Abstract
Bowman-Birk inhibitor (BBI) genes encode serine protease inhibitors that have repetitive cysteine-rich domains with reactive sites for the trypsin or chymotrypsin family. We have identified seven BBI genes from japonica rice (Oryza sativa subsp. japonica var Teqing). All of the genes identified were found in a single cluster on the southern end of the long arm of rice chromosome 1. Four of the seven BBI genes have two repetitive cysteine-rich domains, whereas one has a truncated domain with only one reactive site. We have also identified three novel BBI genes, each of which possesses three repetitive domains instead of two. In situ hybridization analyses indicated that the accumulation of rice BBI transcripts was differentially regulated in germinating embryos and also in the leaves, roots, and flower organs at later developmental stages. Different members of the rice BBI gene family displayed different expression patterns during rice seed germination, and wounding induced the expression of rice BBI transcripts. The three-domain BBIs had higher expression levels than the two-domain BBIs. It was also found that the mRNA of rice BBI genes was present in abundant amounts in scutellar epithelium and aleurone layer cells. RBBI3-1, one of the three-domain RBBI, exhibited in vitro trypsin-inhibiting activity but no chymotrypsin-inhibiting activity. Overexpression of RBBI2-3 in transgenic rice plants resulted in resistance to the fungal pathogen Pyricularia oryzae, indicating that proteinase inhibitors confer resistance against the fungal pathogen in vivo and that they might play a role in the defense system of the rice plant.
Plants have developed defense systems to combat various pathogens throughout their life cycle, from the seed stage until senescence, and it is particularly important that the embryo be kept free from infection. There are several embryonic defense mechanisms, including the production of plant lectins and pathogen-related proteins in response to attacks by pathogens or insects (Swegle et al., 1992; Ye et al., 2001; Guiderdoni et al., 2002). A well-known defense component is Ser protease inhibitors. They are expressed in developing seeds and are thought to play an important role in inhibiting trypsin and chymotrypsin of external origin (Ryan, 1981). Two major Ser protease inhibitors have been studied extensively in plants: Kunitz inhibitors and Bowman-Birk inhibitors (BBIs; Ryan, 1990). BBIs are Cys-rich proteins of about 8 to 16 kD with disulfide bonds and are encoded by a family of related genes. The BBI gene family has been found in both the Fabaceae and the Poaceae. BBIs identified in Fabaceae, such as soybean (Glycine max) and lima bean (Phaseolus lunatus), are 8-kD proteins. They have one BBI domain with two reactive sites for trypsin and the related enzymes, such as chymotrypsin (Birk, 1987). These protease inhibitors are double-headed, with two reactive sites in a single inhibitor molecule. Interestingly, this type of inhibitor displays anticarcinogenic activity (Birk, 1993; Kennedy, 1993).
The BBIs found in barley (Hordeum vulgare) and foxtail millet (Setaria italica) are either 8- or 16-kD proteins (Nagasue et al., 1988; Tashiro et al., 1990). The three-dimensional structures of the 8- and 16-kD BBIs have been explored (Li de la Sierra et al., 1999; Song et al., 1999). The 8-kD inhibitors found in these monocotyledonous plants are single-headed, with only one reactive site in the BBI domain, and inhibit only trypsin. In contrast, the 16-kD inhibitors have two homologous BBI domains with one reactive site in each domain. It is therefore thought that the 16-kD inhibitors evolved from the monocotyledonous 8-kD single-headed inhibitors (Prakash et al., 1996).
The first amino acid sequence of a mature 16-kD BBI protein from rice (Oryza sativa) was reported 15 years ago for the rice bran trypsin inhibitor (RBTI; Tashiro et al., 1987). The cDNA of RBTI was later identified (Chen et al., 1997) and characterized (OsBBPI; Rakwal et al., 2001). Similar to other 16-kD BBIs of monocotyledonous plants, RBTI also has two homologous domains.
We have previously described the biochemical purification of RAFP1, an antifungal protein that is produced by germinating rice seeds and displays inhibitory activity on trypsin (Liu et al., 1994). The N-terminal amino acid sequence of RAFP1 is homologous to the rice BBI, RBTI (Tashiro et al., 1987). In this study, we have synthesized degenerate primers and cloned a family of rice BBI genes. In addition to the previously known types of BBIs, we have identified a novel type of BBI that has a third Cys-rich domain in the N terminus with a potential trypsin-reactive site. Significantly, the genes we have cloned are clustered on the southern end of chromosome I, although they possess different expression patterns. Prokaryotic expression of RBBI3-1 shows that the fusion protein has trypsin-inhibiting activity but no chymotrypsin-inhibiting activity. Transgenic overexpression of the gene RBBI2-3 in rice cv Taipei 309 resulted in strong resistance in the seedling period to the fungal pathogen responsible for rice fungus blast disease, Pyricularia oryzae. The importance of the Ser protease inhibitors in disease resistance will also be discussed.
RESULTS
Cloning of Rice BBI Genes
Two degenerate primers were designed: The 5′ primer was designed based on the N-terminal amino acid sequence of RAFP1 (Liu et al., 1994), whereas the 3′ primer was made according to the sequence of the mature protein of a rice BBI, RBTI (Tashiro et al., 1987). We performed a PCR using rice genomic DNA as the template, with a 450-bp fragment being cloned and sequenced. A clone designated as RBBI8 contained an open reading frame (ORF), and the deduced amino acid sequence was about 74% identical to that of the rice BBI, RBTI (Xie et al., 1996). The RBBI8 gene was expressed in Escherichia coli, and the purified recombinant protein showed strong inhibitory activity against trypsin, as well as slight inhibitory activity against chymotrypsin (Li et al., 1999).
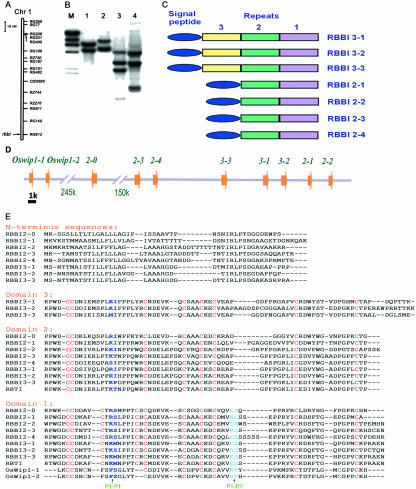
Using a recombinant F2 population of japonica rice (Oryza sativa subsp. japonica cvs Moroberekan and CO39), RBBI8 was found to cosegregate with a single molecular marker RG612 that is located on the distal southern end of rice chromosome 1 (Fig. 1A). Southern-blot hybridization using the RBBI8 gene as a probe showed a multiple band pattern in all three restriction enzymes used in this analysis (Fig. 1B). These results indicate that RBBIs in rice consist of a multigene family of at least four members.
Figure 1.
Molecular cloning of the rice BBI gene family. A, Diagram of rice chromosome 1. The RBBI gene cluster is located on the southern end of this chromosome as indicated by the arrow near the genetic marker, RG612. B, Southern blot using RBBI8 (see “Materials and Methods”) insert (the conserved coding region of RBBI2-3, 450 bp in length) as a probe. Rice genomic DNA was digested with BamHI (1), XhoI (2), BamHI + XhoI (3), and BamHI + EcoRV (4). C, Diagram showing the structural composition of the RBBI family. D, Composition of the RBBI gene cluster on chromosome 1. The arrowhead indicates the directions of the ORFs. Bar = 1 kb. E, Alignment of the conserved Cys-rich domains. Gaps are indicated by a dash. Putative trypsin- and/or chymotrypsin-reactive sites are indicated in blue letters, and the conserved Cys residues are in red.
To clone all of the members of this gene family, a rice bacterial artificial chromosome (BAC) library constructed from the japonica rice var Teqing was screened using RBBI8 as a probe. The four positive BAC clones were found to be the same with respect to patterns of restriction fragments. Fragments from the BAC clone that hybridized with RBBI8 probes were subcloned and sequenced (GenBank accession nos. AJ277468, AJ277469, AJ277470, and AJ277472). Within the BAC clone, a total of seven ORFs were found to encode proteins that are homologous to BBIs from both dicots and monocotyledon plants.
Denomination, Genomic Organization, and Sequence Analysis of the Rice BBI Gene Family
The seven members of the rice BBI gene family were found clustered in a BAC clone (33H2). All of the BBI genes encoded Cys-rich domains. Similar to previously identified BBI proteins, rice BBIs also contain homologous Cys-rich domains. Significantly, these domains are repeated within a protein in most rice BBIs. We have classified our RBBI clones based on the number of the Cys-rich domain repeats (Fig. 1C). As shown in the Figure 1C, those with two repeats were named RBBI2 and those of three repeats were named as RBBI3.
The organization of rice BBI genes in the BAC clone 33H2 is shown in Figure 1D. All of the seven BBI ORFs were tandem clustered on the same strand. The two-domain class members RBBI2-1, RBBI2-2, RBBI2-3, and RBBI2-4 encode proteins of 184, 193, 185, and 190 amino acid residues, respectively (Fig. 1E). The Cys-rich domain composition of the RBBIs of this class has domain 1 with putative double reactive sites at its C terminus and domain 2 with one putative reactive site. The three-domain class members RBBI3-1, RBBI3-2, and RBBI3-3 encode proteins of 251, 259, and 254 amino acid residues, respectively (Fig. 1E). The RBBIs of this class have an additional homologous Cys-rich domain 3 with one putative reactive site in the N terminus.
The RBBI8 gene is identical to the coding region of RBBI2-3, which shows high similarity (from 72.5% to 85.3% identity) to other six members of RBBI gene family at the DNA level. RBTI cDNA (Chen et al., 1997; Rakwal et al., 2001) is identical to the RBBI3-3 gene in the 3′-flanking region (208 bp) and only possesses a 6-bp difference in the coding region (data not shown). The sequence differences between the RBTI cDNA and RBBI3-3 are therefore most likely to represent differences between orthologs of different rice sub-species. Moreover, our sequence data showed that the reported RBTI cDNA sequence (Chen et al., 1997; Rakwal et al., 2001) could be incomplete because it is shorter than the RBBI3-3 transcript in the 5′-terminal region as determined by reverse transcriptase (RT)-PCR and sequencing (M. Liu, J. Chen, and L.-J. Qu, unpublished data).
Three more RBBI genes were further identified from the genomic database of rice after draft sequences of rice genome were released (Goff et al., 2002; Yu et al., 2002). Two of them have one domain, and the other has two domains. The two single-domain RBBI genes were similar to Wip1, a wound-inducible BBI gene found in maize (Zea mays), wheat (Triticum aestivum), barley, and sorghum (Sorghum bicolor; Tiffin and Gaut, 2001), and therefore were designated as Oswip1-1 and Oswip1-2. The one with two domains was similar to the RBBI2s we identified and was therefore designated as RBBI2-0. Interestingly, these 10 RBBI genes were located in a locus of rice chromosome 1 that spanned a region of about 430 kb (Fig. 1D).
Each RBBI interestingly has a highly hydrophobic N-terminal sequence that shows weak homology with the others (Fig. 1E) and that may serve as a signal peptide. The N-terminal sequence for the two-domain members ranges from 52 to 55 amino acid residues in length, but varies between 42 to 45 amino acid residues for the three-domain members (Fig. 1E).
Transcripts of Rice BBIs Can Be Detected in Many Different Tissues
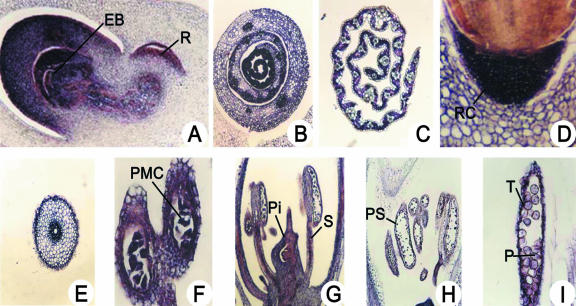
We used in situ hybridization utilizing the coding region of RBBI2-3 as a probe to detect the overall expression patterns of all of the rice BBI gene members in different rice tissues. The data showed that RBBI mRNA could be detected by the antisense probe in many different tissues at different developmental stages (Fig. 2), whereas no signal was detected with the sense probe (data not shown). It was found that mRNA started to accumulate in the embryo (both in the plumule and radicle) 24 h after rehydration. The accumulation was mainly focused in the coleoptiles, the first and the second leaves, and the area around the growing point of the embryo bud with a few transcripts from the mesocotyl to the radicle (Fig. 2A). We detected differential expression of RBBI mRNA in embryonic leaves. Although RBBI genes are expressed in all leaf tissues in the embryo, younger leaves had greater expression compared with older leaves (Fig. 2B). After germination, RBBI transcripts were detected mainly in mesophyll cells within the leaves, with a much more reduced signal in the vascular bundle compared with the embryonic leaves (Fig. 2C). In the radicle, RBBI hybridization signals were concentrated in the meristematic cells close to the radicle cap (Fig. 2D). However, the mature root stem has less expression relative to the root cap, and accumulation was mainly detected in the endodermis and the vascular bundle (Fig. 2E). The expression of the RBBI genes was also observed in different flower organs such as the ovule, style, anther, petals, and pollen sac, and in pollen (Fig. 2, F-I).
Figure 2.
Analysis of RBBI gene expression in rice tissues by in situ hybridization. Expression of RBBI genes was analyzed by in situ hybridization using the conserved coding region of RBBI2-3 (450 bp) as a probe. A, Longitudinal section of an embryo 24 h after rehydration, showing the embryo bud (EB) and radicle (R); B, transverse section of a young leaf at the base of a 6-d-old seedling; C, transverse section of upper part of an old leaf (11 d old); D, radicle tip 24 h after rehydration, showing the radicle cap (RC); E, transverse section of a primary root stem (from 11-d-old plants); F, close-up of the anther at prophase of meiosis, showing pollen mother cells (PMC); G, transverse section of a flower during meiosis, showing the pistil (Pi) and stamens (S); H, transverse section of pollen sacs (PS) at the anaphase of meiosis; and I, close-up of a pollen sac after meiosis showing the tapetum (T) and immature pollen grains (P).
Differential Expression Patterns within the Rice BBI Gene Family
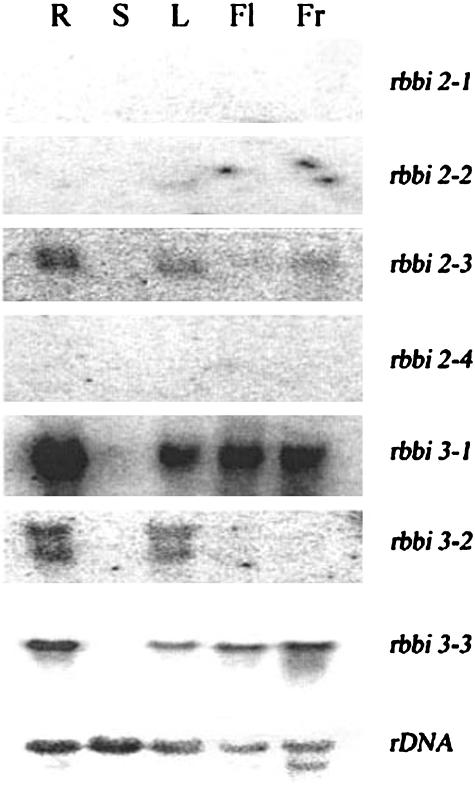
Northern blotting was carried out to study the gene expression patterns of different gene members in different rice organs using the 3′-untranslated region (UTR) of each gene as a probe (Fig. 3). Expression of two of the two-domain class genes, RBBI2-1 and RBBI2-4, was not detected in any of the organs we analyzed. RBBI2-2 was expressed slightly in leaves, but was not detected in other organs. RBBI2-3 was detected in roots and leaves, and in immature rice grains, but was not detected in the stem or in flowers. The three-domain class genes were more abundantly expressed than the two-domain class genes. In particular, RBBI3-1 was highly expressed in roots (Fig. 3).
Figure 3.
Differential expression patterns of RBBI genes. Northernblot analysis of RBBI genes. Gene-specific 3′-UTR probes of RBBI2-1, RBBI2-2, RBBI2-3, RBBI2-4, RBBI3-1, RBBI3-2, RBBI3-3, and an 18S ribosomal RNA probe were used to detect the gene expression in the root (R), stem (S), leaf (L), flower (Fl), and young rice caryopsis (Fr).
RBBIs Are Induced during Germination
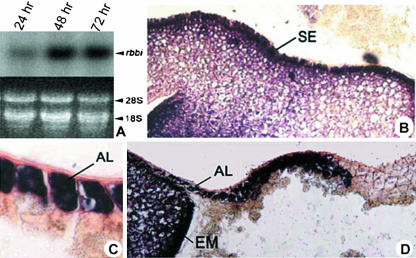
Because the RAFP1 protein was first isolated from germinating seeds, we analyzed the expression pattern of the RBBI genes during germination. RBBI mRNA accumulation could be detected 24 h after imbibition and reached a maximum at 48 h (Fig. 4A).
Figure 4.
Expression pattern of RBBI genes during germination. A, Northern-blot analysis of RBBI genes. Accumulation of the RBBI mRNAs was detected 24, 48, and 72 h after the onset of seed imbibition. In situ hybridization results showing RBBI gene expression in scutellar epithelium (SE; B), aleurone layer (AL; C), and young embryo (EM; D) of imbibed germinating rice seeds. The conserved coding region of the RBBI2-3 gene (450 bp) was used as the probe.
We also analyzed RBBI expression in germinating seeds using in situ hybridization with the probe recognizing all of the RBBI genes. After rehydration of the seed, RBBI transcripts were detected at a high level in the scutellar epithelium that comprises the layer of cells between embryo and endosperm (Fig. 4B). In addition, RBBI mRNA was abundant in aleurone cells (Fig. 4C), but only in the region relatively close to the embryo and not in the entire aleurone cell layer (Fig. 4D). The significance of this interesting expression pattern needs to be further elucidated.
One of the Three-Domain BBIs, RBBI3-1, Inhibits Trypsin
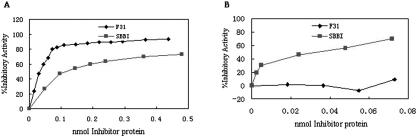
Knowledge of BBIs with three domains was hitherto unknown. The DNA fragment coding for the three-domain of RBBI3-1 was amplified, cloned, and expressed in E. coli to test whether the new three-domain RBBI is active as an inhibitor. The His-Tag fusion protein, designated as F31, was purified before trypsin and chymotrypsin inhibitory assays were performed. F31 showed strong inhibitory activity against trypsin and displayed stronger inhibitory activity than that of the control, commercial soybean BBI (Fig. 5A). However, no inhibitory activity against chymotrypsin was observed (Fig. 5B).
Figure 5.
Titration assay of trypsin- and chymotrypsin-inhibiting activity of F31 and soybean BBI. A, One hundred twenty-two TAME units of trypsin was incubated with increasing amounts of inhibitors in 0.2 mL of 0.05 m Tris-HCl buffer, pH 8.0, at 25°C for 10 min. Trypsin activity was then determined. The reaction volume was 1 mL. B, Chymotrypsin (2.0 μg) was incubated with increasing amounts of inhibitors in 0.2 mL of 0.05 m Tris-HCl buffer, pH 7.5, at 25°C for 10 min. Chymotrypsin activity was then determined. The reaction volume was 1 mL.
Expression of BBI Genes Is Wound Inducible
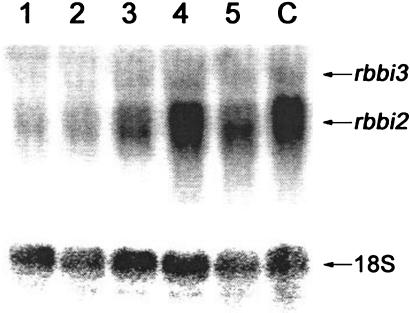
The coding region of RBBI2-3 was used to study the inducible expression of RBBI genes in rice leaves by northern blotting. Because the probe sequence is a conserved region showing 72.5% to 85.3% sequence identity to other six members of RBBI family at the DNA level, the detected signal is actually a mixed signal. The experimental results show that the accumulation of some RBBI transcripts (especially the two-domain transcripts) is wound inducible (Fig. 6). This is consistent with a report that a rice BBI gene OsBBPI (i.e. RBBI3-3) responded to cut, jasmonic acid, and ethylene signals (Rakwal et al., 2001). The probe used in that report consisted of a 259-bp conserved coding sequence and a 238-bp 3′ non-coding region of RBBI3-3, and showed 96% and 83% identity to the same regions of RBBI3-1 and RBBI2-3, respectively. Moreover, the 259-bp coding sequence of the probe was 74%, 78%, 81%, and 84% identical to the comparable coding regions of RBBI2-2, RBBI2-1, RBBI3-2, and RBBI2-4, respectively. Therefore the detected wound-induced signals in that report could also be a mixture, at least of RBBI2-3, RBBI3-1, and RBBI3-3 (Rakwal et al., 2001). These results suggest that RBBIs may be involved in the defense response against pathogens and/or insects.
Figure 6.
RBBI genes are wound inducible. The conserved region of RBBI2-3 (350 bp) was used as a probe to detect transcripts of all of the RBBI2 and RBBI3 members after the onset of wound induction. 1 through 5, Wound-treated samples after 0, 0.5, 1, 4, and 12 h of culture, respectively, in Murashige and Skoog liquid medium; C, the wound-treated sample after a 4-h culture in distilled water as control.
Overexpression of RBBI2-3 in Rice cv Taipei 309 Leads to Resistance to the Rice Blast Fungus Pathogen
We have transformed the rice cv Taipei 309 with RBBI2-3 to test the role of RBBI genes in plant defense mechanisms. The rice cv Taipei 309 was chosen as a host for this experiment because this cultivar is known to be susceptible to various pathogens. We transformed embryonic callus of rice cv Taipei 309 with RBBI2-3 by particle bombardment, and two transgenic lines, T137-2 and T191-2, were obtained. Integration of the transgene into the host rice genome was confirmed by PCR and Southern blot (data not shown). Two independent groups performed the resistance experiments simultaneously. The Beijing group tested two isolates of P. oryzae, Zhong 10-8-14 and Sichuan 26, which are common in southern China and severely infect rice cv Taipei 309. The Kunming group tested four isolates of P. oryzae (i.e. 96-4-1a, 96-4-2a, 96-8-1a, and 96-10-1a) that are common in southwest China. For each resistance test, 100 seeds of each transgenic line, together with the nontransgenic rice cv Taipei 309 as a control, were planted first on Murashige and Skoog medium and then transferred to soil. Rice blast fungus pathogen P. oryzae was inoculated onto rice leaves. Two days after inoculation, semiquantitative RT-PCR was performed. Six to 7 d after inoculation, the resistance of the plants against the pathogens was evaluated.
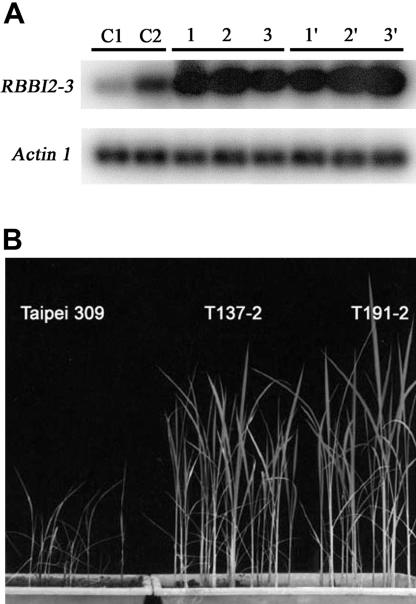
Although 2,000 kilometers apart, the two independent groups obtained very similar results (Table I). The semiquantitative RT-PCR analysis showed a higher expression of RBBI2-3 in transgenic rice leaves relative to controls (Fig. 7A). As expected, analysis of pathogen effects showed that untransformed rice plants of rice cv Taipei 309 were severely infected, showing many typical spindle-shaped lesion spots on the leaves and a severely retarded growth (Fig. 7B). However, the transgenic T137-2 and T191-2 grew healthy and normally after the inoculation of fungal pathogen. Not even a single small lesion spot was found on all of the leaves of the transgenic seedlings, indicating that the transgenic lines were highly resistant (resistance level 0; Mackill and Bonman, 1992) to all isolates of the blast fungus pathogen tested (Fig. 7B).
Table I.
Resistance test of transgenic rice plants against the rice fungus pathogen P. oryzae
| Pathogen Isolates | cka | T137-2 | T191-2 |
|---|---|---|---|
| Race 033 | |||
| 96-4-1a | ss | HR | HR |
| 96-4-2a | ss | HR | HR |
| Race 335 (96-8-1a) | ss | HR | HR |
| Race 317 (96-10-1a) | ss | HR | HR |
| Zhong 10-8-14 | ss | HR | HR |
| Sichuan -26 | ss | HR | HR |
a ck, Untransformed Taipei 309 as control; ss, highly susceptible (level 5); HR, highly resistant (level 0).
Figure 7.
A, Semiquantitative RT-PCR analysis of RBBI2-3 expression 2 d after inoculation. C1, Untransformed rice cv Taipei 309 before inoculation; C2, untransformed rice cv Taipei 309 after inoculation; 1, 2, and 3, three randomly picked rice plants of T137-2 after inoculation; 1′, 2′, and 3′, three randomly picked rice plants of T191-2 after inoculation. B, Transgenic rice cv Taipei 309 plants are resistant to rice blast fungus pathogens. Non-transformed control wild-type rice cv Taipei 309 plants were grown next to the transgenic rice cv Taipei 309 (T137-2 and T191-2) homozygous for the RBBI2-3 transgene. Plants were infected with one of six isolates (Zhong 10-8-14, Sichuan 26, 96-4-1a, 96-4-2a, 96-8-1a, and 96-10-1a) of P. oryzae. High resistance of both T137-2 and T191-2 seedlings was observed against the infection of all of the six isolates.
DISCUSSION
A genomic clone containing seven full-length rice BBI genes is reported in this study. The genomic organization of the RBBI gene family became clear after three more RBBI genes were found in the databases of draft rice genome sequences (Fig. 1D). Among the different RBBI members, the most obvious difference is the number of domains a gene contained. We report here for the first time that rice BBIs contain one, two, or three BBI domains, although there are reports that monocot (such as wheat) BBIs possess either one or two domains (Odani et al., 1986). The standard mechanism, exhibited by inhibitors when they react with cognate proteinases, indicates that the P1 residue in the reactive loop determines specificity of the inhibitor (Laskowski and Kato, 1980). Several different P1 residues were detected in the RBBI domains. Those Wip1-like RBBIs, single-domain inhibitors, have Phe as the N-terminal P1 residue, indicating that these inhibitors tend to inhibit chymotrypsin-like enzymes. The N-terminal P1 residue for the multidomain RBBIs is Arg or Lys in most cases and shows inhibiting activities against trypsin-like enzymes (Fig. 1E) as determined inhibitory activity assays (Fig. 5). However, some P1 residues are changed from alkaline amino acid residues to others. For example, the P1 residue of the first domain of RBBI3-3 is changed to Met, which theoretically may change its inhibitory activity to target chymotrypsin-like enzymes. In another case, the P1 of the second domain of RBBI2-4 is changed to Ser, making the inhibitor more likely to inhibit elastase-like enzymes (Fig. 1E). More varieties are found in other residues of the reactive loops, indicating that the effectiveness of those proteinase-inhibiting domains against a given proteinase of external origin would vary. Evidence showing that effectiveness of inhibition could vary enormously has been reported (Belitz et al., 1982; Christeller and Shaw, 1989). Recent data has shown that the effectiveness of inhibitory activity against the same class of proteinase from the same insect species could vary enormously (Volpicella et al., 2003). Therefore, in response to changes of proteinases of external origin, an adequate number of BBI domains with sufficient sequence variations within the RBBI family would serve as a reservoir from which more effective inhibitors would evolve from a process of selection.
In the novel three-domain BBI genes, an additional domain is present that would contribute to the development of new proteinase-inhibiting activity. Four reactive-like sites were identified in the F31 protein in this study (Fig. 1E, blue letters in RBBI3-1 sequence), of which two P1 residues are the trypsin-inhibitory R and K. The other two P1 residues are two Glu residues (E), whose inhibitory specificity has not been reported. High trypsin-inhibitory activity was detected as expected (Fig. 5), indicating that the two P1 residues function normally. However, chymotrypsin-inhibitory (Fig. 5), subtilisin-inhibitory, or papain-inhibitory activity was not detected (data not shown), suggesting that the two Glu residues in the P1 sites do not correspond to the proteinase-inhibitory activity we have known and investigated thus far. The possibility that new inhibitory specificities have evolved in these three-domain inhibitors needs to be further clarified by biochemical means.
Our detailed northern blotting and in situ hybridization analysis suggested that RBBIs are both developmentally regulated and induced in response to wound or pathogen attacks. Similar regulatory patterns were found in plant defensive proteins or toxic secondary compounds (Ryan, 1990; Molyneux and Ralphs, 1992). The RBBIs of rice are expressed in an effective and economical way. First, not all of the members of RBBI genes are expressed equally. The three-domain genes generally showed higher expression than the two-domain genes (Fig. 3). It is more likely that the three-domain genes are able to provide more combinations of different inhibitory activities when the same amount of protein molecules is synthesized. Second, the expression level of the RBBI genes is higher in those tissues more likely to experience attack by predators or pathogens, e.g. rehydrated embryo, young leaves, root cap, and flower organs. It is reasonable to speculate that higher expression of RBBIs in these tissues would serve as an effective protection mechanism against attacks by predators or pathogens. One interesting finding is the detection of high-level expression of RBBIs in aleurone cells, i.e. expression in the cells surrounding the endosperm and embryo (Fig. 4, C and D). Moreover, the expression of RBBIs in these tissues is temporally regulated, e.g. the RBBI mRNA level in the embryo peaked up at 48 h after rehydration (Fig. 4A) and then dropped precipitously afterward (data not show). Third, RBBI genes are induced by wound or pathogen attack (Figs. 6 and 7A). It was recently reported that the expression of RBBI3-3 could be induced by cut and jasmonic acid (Rakwal et al., 2001). It is generally accepted that wounding or attack by predators or pathogens will trigger plant defense responses that may include inhibitory reactions or even the production of toxic proteins or chemical compounds (Ryan, 1990; Baldwin and Preston, 1999). On the basis of the three points mentioned above, it is reasonable to suggest that plants should have optimized their tradeoffs with respect to the allocation of resources to growth and development by choosing an effective way of expressing plant defense proteins such as BBIs.
The sequence divergence, multiple inhibitory activities, and inducible expression pattern suggest that RBBIs may serve as potential effective defense proteins in vivo. It has long been known that proteinase inhibitors could be induced by pathogens or pathogen/plant cell wall-derived elicitors (Roby et al., 1987; Rickauer et al., 1989; Cordero et al., 1994; McGurl et al., 1995; Choi et al., 2000; Pernas et al., 2000). Furthermore, in vitro experiments showed that growth of fungi or activities of proteinases from fungi were inhibited by proteinase inhibitors (Senser et al., 1974; Mosolov et al., 1976, 1979; Brown and Ryan, 1984; MacGibbon and Mann, 1986; Xie et al., 1996). Meanwhile, higher levels of trypsin-inhibitory activity were found in tomato (Lycopersicon esculentum) and wheat varieties with higher resistances against pathogenic fungi (Peng and Black, 1976; Yamaleev et al., 1980). However, although there is an increasing amount of direct evidence supporting the defensive roles of proteinase inhibitors against insects and other herbivores, the role played by proteinase inhibitors in fungal resistance is only supported by indirect evidence (Ryan, 1990).
The transgenic data of this study demonstrates that proteinase inhibitors play an important role in the defense response of rice against fungal infection. It is the first direct evidence that proteinase inhibitors could arrest fungal invasion and inhibit the growth of the fungus not only in vitro (Xie et al., 1996) but also in vivo (Fig. 7B), possibly by inhibiting proteolysis and thereby limiting availability of amino acids. Our data also showed that the inhibiting effect of proteinase inhibitor against proteinase in vitro was dosage dependent. A higher percentage of inhibitory effect was observed when higher concentrations of RBBI3-1 were used to incubate with a given concentration of proteinase (Fig. 5A). Although the mRNA of RBBI2-3 increased after inoculation in susceptible plants, more than a 10-fold amount of RBBI2-3 mRNA was found accumulated in transgenic rice leaves, driven by the 35S promoter (Fig. 7A). This data suggests that a high concentration of proteinase inhibitors is required to inhibit growth and multiplication of the pathogens. Pathogenic fungi infect their host by first secreting hydrolytic enzymes outside of their bodies to hydrolyze complex substrates into small organic molecules (Campbell et al., 1999). It is reasonable to suggest that the overexpression of RBBI2-3 protein in transgenic rice may inhibit the function of proteinases secreted by P. oryzae through a similar biochemical mechanism like RBBI3-1. If different RBBI proteins have different inhibitory activities, it may also be expected that overexpression of the RBBI member exhibiting the highest inhibitory activity in transgenic rice would result in the highest resistance against pathogen.
Two possible signal pathways will be triggered when a plant is infected by a pathogen: one related to wound through the penetration and colonization of the pathogen, and the other through direct molecular recognition of the pathogen (Cordero et al., 1994). The fact that RBBI genes in this study are induced by wounding, together with data from other studies (Rakwal et al., 2001), suggests that the wound-related signal pathway is possibly involved in increasing the expression of RBBIs in rice plants when a pathogen attacks. Further evidence, however, needs to be presented.
MATERIALS AND METHODS
Screening of a BAC Library and Analysis of the Positive Clones
The filters and clones of a rice (Oryza sativa) BAC library, generated from O. sativa subsp. japonica var Teqing, were kindly provided by Dr. Hongbing Zhang (University of Texas A & M, College Station). The filters were immersed in a hybridization buffer (50% [v/v] formamide, 0.12 m Na2HPO4, pH 7.2, 0.25 m NaCl, and 7% [w/v] SDS). After 30 min of prehybridization at 43°C, the filters were placed into a fresh hybridization buffer for hybridization. The RBBI8 probe, a fragment size of about 450 bp, was radioactively labeled with [32P]dCTP (NEN, Boston) using a random primer labeling kit (Promega, Madison, WI) according to the manufacturer's instructions. The probe was denatured by boiling for 2 min and then added to the hybridization buffer. After an 18-h hybridization period at 43°C, the filters were first rinsed twice with 2× SSC and then washed for 15 min each in 2× SSC/0.1% (w/v) SDS, 0.5× SSC/0.1% (w/v) SDS, and 0.1× SSC/1% (w/v) SDS at 43°C. The washed filters were exposed and analyzed using a Phos-phorImager IS445 (Molecular Dynamics, Sunnyvale, CA). The positive BAC clone DNA was then digested by EcoRV, HindIII, or XbaI, and a Southern blot was performed with a Zeta-GT Probe nylon membrane (Bio-Rad, Hercules, CA) using the same probe used to screen the BAC clone library. The positive DNA fragments were recovered by Qiaquick Gel Extraction Kit (Qiagen, Valencia, CA) and subcloned into pBlueScript SK+ (Stratagene, La Jolla, CA). The plasmid DNA was purified using the Wizard Plus Minipreps DNA purification kit (Promega) and sequenced with an ABI 377 DNA sequencer with a Big Dye Primer Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA). Identified sequences were then searched for putative ORFs using DNASIS programs (Hitachi, Tokyo). The deduced amino acid sequences from these putative ORFs were investigated for similarity to proteins in the EMBL and SWISSPRO databases using the BLAST search program (Altschul et al., 1997). The amino acid sequence alignment of rice BBIs was made using ClustalW (Thompson et al., 1994). The GenBank accession numbers of these genes are as follows: AJ277468 (RBBI2-1, RBBI2-2, RBBI3-1, and RBBI3-2), AJ277469 (RBBI3-3), AJ277470 (RBBI2-3), and AJ277472 (RBBI2-4), respectively.
Northern-Blot Analysis
Total RNA was isolated from rice leaves, roots, flowers, and immature rice grains either using an RNA Extraction Kit (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instruction or by the guanidium thiocyanate method (Chomczynski and Sacchi, 1987). The total RNA samples (10 μg lane–1) were separated on 1% (w/v) agarose gels and were transferred to a Hybond-N+ nylon membrane (Amersham Biosciences). After a 0.5-h prehybridization, the filters were hybridized in the hybridization buffer (50% [v/v] formamide, 0.12 m Na2HPO4, pH 7.2, 0.25 m NaCl, and 7% [w/v] SDS) at 43°C for 22 h. Then the filters were first rinsed twice with 2× SSC and washed for 15 min each in 2× SSC/0.1% (w/v) SDS, 0.5× SSC/0.1% (w/v) SDS, and 0.1× SSC/1% (w/v) SDS at 43°C, respectively. The insert of prbbi8 (i.e. the coding region of RBBI2-3 and 450 bp in length) was radioactively labeled with [32P]dCTP (NEN) using a random primer labeling kit (Promega) according to the manufacturer's instructions. The gene-specific primers that are from the 3′-UTR sequences of the seven members of the rice BBI gene family were used to make probes for northern hybridization. The 3′-UTR sequences of RBBIs were amplified by PCR from the genomic DNA using gene-specific primers (RBBI2-1, 5′-ACT GAT TAA CTA TAG CTA GC-3′ and 5′-CTA AAG TTG CAC TTT TCT GA-3′; RBBI2-2, 5′-GAA GAG AAC TAC TTC TCT GT-3′ and 5′-C TAC CTG CTT TCC ACC GGA-3′; RBBI2-3, 5′-CTG CAG ATC GAT ATG TAT GAT-3′ and 5′-TGA CTG ATG CGC CTA TGG-3′; RBBI2-4, 5′-ATG AAT AGC GGC AAT ATG GC-3′ and 5′-TCA CTT GTG TTC GTC GGG TG-3′; RBBI3-1, 5′-TA GCT ACA GAT GAT CGA TGA-3′ and 5′-CAA TAT AAG TGT CCT CTC AG-3′; RBBI3-2, 5′-TCC TTG CAG ATA TGA TCG AT-3′ and 5′-AC AAG GTT AAG GTC AAA CTC-3′; and RBBI3-3, 5′-GCT ACA GAT GAT CGA TGA TCG-3′ and 5′-AC CCA CAA CAT CTT CTA ACG-3′). The PCR fragments were cloned into a pBlueScript vector, and the plasmids were used to make radioactive probes for the northern-blot filters by PCR. The PCR reactions containing cDNA: 1× polymerase buffer (Promega); 2.5 mm MgCl2; 200 μm of each of dATP, dGTP, and dTTP; 100 μm [32P]dCTP (NEN); 0.1 μm of each primer; and 2 units of TaqDNA polymerase (Promega) were performed in a final volume of 20 μL. The PCR reactions were carried out for 30 cycles (94°C, 40 s; 58°C, 30 s; and 72°C, 30 s).
In Situ Hybridization
Rice materials including germinating seeds (24, 48, and 72 h after dehydration), 6-d-old seedlings, and 11-d-old seedlings were collected. Rice tissues were fixed with formaldehyde and were embedded in paraplast. The tissues were sliced into 5-μm-thick slices and mounted on poly l-Lys-coated glass slides. These were then rinsed in 10 mm Tris-HCl, pH 8.0, and incubated in 1% (w/v) bovine serum albumin in the same buffer for 1 h. After acetylation, sections were dehydrated with a graded series of ethanol washes and air-dried. The probe was labeled with digoxigenin (DIG) and hydrolyzed in hydrolysis carbonate buffer (60 mm Na2CO3 and 40 mm NaHCO3, pH 10.2) before it was used. After prehybridization, a hybridization solution (0.3 m NaCl, 10 mm Tris-HCl, pH 7.5, 5 mm EDTA, 1× Denhardt's solution, 50% [v/v] formamide, 2 mg mL–1 tRNA, and 200 units mL–1 RNase inhibitor) containing denatured (DIG)-labeled sense or antisense probe -RNA was added. The slides were incubated overnight in a humid box at 45°C. After two washing steps with 0.2× SSC at 55°C for 30 min each, slides were incubated with 20 μg mL–1 RNase A (Promega) at 37°C for 30 min, followed by washing with 0.2× SSC at 55°C for 1 h. Immunological detection of DIG-labeled RNA probe was performed with anti-DIG antibody conjugated with alkaline phosphatase (Roche Diagnostics, Mannheim, Germany) according to the supplier's protocol. Staining was obtained using the substrates nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate. Slides were analyzed under bright field microscopy (DM RE HC microscope, Leica, Wetzlar, Germany) equipped with a CCD camera (Eastman Kodak, Rochester, NY). The probe used for the in situ hybridizations was the conserved coding region of the RBBI2-3 gene (450 bp).
Constructs for Prokaryotic Expression
The pBS-rbbi21K containing RBBI3-1 was used as the template to amplify the three-domain fragment. Fifty picomoles each of the 5′ primer (5′-GGC GGA TCC GCA CCA CCA CGC CCG CCC-3′) and 3′ primer (5′-TGT AAG CTT CTA GTT CTC CGC TCG GGG-3′), together with 10 units of thermostable pfu DNA polymerase (Stratagene) was used to prime a 100-μL PCR (95°C, 1 min; 55°C, 45 s; and 72°C, 1 min, 35 cycles). The amplified 660-bp-long fragment that codes for the three BBI domains of RBBI3-1 was purified and then digested with BamHI and HindIII, which were introduced by the two primers (bold sequences) before being ligated into pET28a (Novagen, Madison, WI). Competent cells of Escherichia coli strain BL21 (DE3) were transformed, and recombinant plasmids were screened and sequenced for verification. The recombinant plasmid was designated as pET28-F31.
Expression, Refolding, and Purification of the Fusion Protein F31
The cells were first cultured overnight in 10 mL of Luria-Bertani medium (50 mg mL–1 kanamycin) at 37°C. Five milliliters of the overnight culture was transferred into 1,000 mL of Luria-Bertani medium and allowed to continue to grow until A600 reached 0.4 to 0.6. Isopropylthio-β-galactoside was added to 0.5 mm and allowed to induce for 3 to 6 h. The bacteria were harvested and suspended in 50 mL of cell lysis buffer (50 mm Tris-HCl and 100 mm NaCl, pH 8.0) before being sonicated in an ice-water slurry (30 s of sonication alternated with 30 s of cooling, six times).
Most of the three-domain fusion protein F31 was not soluble in our experiments, and therefore inclusion bodies were harvested. Collection of inclusion bodies, protein refolding, and purification were carried out as described before (Li et al., 1999). The purity of the proteins was checked by SDS-PAGE.
Inhibitory Activity Assay of the Fusion Proteins F31
Inhibitory activities against trypsin 1:250 (Difco Laboratories, Detroit) and chymotrypsin (C4129, Sigma-Aldrich, St. Louis) were determined by incubating purified fusion proteins together with the enzymes at 25°C for 5 min. The remaining trypsin activity was measured with TAME (T4626, Sigma-Aldrich) as described previously (Hummel 1959). The remaining chymotrypsin activity was measured with ATEE (A6751, Sigma-Aldrich; Schwert and Takenaka, 1955). The absorbency at 247 nm (for ATME) and 237 nm (for ATEE) was measured with a GBC Cintra 10e UV-Visible spectrometer (GBC, Melbourne, Australia). Commercial soybean BBI (T9777, Sigma-Aldrich) was assayed in parallel as positive controls.
Wound-Inducibility Analysis
Rice leaves were cut into 0.5-cm2 pieces and placed into an Murashige and Skoog liquid medium and distilled water for 0.5, 1, 2, 4, and 12 h. RNA samples were isolated from the treated leaves, and northern blotting was performed as described.
Rice Transformation, Pathogen Inoculation, and Resistance Evaluation
Rice cv Taipei 309 was chosen as a host onto which to transform the RBBI2-3 gene that was cloned into properly digested pMON310 and driven by the 35S cauliflower mosaic virus promoter. Rice callus was prepared from immature embryos, and transformation by particle bombardment was carried out as described previously (Zheng et al., 1997). The regenerated shoots were selected by streptomycin, and 20 lines of T1 generation seeds were obtained. The T1 transgenic rice plants were grown to the four-leaf stage in a greenhouse.
The conidiospores of Pyricularia oryzae were suspended in 0.02% (v/v) Tween 20 to a concentration of 30 to 50 spores/100× viewing field and were evenly sprayed onto rice leaves using an N-1 type inoculation device. After inoculation, the plants were kept at 26°C for 24 h with 100% humidity before being transferred to a humid greenhouse. The development of disease symptoms was checked 6 to 7 d later, and the resistance level was evaluated according to the International Rice Research Institute evaluation standard (Mackill and Bonman, 1992).
RBBI2-3 Expression Analysis by Semiquantitative RT-PCR
Total RNA was extracted from leaves of transgenic and control rice plants 48 h after inoculation using the previously described method. After DNase I treatment, 5 μg of the total RNA was used to synthesize the first strand of cDNA with an oligo(dT) and RT SuperscriptII as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Two microliters of the first strand of cDNA was used as a template for semiquantitative RT-PCR. The PCR reaction containing cDNA 1× polymerase buffer (Promega), 2.5 mm MgCl2, 200 μm of each of dNTP, 0.1 μm of each primer, and 2 units of TaqDNA polymerase (Promega) was performed in a final volume of 100 μL. Primers specific for RBBI2-3 are as above, whereas the primers for the internal control Act 1 gene are 5′-CTG ACG GAG CGT GGT TAC TCA TTC-3′ and 5′-GCT AGG AGC AAG GCA GTG ATC TTC-3′. To ensure that the amplification reaction was within linear range, the PCR reactions were carried out for 20 cycles. Five microliters of the RT-PCR reaction mixtures was loaded on a 0.8% (w/v) agarose gel, transferred to Hybond N+ nylon membranes (Amersham Biosciences), and hybridized with radioactive-labeled corresponding probes using the procedure detailed previously.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Dr. S. McCouch (Cornell University, Ithaca, NY) for help with the mapping of the marker RBBI8 and Dr. Hongbing Zhang (University of Texas A & M, College Station) for kindly providing the rice BAC library. We sincerely thank Prof. XingWang Deng (Yale University, New Haven, CT) and Dr. Hongwei Guo (University of California, Los Angeles) for critical review, comments, and valuable discussions and Dr. Matthew Terry (University of Southampton, UK) for help with the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.024810.
This work was supported by the National High-Tech Program of China (project no. GN 863-01-101-02-02 and 2001AA222261 to L.-J.Q.), by the National Natural Science Foundation (project no. GN 39830020 to H.G.), by the State Key Basic Research and Development Plan (project no. GN G1999011602), and by the Rockefeller Foundation (grant no. GN#97003 to Z.L.C.).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Preston CA (1999) The eco-physiological complexity of plant responses to insect herbivores. Planta 208: 137-145 [Google Scholar]
- Belitz H-D, Lynen F, Weder JKP (1982) Comparative studies on the inhibitory action of some legume seeds, potato tubers, and bran against human and bovine proteinases. Z Lebensm Unters Forsch 174: 442-446 [DOI] [PubMed] [Google Scholar]
- Birk Y (1987) Proteinase inhibitors. In A Neuroberger, K Brocklehurst, eds, Hydrolytic Enzymes. Elsevier Science Publishers, Amsterdam, pp 257-300
- Birk Y (1993) Protease inhibitors of plant origin and role of protease inhibitors in human nutrition. In W Troll, AR Kennedy, eds, Protease Inhibitors as Cancer Chemopreventive Agents. Plenum Press, New York, pp 97-106
- Brown WE, Ryan CA (1984) Isolation and characterization of a wound-induced trypsin inhibitor from alfalfa leaves. Biochemistry 23: 3418-3422 [DOI] [PubMed] [Google Scholar]
- Campbell NA, Reece JB, Mitchell LG (1999) Fungi: Mycology. In Biology, Ed 5. Benjamin/Cummings, Essex, UK
- Chen PW, Chow SH, Chen LJ (1997) Nucleotide sequence of a cDNA (accession no. U76004) encoding rice Bowman-Birk proteinase inhibitor (PGR 97-015). Plant Physiol 113: 6639046601 [Google Scholar]
- Choi D, Park JA, Seo YS, Chun YJ, Kim WT (2000) Structure and stress-related expression of two cDNAs encoding proteinase inhibitor II of Nicotiana glutinosa L. Biochim Biophys Acta 1492: 211-215 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single step method of RNA isolation by acid guanidium thiocyanate-phenol chloroform extraction. Anal Biochem 162: 156-159 [DOI] [PubMed] [Google Scholar]
- Christeller JT, Shaw BD (1989) The interaction of a range of serine proteinase inhibitors with bovine trypsin and Costelytra zealandica trypsin. Insect Biochem 19: 233-241 [Google Scholar]
- Cordero MJ, Raventos D, San Segundo B (1994) Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: systemic wound-response of a monocot gene. Plant J 6: 141-150 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100 [DOI] [PubMed] [Google Scholar]
- Guiderdoni E, Cordero MJ, Vignols F, Garcia-Garrido JM, Lescot M, Tharreau D, Meynard D, Ferriere N, Notteghem JL, Delseny M (2002) Inducibility by pathogen attack and developmental regulation of the rice Ltp1 gene. Plant Mol Biol 49: 683-699 [DOI] [PubMed] [Google Scholar]
- Hummel BC (1959) A modified spectrophotometric determination of chymotrypsin, trypsin, and thrombin. Can J Biochem Physiol 37: 1394-1399 [PubMed] [Google Scholar]
- Kennedy AR (1993) Anticarcinogenic activity of protease inhibitors. In W Troll, AR Kennedy, eds, Protease Inhibitors as Cancer Chemopreventive Agents: Plenum Press, New York, pp 9-64
- Laskowski MJ, Kato I (1980) Protein inhibitors of proteinases. Annu Rev Biochem 49: 593-626 [DOI] [PubMed] [Google Scholar]
- Li N, Qu L-J, Liu Y, Li Q, Gu H, Chen ZL (1999) The refolding, purification, and activity analysis of a rice Bowman-Birk inhibitor expressed in Escherichia coli. Prot Exp Purif 15: 99-104 [DOI] [PubMed] [Google Scholar]
- Li de la Sierra I, Quillien L, Flecker P, Gueguen J, Brunie S (1999) Dimeric crystal structure of a Bowman-Birk protease inhibitor from pea seeds. J Mol Biol 285: 1195-1207 [DOI] [PubMed] [Google Scholar]
- Liu H, Gu H, Chen X, Pan NS, Chen ZL (1994) Isolation, purification and characterization of an antifungal protein from rice. High Tech Lett 4: 22-26 [in Chinese with English abstract] [Google Scholar]
- MacGibbon DB, Mann JD (1986) Inhibition of animal and pathogenic fungal proteases by phloem exudate from pumpkin fruits (Cucurbitaceae). J Sci Food Agric 37: 515-522 [Google Scholar]
- Mackill DJ, Bonman JM (1992) Inheritance of near-isogenic lines of rice. Phytopathology 82: 746-749 [Google Scholar]
- McGurl B, Mukherjee S, Kahn M, Ryan CA (1995) Characterization of two proteinase inhibitor (ATI) cDNAs from alfalfa leaves (Medicago sativa var. Vernema): the expression of ATI genes in response to wounding and soil microorganisms. Plant Mol Biol 27: 995-1001 [DOI] [PubMed] [Google Scholar]
- Molyneux RJ, Ralphs MH (1992) Plant toxins and palatability to herbivores. J Range Manage 45: 13-18 [Google Scholar]
- Mosolov VV, Loginova MD, Fedurkina NV, Benken II (1976) The biological significance of proteinase inhibitors in plants. Plant Sci Lett 7: 77-80 [Google Scholar]
- Mosolov VV, Loginova MD, Malova EL, Benken II (1979) A specific inhibitor of Collectotrichum lindemunthianum protease from kidney bean (Phaseolus vulgaris) seeds. Planta 144: 265-269 [DOI] [PubMed] [Google Scholar]
- Nagasue A, Fukamachi H, Ikenaga H, Funatsu G (1988) The amino acid sequence of barley rootlet trypsin inhibitor. Agric Biol Chem 52: 1505-1514 [Google Scholar]
- Odani S, Koide T, Ono T (1986) Wheat germ trypsin inhibitors: isolation and structural characterization of single-headed and double-headed inhibitors of the Bowman-Birk type. J Biochem 100: 975-983 [DOI] [PubMed] [Google Scholar]
- Peng JH, Black LL (1976) Increased proteinase inhibitor activity in response to infection of resistant tomato plants by Phytophthora infestans. Phytopathology 66: 958-963 [Google Scholar]
- Pernas M, Sanchez-Monge R, Salcedo G (2000) Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett 467: 206-210 [DOI] [PubMed] [Google Scholar]
- Prakash B, Selvaraj S, Murthy MRN, Sreerama YN, Rao DR, Gowda LR (1996) Analysis of the amino acid sequences of plant Bowman-Birk inhibitors. J Mol Evol 42: 560-569 [DOI] [PubMed] [Google Scholar]
- Rakwal R, Agrawal GK, Jwa N-S (2001) Characterization of a rice (Oryza sativa L.) Bowman-Birk proteinase inhibitor: tightly light regulated induction in response to cut, jasmonic acid, ethylene and protein phosphatase 2A inhibitors. Gene 263: 189-198 [DOI] [PubMed] [Google Scholar]
- Rickauer M, Fournier F, Esquerre-Tugaye M-T (1989) Induction of proteinase inhibitors in tobacco cell suspension culture by elicitors of Phytophthora parasitica var. nicotianae. Plant Physiol 90: 1065-1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roby D, Toppan A, Esquerre-Tugaye M-T (1987) Cell surfaces in plant micro-organism interactions: VIII. Increased proteinase inhibitor activity in melon plants in response to infection by Colletotrichum lagenarium or to treatment with an elicitor fraction from this fungus. Physiol Mol Plant Physiol 30: 453-460 [Google Scholar]
- Ryan CA (1981) Proteinase inhibitors. In A Marcus, ed, The Biochemistry of Plants, Vol 6. Academic Press, New York, pp 351-370 [Google Scholar]
- Ryan CA (1990) Proteinase inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytophathol 28: 425-449 [Google Scholar]
- Schwert GW, Takenaka Y (1955) A spectrophotometeric determination of trypsin and chymotrypsin. Biochem Biophys Acta 16: 570-575 [DOI] [PubMed] [Google Scholar]
- Senser F, Belitz H-D, Kaiser K-P, Santarius K (1974) Suggestion of a protective function of proteinase inhibitors in potatoes: inhibition of proteolytic activity of microorganisms isolated from spoiled potato tubers. Z Lebensm Unters Forsch 155: 100-101 [Google Scholar]
- Song HK, Kim YS, Yang JK, Moon J, Lee JY, Suh SW (1999) Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 Å resolution. J Mol Biol 293: 1133-1144 [DOI] [PubMed] [Google Scholar]
- Swegle M, Kramer KJ, Muthukrishnan S (1992) Properties of barley seed chitinases and release of embryo-associated isoforms during early stages of imbibition. Plant Physiol 99: 1009-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M, Hashino K, Shiozaki M, Ibuki F, Maki Z (1987) The complete amino acid sequence of rice bran trypsin inhibitor. J Biochem 102: 297-306 [DOI] [PubMed] [Google Scholar]
- Tashiro M, Asao T, Hirata C, Takahashi K, Kanamoru M (1990) The complete amino acid sequence of a major trypsin inhibitor from seeds of foxtail millet (Setaria italica). J Biochem 108: 669-672 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Gaut BS (2001) Molecular evolution of the wound-induced serine protease inhibitor Wip1 in Zea and related genera. Mol Biol Evol 18: 2092-2101 [DOI] [PubMed] [Google Scholar]
- Volpicella M, Ceci LR, Cordewener J, America T, Gallerani R, Bode W, Jongsma MA, Beekwilder J (2003) Properties of purified gut trypsin from Helicoverpa zea, adapted to proteinase inhibitors. Eur J Biochem 270: 10-19 [DOI] [PubMed] [Google Scholar]
- Xie M, Chen X, Qu L-J, Liu H, Gu H, Chen ZL (1996) Molecular cloning and sequence analysis of a new gene encoding rice proteinase inhibitor. Acta Bot Sin 38: 444-450 [in Chinese with English abstract] [Google Scholar]
- Yamaleev AM, Mukhsionov U Kh, Isaev RF, Yamaleeva AA, Krivchenko VI (1980) The activity of inhibitors of protease and the resistance of wheat to the causal agent of hard smut. Skkh Biol 15: 143-144 [Google Scholar]
- Ye XY, Ng TB, Tsang PW, Wang J (2001) Isolation of a homodimeric lectin with antifungal and antiviral activities from red kidney bean (Phaseolus vulgaris) seeds. J Protein Chem 20: 367-375 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79-92 [DOI] [PubMed] [Google Scholar]
- Zheng HH, Li Y, Yu Z, Li W, Chen M, Ming X, Casper R, Chen ZL (1997) Recovery of transgenic rice plants expressing the rice dwarf virus outer coat protein gene (S8). Theor Appl Genet 94: 522-527 [Google Scholar]