of its own promoter also is lethal, suggesting a general role for the enzyme in regulating development in legumes (Woo et al., 1999). A less deleterious phenotype was achieved in alfalfa by expressing PsUGT1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter, which is expressed constitutively within the root cap meristem but is not regulated in correlation with the induction of root cap turnover. Thymidine labeling of DNA in roots synchronized by the use of 5-aminouracil revealed that the time required to complete a cell cycle was increased by 2-fold, which correlated with a 2-fold reduction in rate of root growth. The results were consistent with the hypothesis that PsUGT1 plays a critical role in the regulation of cell cycle in higher plants by reversible GlcUA conjugation of an unknown acceptor molecule (Woo et al., 1999). In the current study, predictions of this hypothesis were examined using alfalfa and Arabidopsis as complementary model systems.
RESULTS
Altered Adventitious Root Development in Response to Expression of CaMV35S::PsUGT1 Antisense mRNA
Alfalfa plants expressing CaMV35S::PsUGT1 antisense mRNA were sterile, so stem cuttings were propagated for further analysis of root development (as done by Woo et al. [1999]). A detailed analysis was conducted at each time point during adventitious root development from stem cuttings of wild-type and transgenic plants. For wild-type stem cuttings, root initials were visible in 7 d (Fig. 1A); by d 13, several long roots (>5 cm) were present. In contrast, visible root initials developed at the base of antisense stem cuttings only after 11 d (Fig. 1B), and they were fewer in number and length. These differences in root morphology were maintained in later time points over the course of 4 weeks (data not shown).
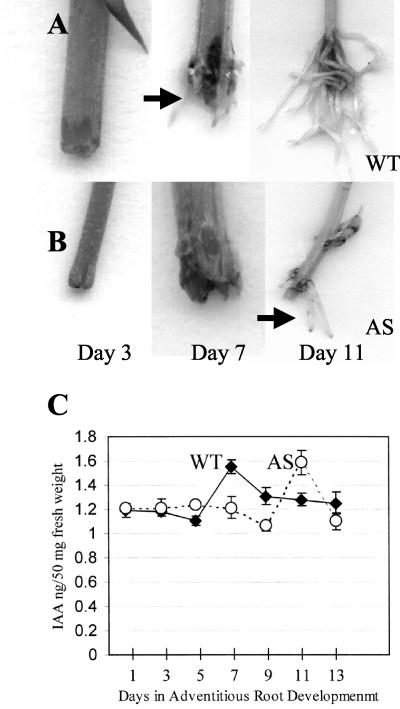
Figure 1.
Correlation between time of emergence of adventitious roots and a transient increase in free IAA at the site of adventitious root emergence: Adventitious roots (arrow) emerge at d 7 from wild-type stem cuttings (WT; A) and at d 11 from stem cuttings of plants expressing CaMV35S::PsUGT1 antisense mRNA (AS; B). C, Emergence of roots from both antisense (dotted line) and wild-type plants (solid line) was correlated with a transient increase in free IAA at d 7 and 11, respectively. IAA levels were quantified using gas chromatography/mass spectrometry (GC/MS) analysis of tissue from the base of stem cuttings; values represent means and sds from three independent experiments.
Correlation between Adventitious Root Initiation and a Transient Increase in Indole Acetic Acid (IAA) Accumulation
A 33% increase in endogenous free IAA levels occurred both in wild-type plants and in plants expressing CaMV35S::PsUGT1 antisense mRNA, at the point of adventitious root development (Fig. 1C). The increase in IAA occurred at d 7 in wild-type stem cuttings, but not until d 11 in plants expressing CaMV35S::PsUGT1 antisense mRNA. By d 9 in the wild-type stem cuttings and d 13 in the antisense stem cuttings, the free IAA levels decreased to background levels.
Altered Lateral Root Development in Response to Expression of CaMV35S::PsUGT1 Antisense mRNA
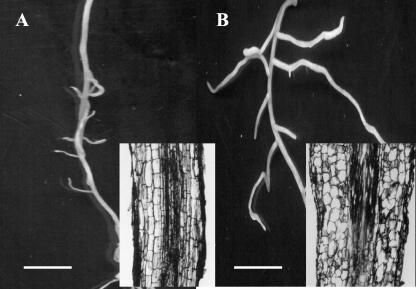
Lateral root growth in wild-type root clones (Fig. 2A) was not as extensive as in clones expressing CaMV35S::PsUGT1 antisense mRNA (Fig. 2B). The number of lateral roots also was increased slightly in plants expressing CaMV35S::PsUGT1 antisense mRNA (5.9 per cm) compared with wild-type roots (4.5 per cm). Differences in cell shape and organization between wild-type (Fig. 2A, inset) and antisense plants (Fig. 2B, inset) were also evident. In roots of plants expressing antisense mRNA, the cells were rounded rather than elongated as in wild type.
Figure 2.
Altered lateral root emergence and morphology in alfalfa plants expressing CaMV35S::PsUGT1 antisense mRNA. A, In wild-type adventitious roots, lateral roots emerged at an average number of 4.5 per centimeter; inset, in longitudinal section, root cells were elongated and arranged in parallel files; B, in roots of antisense plants, lateral roots emerged at an average number of 5.9 per centimeter; inset, in longitudinal section, the cells appear rounded and not elongated. Bars = 3 mm.
Delayed Cell Cycle in Suspension Cultures of Transgenic Alfalfa Expressing CaMV35S::PsUGT1 Antisense mRNA
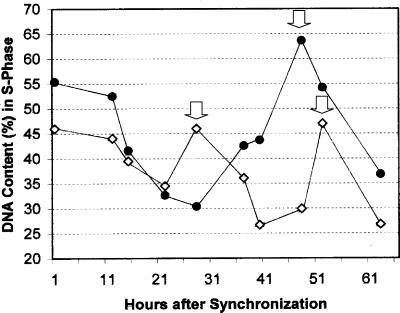
The cell division cycle was measured by checking DNA contents in nuclei isolated from alfalfa cell suspension culture using flow cytometry. Alfalfa cell suspension cultures were treated by 5-aminouracil (5-AU) to synchronize DNA synthesis during cell division. Flow cytometry revealed that the duration of cell cycle between the first and second S phases was about 24 h in wild-type cell cultures and 48 h in cell suspension cultures derived from transgenic alfalfa expressing CaMV35S::PsUGT1 antisense mRNA (Fig. 3).
Figure 3.
Increased duration of the cell cycle in alfalfa cell suspension cultures derived from plants expressing CaMV35S::PsUGT1 antisense mRNA. Durations of cell cycle between S phases were 24 and 48 h in cell suspension cultures of wild-type (white squares) and antisense plants (black circles), respectively. Peaks of DNA synthesis are denoted with arrows.
Generation and Analysis of Transgenic Arabidopsis Expressing PsUGT1::uidA
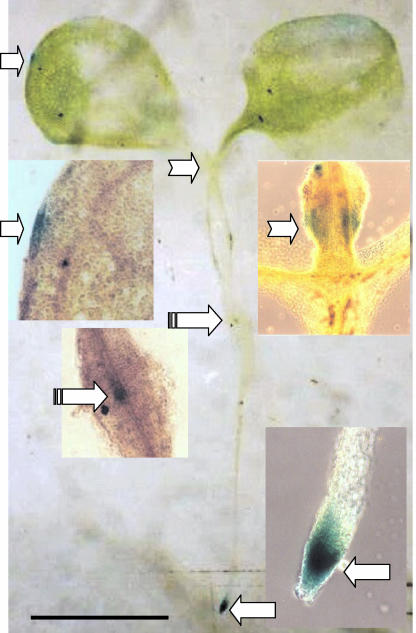
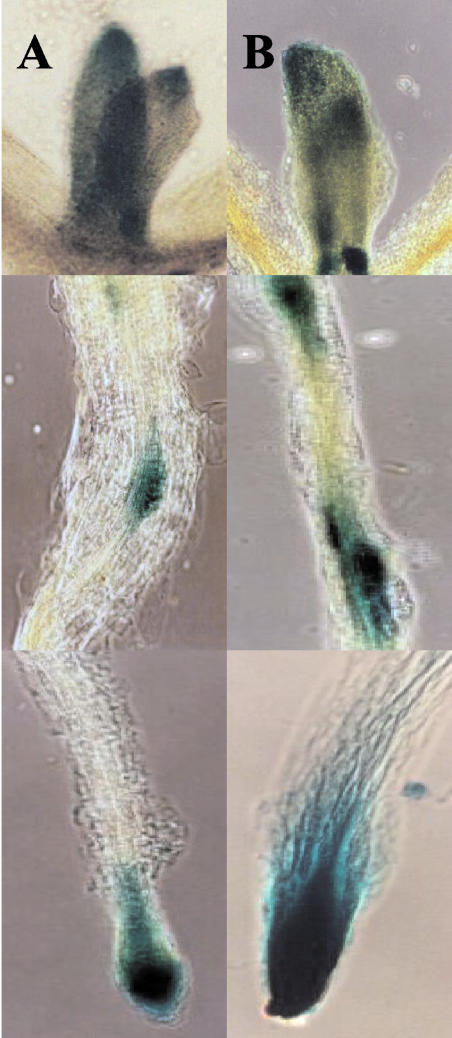
The uidA gene was used as a reporter to compare PsUGT1 expression patterns in Arabidopsis with those in pea and alfalfa. Recovery of transgenic plants expressing PsUGT1::uidA was very low (data not shown). An Arabidopsis seedling expressing the PsUGT1::uidA reporter gene demonstrated that, as in alfalfa, expression was localized to regions undergoing active cell division including the root apical meristem, the root cap meristem, and root primordia (Fig. 4). The highest activity occurred in the root apical meristem, with strong expression also occurring in the root cap meristem. Expression based on β-glucuronidase (GUS) staining also was observed in leaf primordia, at the tips of cotyledons, and in the transition zone between the hypocotyl and the root (Fig. 4).
Figure 4.
Expression of PsUGT1::uidA in Arabidopsis. Localized expression occurred in the outer tip of leaves (right arrows); leaf primordia (notched arrows); transition zone between hypocotyls and root (banded arrows); and the root apical and root cap meristems (left arrows). In the enlargement, the tissue was cleared to show the staining better. Bar = 3.5 mm.
Generation and Analysis of Transgenic Arabidopsis Expressing CaMV35S::PsUGT1
Fertile transgenic Arabidopsis ecotype Columbia plants that constitutively express PsUGT1 under the control of the CaMV35S promoter were developed. Among 19 independent lines expressing CaMV35S:: PsUGT1, several exhibited high levels of PsUGT1 transcript expression in leaves (not shown) and were used to examine the impact of PsUGT1 expression on development.
Reduced Life Cycle in Arabidopsis Plants Expressing PsUGT1
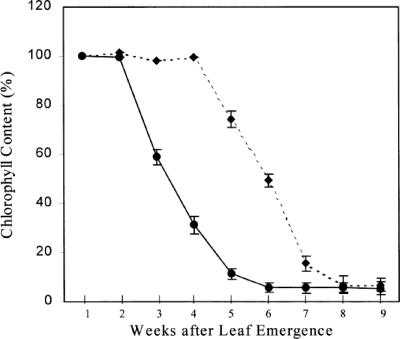
Columbia expressing CaMV35S::PsUGT1 required 6 to 7 d of cold treatment before germination compared with 2 to 3 d of cold treatment for wild-type seedlings. Under controlled environmental conditions, wild-type Columbia produced flowers 4 to 5 weeks after germination, and most plants were dead within 6 to 7 weeks (data not shown). In contrast, plants expressing CaMV35S::PsUGT1 produced flowers 3 to 4 weeks after germination and most plants were dead within 5 weeks (not shown). The differences in the onset of senescence between plants expressing CaMV35S::PsUGT1 and wild-type Columbia were quantified by measuring loss of chlorophyll from leaves (Fig. 5). Wild-type leaves started to lose chlorophyll by 5 weeks after germination, and loss was complete by 8 weeks (Fig. 5, dotted line). In contrast, leaves of plants expressing CaMV35S::PsUGT1 began to lose chlorophyll by 3 weeks after germination, and loss was complete by 6 weeks (Fig. 5, solid line).
Figure 5.
Quantitative assessment of senescence based on loss of chlorophyll over time in leaves of wild-type plants (dotted line) and transgenic plants expressing PsUGT1 (solid line).
Lack of Effect of Ethylene on Early Senescence in Arabidopsis Expressing CaMV35S::PsUGT1
To examine the possibility that the early senescence phenotype in plants expressing CaMV35S::PsUGT1 was related to ethylene accumulation, plants were treated with silver nitrate, which prevents ethylene action (Reid, 1987), and aminoethoxyvinyl-Gly (AVG), which inhibits ethylene biosynthesis (Mckeon and Yang, 1987). Treatment with silver nitrate and AVG had no influence on the early senescence phenotype. The life cycle of CaMV35S::PsUGT1 plants was the same duration either with or without the treatment to inhibit ethylene production (as in Fig. 5).
Colocalization of Expression of DR5:uidA and PsUGT1:uidA in Arabidopsis Seedlings
The auxin-responsive reporter DR5::uidA was transformed into wild-type plants and lines expressing CaMV35S::PsUGT1 and was stained for GUS activity 3 and 7 d after seed germination. In 3-d-old wild-type seedlings, histochemical staining was localized to the emerging leaf primordia, the transition zone between the hypocotyl and the root, and the root apex, as reported by Hinmanen et al. (2002) and Avsian-Kretchmer et al. (2002; data not shown). The expression of the DR5::uidA reporter (Fig. 6) colocalized with expression of PsUGT1::uidA (Fig. 4).
Figure 6.
Altered auxin responsiveness in plants expressing CaMV35S::PsUGT1, demonstrated by the synthetic auxin-responsive reporter DR5::uidA. A, In wild-type Columbia 7 d after germination, expression of the DR5::uidA (top to bottom) is localized to the leaf primordium and the tip of the leaf, with no expression in roots except as sites of lateral root initiation and the primary root apex. B, Ectopic PsUGT1 expression lines 7 d after germination also exhibit expression of DR5::uidA (top to bottom) in the leaf primordium and the tip of the leaf, and there is increased expression at sites surrounding lateral root primordia and throughout the primary root apex.
In 7-d-old wild-type seedlings, the expression of the DR5::uidA reporter also was localized to leaf primordia, the tips of the older leaves where auxin is synthesized (Fig. 6A), the transition zone between the hypocotyl and the root (not shown), lateral root initiation sites, and the root apex (Fig. 6A). Little or no expression occurred in the vascular system or elsewhere within root tissue. In 3-d-old seedlings expressing CaMV35S::PsUGT1, histochemical staining in response to the DR5::uidA reporter was enhanced at all sites of localized expression, especially in the root apex and lateral root primordia (not shown). Similarly, in roots of 7-d-old seedlings expressing CaMV35S::PsUGT1, histochemical staining as measured by the DR5::uidA reporter was increased in tissue surrounding sites of lateral root initiation, and enhanced staining was observed in the root apex and adjacent cells (Fig. 6B).
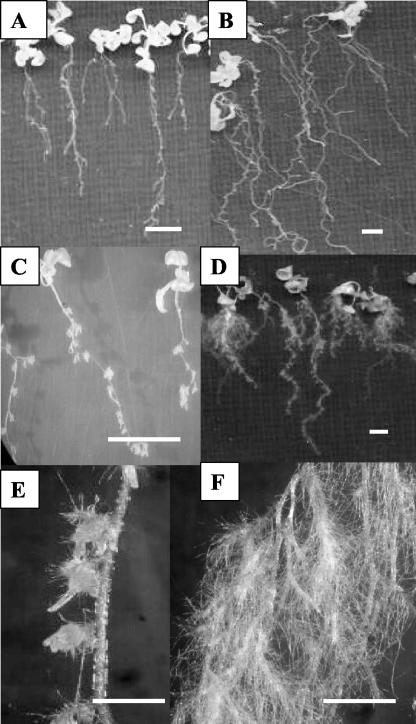
Altered Phenotypic Responses of CaMV35S::PsUGT1-Expressing Roots to Exogenous Naphthalene Acetic Acid (NAA)
To examine whether the capacity to respond to exogenous auxin changes in parallel with the observed changes in endogenous auxin distribution, wild-type and CaMV35S::PsUGT1 plants were treated with IAA, 2,4-dichlorophenoxyacetic acid (2,4-D), and NAA at 10–6 to 10–10 m (Fig. 7). Responses were identical, with two notable exceptions: NAA at a concentration of 10–8 m caused mild inhibition of root growth (Fig. 7A) and at 10–7 m strongly inhibited root growth (Fig. 7, C and E) of wild-type roots. In contrast, roots of plants expressing CaMV35S::PsUGT1 exhibited dramatic changes in morphology including root “waving” and “curling” (Fig. 7B) with treatments of 10–8 m NAA and enhanced root hair development (Fig. 7, D and F) in response to 10–7 m NAA.
Figure 7.
Phenotype effects of NAA treatment implant expressing CaMV35S::PsUGT1. A, No effect from treatment of 10–8 m NAA in wild-type roots. B, Treatment of 10–8 m NAA caused root curling/waving in ectopic expression lines. C and E, Treatment of 10–7 m NAA inhibited root growth and enhanced lateral root development in wild-type plants. D and F, Treatment of 10–7 m NAA enhanced lateral root development and root hair growth in ectopic expression lines. Bars in A through D = 10 mm. Bars in E and F = 2.5 mm.
DISCUSSION
A wide variety of products in plants are reversibly conjugated by sugars including GlcUA, by the action of dozens of individual glycosyltransferase (GT)-encoding genes, many of which exhibit activity on a range of substrates (Vogt and Jones, 2000). To our knowledge, PsUGT1 is the first such enzyme whose inhibition produces a lethal phenotype (e.g. expression of PsUGT1 antisense mRNA under the control of its own promoter is lethal; Woo et al., 1999), suggesting that its normal substrate plays a key role in plant growth and development and that other GTs are unable to compensate for its activity. The effect of PsUGT1 appears to be dosage dependent, based on the finding that a milder phenotype was achieved using the CaMV35S promoter instead of the PsUGT1 promoter for expression of antisense mRNA. In previous studies, inhibiting PsUGT1 expression by expressing CaMV35S::PsUGT1 antisense mRNA in alfalfa resulted in a 24-h delay in completion of the cell cycle in root tips. The close temporal and spatial correlation between cell division in the root cap meristem and PsUGT1 expression suggested that the PsUGT1 substrate is a factor controlling cell cycle (FCC; Woo et al., 1999).
In the current study, an effect on duration of the cell cycle in cell suspension cultures from clonal alfalfa expressing CaMV35S::PsUGT1 antisense mRNA paralleled its effect on the cell cycle in roots. This result also is consistent with the hypothesis that FCC activity influences cell division resulting in a 24-h delay in the time required to complete a cell cycle in cultured cells as well as in organized tissue. Additional effects that occurred in alfalfa expressing CaMV35S::PsUGT1 antisense mRNA included delayed adventitious root initiation and development, altered lateral root initiation and development, and a loss of the normal elongated shape of cells in the root. Each of these phenotypes is characteristic of changes that can occur in response to changes in endogenous or exogenous auxin levels (Okada et al., 1991; Bennett et al., 1996; Garbers et al., 1996; Ruegger et al., 1997; Christensen et al., 2000; Benjamins et al., 2001; Casimiro et al., 2001; Rashotte et al., 2001; Marchant et al., 2002). For example, antisense suppression of ABP1 encoding a putative auxin receptor causes slow proliferation, eliminates auxin-induced cell elongation, and reduces cell division in tobacco (Nicotiana tabacum) cells (Chen et al., 2001). The possibility that a change in auxin activity occurs in correlation with altered PsUGT1 expression was tested directly. The results confirmed that the delay in onset of adventitious root development in stem cuttings corresponds to a delay in auxin accumulation at the base of the stem cuttings.
If FCC acts to directly or indirectly influence auxin activities resulting in altered cell division, then plants with altered PsUGT1 expression would be predicted to exhibit altered auxin levels and/or distribution during development. Unfortunately, detailed studies of PsUGT1 effects in alfalfa are hampered because overexpression yields no obvious phenotypes. Expression of PsUGT1 antisense mRNA under the control of its own promoter is lethal, and alfalfa expressing CaMV35S::PsUGT1 antisense mRNA are infertile (Woo et al., 1999). However, powerful new tools to monitor cellular auxin levels in situ have been developed in Arabidopsis (e.g. Ulmasov et al., 1997; Avsian-Kretchmer et al., 2002). Therefore its potential use as an alternative model to study PsUGT1 was evaluated by expressing uidA under the control of PsUGT1 promoter. As in alfalfa, expression in Arabidopsis was localized at sites of cell division including lateral root primordia, the root cap meristem, and the root apical meristem (Woo et al., 1999). In addition, expression of PsUGT1::uidA in Arabidopsis seedlings occurred at the outer edge of leaves, leaf primordia, and the transition zone between the hypocotyl and the root. This spatial pattern was virtually identical to GUS expression patterns that have been reported to occur using promoters of genes that function in the cell cycle and promoters such as DR5, which are induced in response to the presence of auxin (e.g. Ulmasov et al., 1997; den Boer and Murray, 2000a, 2000b; Hinmanen et al., 2002).
Whereas reduced PsUGT1 expression in alfalfa caused a delay in development, ectopic expression of PsUGT1 in Arabidopsis caused the reverse effect. The rate of development was increased, resulting in a faster life cycle than normal. Wild-type plants exhibited senescence after 6 to 8 weeks, but plants expressing CaMV35S::PsUGT1 were completely senescent within 5 weeks. Increased ethylene, whose concentrations can increase in response to a variety of stimuli including stress, can cause a similar induction of early senescence (Bleecker and Kende, 2000). The fact that both AVG and silver nitrate, which inhibit ethylene action by distinct mechanisms, failed to reverse the early senescence phenotype does not support the hypothesis that ethylene is responsible.
When CaMV35S::PsUGT1 was co-expressed with the auxin-responsive promoter DR5::uidA reporter in Arabidopsis, an increased GUS staining consistent with a change in distribution of auxin was observed, especially in root systems. Instead of a tight localization of GUS staining only within actively dividing cells, staining was enhanced throughout tissues surrounding sites of lateral root initiation, with particularly intense staining in the root apex and adjacent cells. Wild-type and transgenic roots also exhibited divergent responses to specific concentrations of exogenous NAA. An effect of PsUGT1 on auxin synthesis, stability, uptake, localization, and/or transport would account for all of the observed changes in cell division, growth, and development in alfalfa and Arabidopsis. Given the myriad roles played by auxin in plant development, a direct role in regulating cellular auxin levels would also account for the fact that suppression of its inducible, meristem-localized activity by antisense mRNA expression under control of its own promoter is lethal (Woo et al., 1999).
Several factors argue against the possibility that auxin is FCC, the unknown substrate that is reversibly conjugated by PsUGT1 in meristematic cells during mitosis. First, to our knowledge, auxin conjugation with GlcUA has not been reported to occur in plant tissues (Ljung et al., 2002; Zazimalova and Napier, 2003). Second, the changes in phenotype that occurred were not always directly correlated with predicted effects of a straightforward increase or decrease in available auxin in particular tissues. Thus, ectopic expression of PsUGT1 in Arabidopsis yielded an early senescence phenotype that would be predicted for increased auxin levels. However, alfalfa with reduced PsUGT1 exhibited increased lateral root formation, which also is associated with augmented auxin levels. In addition, levels of auxin in wild-type and antisense alfalfa stem cuttings were similar, but the time required for accumulation was delayed. Finally, preliminary chemical analysis of FCC indicates that its properties are consistent with those of flavonoids rather than with IAA and other auxins, and the predicted PsUGT1 protein contains the signature sequence of flavonoid-binding proteins (Woo et al., 1999).
An alternative model is that FCC might play a role in regulated uptake of auxin into specific cells. Interestingly, flavonoids have been implicated in regulating endogenous auxin levels by effects on polar auxin transport (Jacobs and Rubery, 1988; Murphy et al., 2000; Brown et al., 2001; Peer et al., 2001). The proposed mechanism involves reversible blocking of auxin transport by inhibition of the auxin efflux carrier complex. In our system, this model would implicate FCC as the active endogenous compound that regulates functional activity of auxin by modulating its movement and/or uptake. Thus, PsUGT1 would be predicted to control auxin levels by modulating the activity of FCC through conjugation. Under- or overexpression of the conjugating enzyme would result in deregulated auxin transport and/or uptake that would be predicted to cause diverse changes similar to the observed effects of PsUGT1 on regulation of cell cycle, gene expression, growth, development, and life cycle. This model can be tested once the structural characterization of FCC is complete; these studies are under way.
CONCLUSIONS
The results of this study demonstrate a critical role for a specific GT in growth and development in diverse plant species. Such genes potentially provide an attractive target for improved crop production (Vogt and Jones, 2000). With ever-accumulating evidence that flavonoids have many important functions in the human body, such as estrogen metabolism and anticancer activities (Lopez-Lazaro and Akiyama, 2002), plants with GT-induced changes in flavonoid activity also may serve as resources to augment human health (Folts, 2002; Steinberg et al., 2003).
MATERIALS AND METHODS
Plasmid and Bacterial Strains
For development of alfalfa (Medicago sativa) plants expressing PsUGT1 antisense mRNA, the Escherichia coli uidA gene encoding GUS was removed from the pBI121 vector using SstI and BamHI. The cDNA (+730 to +1,510) from PsUGT1 was ligated into the SstI and BamHI sites of pBI121 without the uidA gene, which resulted in the insertion of the PsUGT1 sequence into pBI121, in the opposite orientation, as presented by Woo et al. (1999). The resulting construct expressing the CaMV35S::PsUGT1 antisense mRNA was transformed into E. coli HB101 and then into Agrobacterium tumefaciens LBA4404 by triparental conjugation.
For ectopic expression of PsUGT1 in Arabidopsis, the pBI121 vector was linearized using EcoRI, and the EcoRI site was filled by Klenow fragment. The E. coli uidA gene encoding GUS was removed from pBI121 using BamHI. PsUGT1 DNA (+1 to +1,578) was generated by PCR using PsUGT-2 (BamHI) primer (ACTACGGATCCTTTCTTGTGGTAATTAGTTCTGC) and PsUGT-3 primer (TACTAAGATACAACAAAAGCTAG). After BamHI digestion, PsUGT1 DNA was ligated into pBI121 (BamHI and blunt end). The resulting construct was transformed into E. coli HB101 and then into A. tumefaciens ASE by triparental conjugation. For localization of PsUGT1 expression, the PsUGT1 promoter (–867 to +450) was cloned into HindIII and BamHI sites of pBI121. The resulting vector was transformed into E. coli HB101 and then into A. tumefaciens ASE by triparental conjugation.
Isolation and Derivatization of Free IAA from Alfalfa Stems
Free IAA was isolated and pentafluorobenzyl-derivatized according to Prisnsen et al. (2000). Alfalfa stem cuttings were grown in a controlled environmental chamber with continuous light. Three independent sets of stem cuttings that developed adventitious roots were collected at d 0, 3, 5, 7, 9, 11, and 13. The free form of IAA was extracted from the base of stems (2 mm) at the site of adventitious root emergence. For each sample, five to six stem sections (200 mg fresh weight) were collected in liquid nitrogen and ground with 2 mL of ice-cold extraction buffer consisting of methanol containing 0.02% (w/v) sodium diethyldithiocarbamate. Because previous GC/MS analysis showed that alfalfa stem tissue contains no detectable indole propionic acid (IPA), 100 ng of IPA was added to the sample as an internal standard. The IPA-spiked ground samples were extracted at 4°C for 2 h in dark. After centrifugation (24,000g, 4°C, 10 min), the supernatant was collected, loaded onto a disposable C18 cartridge (Bond Elut, Varian, Harbor City, CA) that had been previously activated with methanol, and washed with water. After loading the sample, non-retained material was removed by further washing with water and the IAA-containing fraction was eluted with 90% methanol. The eluate was dried in a vacuum evaporator, and the dried sample was redissolved in 400 μL of acetone containing 0.02% (w/v) sodium diethyldithiocarbamate.
Quantification of Free IAA by GC/MS Analysis
Fifty microliters of the partially purified IAA-containing sample in acetone was treated with 1 μL of 1-ethylpiperidine and 5 μL of α-bromopentafluorotoluene at 60°C for 45 min, after which the sample was dried under a nitrogen stream and resuspended in 20 μL of methanol (Edlund et al., 1995). One microliter of the derivatized sample was injected (splitless mode; 6890A GC, Agilent Technologies, Palo Alto, CA) onto a medium polarity fused silica capillary column (HP-5MS, 30-m, 0.32-mm-i.d., 0.25-μm-film thickness column) with the injector port and GC/MS transfer line at 250°C and the GC oven at 80°C for 1 min after injection, and then ramped to 250°C at 15°C min–1 to a plateau at 250°C. The end of the column was inserted into the chemical ionization source (methane reagent gas) of a time-of-flight mass spectrometer (GCT, Micromass, Milford, MA). The mass spectrometer was scanned (negative ion mode) from m/z 100 to 1,000 every second, and off-line, selected ion chromatograms were reconstructed for the signals at m/z 174.2 and 188.2 corresponding to the carboxylate fragment anions of IAA-PFB and IPA-PFB, respectively. Peaks were observed at 13.05 min for IAA-PFB and 13.42 min for IPA-PFB. The selected ion chromatograms were integrated using the MassLynx Quantitation software provided with the GCT. A standard curve was created from a series of standard samples containing the same amount of IPA and increasing amounts of IAA, by plotting the relative IAA response (area of m/z 174.2 peak/area of m/z 188.2 m/z peak) on the ordinate and concentration of IAA on the abscissa. The IAA content of biological samples was determined by interpolation of the relative IAA response from the standard curve. For all experiments, three sets of triplicate samples were prepared, and the data were presented as average values.
Cell Cycle Analysis in Alfalfa Cell Suspension Culture by Flow Cytometer
Transgenic alfalfa plants expressing the CaMV35S::PsUGT1 antisense mRNA were developed as described previously (Fang and Hirsch, 1998; Woo et al., 1999). Leaf discs from wild-type or transgenic plants were used to generate cell suspension cultures, which were maintained in appropriate media with or without antibiotics. Alfalfa cell suspension cultures were maintained in the dark. Alfalfa cell suspension cultures were treated by 12 mm 5-amino uracil for 2 h to synchronize mitosis (Jakob and Trosko, 1965). After removing 5-amino uracil, the cultures were collected for analysis after 0, 12, 15, 22, 28, 37, 40, 48, 52, and 63 h. Cells were washed with water, and nuclei were isolated from cells using a Percoll gradient. DNA contents in nuclei were measured by a flow cytometer (FACScan Analytic, BD Biosciences, San Jose, CA). Total DNA content in the diploid state, including G1, G2, and S phases, is 100%.
Plant Transformation, Selection, and Analysis of Transgenic Plants
Both wild-type Columbia and Arabidopsis cv Columbia (DR5::uidA; kindly provided by Tom J. Guilfoyle, University of Missouri, Columbia) were transformed with CaMV 35S::PsUGT1. Four-week-old seedlings were transformed by vacuum infiltration with A. tumefaciens ASE (CaMV 35S::PsUGT1) as described previously (Clough and Bent, 1998).
Seeds from regenerated plants were germinated on Murashige and Skoog medium containing kanamycin. For selection of transgenic plants, kanamycin-resistant primary plants were analyzed for the presence of the transgene by PCR using Taq polymerase. Template DNA was obtained from leaves. PsUGT-2 primer and PsUGT-3 primer, which amplify 1,578-bp PsUGT1 DNA present only in transgenic plants, were used for PCR analysis (Fig. 5).
Transgenic plants were under maintained in a controlled environmental chamber with 16 h of light (mixed fluorescent bulbs) at 21°C and 8 h of dark at 19°C and were carried through five generations before analysis of phenotype. Because transgenic plants required a 7-d cold treatment for germination, both wild-type and transgenic seeds were cold-treated at 4°C for 7 d.
For RT-PCR analysis, total RNA was isolated from leaves of wild-type and transgenic plants. First-strand cDNA was synthesized using PsUGT-3 as primer. The PsUGT-2 primer and PsUGT-3 primer were used for RT-PCR. The PCR product was fractionated on 1% (w/v) agarose gel and blotted onto nylon membrane. 32P-labeled probes were generated by random priming using PsUGT1 DNA. Blot hybridization was followed as in standard protocols.
Phenotype Analysis and Measurement of Chlorophyll Content
The life cycles of transgenic plants were measured by observing leaf senescence up to 9 weeks after germination. Chlorophyll content was measured according to Lichtenthaler (1987). One gram of fresh, green leaves was boiled in 95% (v/v) ethanol at 80°C for 5 min. During boiling, the container was wrapped with aluminum foil to prevent photobleaching. Absorbance was measured at 662 nm for chlorophyll a and at 644 nm for chlorophyll b. Chlorophyll content was calculated as follows.
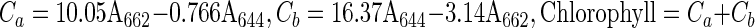 |
Treatment of AgNO3 and AVG to Inhibit Ethylene Activity and Synthesis
To inhibit the action of ethylene, AgNO3 (1–100 mg L–1 medium) was used. To prevent ethylene synthesis, 10–5 to 10–8 m AVG was added to growth medium. One-week-old seedlings were transferred to petri dishes containing AgNO3- or AVG-treated medium. Seedlings were observed daily, for up to 2 months. Six-week-old cultures were photographed.
Histochemical GUS Assays
Three- to 7-d-old seedlings were stained by 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, by standard procedures. Samples were observed and photographed using a stereomicroscope or light microscope.
Treatment of IAA, 2,4-D, and NAA to Arabidopsis Seedlings
Seeds were germinated in auxin-free medium composed of Murashige and Skoog minimal organics medium (Sigma-Aldrich, St. Louis) solidified with 1.5% (w/v) agar (type E, Sigma-Aldrich). The Suc content within the medium was 0.3% (w/v). The plates were kept in the dark, at 4°C for 7 d. Afterward, the plates were transferred to a growth chamber (20°C, 100% relative humidity, and 16-h/8-h light/dark cycle; Conviron, Winnipeg, Manitoba, Canada). One-week-old seedlings were transferred to petri dishes containing the same medium, plus IAA, 2,4-D, or NAA as indicated.
For assays of root waving/curling, seedlings grown on medium in square petri dishes were positioned vertically. Three-week-old cultures were evaluated by direct observation, and phenotypes were documented photographically.
Acknowledgments
We thank Fushi Wen for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026278.
This work was supported by the National Science Foundation (to M.C.H. and H.-H.W.), by the Department of Energy, Division of Energy Biosciences (to M.C.H. and H.-H.W.), by the University of Arizona College of Agriculture and Life Sciences Experiment Station, and by the National Institutes of Health (grant no. NCCAMSPSO AT00151 to the Center for Dietary Supplement Research Botanicals at University of California, Los Angeles to A.M.H. [director, Agricultural Botany Core] and H.-H.W. [junior investigator]). The National Science Foundation is acknowledged for partial support for purchase of mass spectrometric instrumentation (CHE grant no. 007829).
References
- Avsian-Kretchmer O, Cheng J-C, Chen L, Moctezuma E, Sung ZR (2002) Indole acetic acid distribution coincides with vascular differentiation pattern during Arabidopsis thaliana leaf ontogeny. Plant Physiol 130: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PW (1975) The root cap. In JG Torrey, DT Clarkson, eds, The Development and Function of Roots. Academic Press, London, pp 21–54
- Barlow PW (2003) The root cap. J Plant Growth Regul (in press)
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schultz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Breyne P, Zabeau M (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol 4: 136–142 [DOI] [PubMed] [Google Scholar]
- Brigham LA, Woo H-H, Wen F, Hawes MC (1998) Meristem-specific suppression of mitosis and a global switch in gene expression in the root cap of pea by endogenous signals. Plant Physiol 118: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis thaliana. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM (2001) ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D (2000) Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- den Boer BGW, Murray JAH (2000a) Triggering the cell cycle in plants. Trends Cell Biol 10: 245–250 [DOI] [PubMed] [Google Scholar]
- den Boer BGW, Murray JAH (2000b) Control of plant growth and development through manipulation of cell cycle genes. Curr Opin Biotechnol 11: 138–145 [DOI] [PubMed] [Google Scholar]
- Doonan J, Forbert P (1997) Conserved and novel regulators of the plant cell cycle. Curr Opin Cell Biol 9: 824–830 [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklof S, Sundberg B, Moritz T, Sandberg G (1995) A microscale techniques for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol 108: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Hirsch AM (1998) Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol 116: 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folts JD (2002) Potential health benefits from the flavonoids in grape products on vascular disease. Adv Exp Med Biol 505: 95–111 [DOI] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruere J, Bernasconi P, Soll D (1996) A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J 15: 2115–2124 [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Bengough GA, Cassab G, Ponce G (2003) Root cap and rhizosphere. J Plant Growth Regul (in press)
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X (2000) The role of root border cells in plant defense. Trends Plant Sci 5: 128–132 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Lin H-J (1990) Correlation of pectolytic enzymes activity with the programmed release of cells from root caps of pea. Plant Physiol 94: 1855–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinmanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH (1988) Naturally occurring auxin transport regulators. Science 24: 346–349 [DOI] [PubMed] [Google Scholar]
- Jakob KM, Trosko JE (1965) The relation between 5-aminouracil-induced mitotic synchronization and DNA synthesis. Exp Cell Res 40: 56–67 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 350–382 [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of IAA in Arabidopsis thaliana. Plant Mol Biol 50: 309–332 [DOI] [PubMed] [Google Scholar]
- Lopez-Lazaro M, Akiyama M (2002) Flavonoids as anticancer agents: structure-activity relationship study. Curr Med Chem Anti Cancer Agents 2: 691–714 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Meszaros T, Miskolczi P, Deak M, Feher A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M et al. (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklof J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckeon TA, Yang S-F (1987) Biosynthesis and metabolism of ethylene. In PJ Davies, ed, Plant Hormones and Their Role in Plant Growth and Development. Martinus Nijhoff Publishers, Dordrecht, The Netherlands, pp 94–112
- Murphy A, Peer WA, Taiz L (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211: 315–324 [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol 126: 536–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S, Glab N, Inzé D, Bergounious C (2000) Chemical inhibitors: a tool for plant cell cycle studies. FEBS Lett 476: 78–83 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Prisnsen E, Van Laer S, Oden S, Van Onckelen H (2000) Auxin analysis. In GA Tucker, JA Roberts, eds, Plant Hormone Protocols, Methods in Molecular Biology, Vol 141. Humana Press, Totowa, NJ, pp 49–65 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MS (1987) Ethylene in plant growth, development, and senescence. In PJ Davies, ed, Plant Hormones and Their Role in Plant Growth and Development. Martinus Nijhoff Publishers, Dordrecht, The Netherlands, pp 257–279
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday GK, Estelle M (1997) Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A, Braun M (1996) The root cap: structure and function. In Y Waisel, A Eshel, U Kafkafi, eds, Plant Roots: The Hidden Half, Ed 2. Marcel Dekker, New York, pp 31–49
- Steinberg FM, Bearden MM, Keen CL (2003) Cocoa and chocolate flavonoids: implications for cardiovascular health. J Am Diet Assoc 103: 215–223 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Woo HH, Brigham LA, Hawes MC (1994) Primary structure of the mRNA encoding a 16.5-kDa ubiquitin-conjugating enzyme of Pisum sativum. Gene 148: 369–370 [DOI] [PubMed] [Google Scholar]
- Woo HH, Brigham LA, Hawes MC (1995) Molecular cloning and expression of mRNAs encoding H1 histone and an H1 histone-like sequences in root tips of pea (Pisum sativum L.). Plant Mol Biol 28: 1143–1147 [DOI] [PubMed] [Google Scholar]
- Woo HH, Hawes MC (1997) Cloning of genes whose expression is correlated with mitosis and localized in dividing cells in root caps of Pisum sativum L. Plant Mol Biol 35: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Woo HH, Kuleck G, Hirsch AM, Hawes MC (2002) Flavonoids: signal molecules in plant development. In BS Buslig, JA Manthey, eds, Flavonoids in Cell Function, Advances in Experimental Medicine and Biology, Vol 505. Kluwer Academic/Plenum Publishers, New York, pp 51–60 [DOI] [PubMed] [Google Scholar]
- Woo HH, Orbach M, Hirsch AM, Hawes MC (1999) Meristem-localized inducible expression of an UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell 11: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazimalova E, Napier RM (2003) Points of regulation for auxin regulation. Plant Cell Rep 21: 625–634 [DOI] [PubMed] [Google Scholar]