Abstract
We have isolated a cDNA (GhWBC1) from cotton (Gossypium hirsutum) that encodes an ATP-binding cassette transporter of the WBC (white/brown complex) subfamily. Members of this subfamily are half-sized transporters and are reported to mediate lipid and drug excretion in human (Homo sapiens). GhWBC1 is highly expressed in developing fiber cells, but transcripts were also detectable in other tissues except roots. The transcript level peaked in rapidly expanding fibers from 5 to 9 DPA and then decreased. The GhWBC1 expression was weak in fiber cells of an li (ligon-lintless) mutant, which is defective in fiber cell elongation. These data indicate that GhWBC1 gene expression correlates with cotton fiber elongation. Transient expression of enhanced green fluorescence protein-GhWBC1 fusion protein in onion (Allium cepa) epidermal cells revealed plasma membrane localization. The GhWBC1 cDNA driven by a constitutive 35S promoter was introduced into Arabidopsis. About 13% of the transformants produced short siliques (SSs), whereas others had normal siliques (long siliques [LSs]). In siliques of SS lines, most embryos were severely shriveled, and only several seeds per silique could be found at maturity. The transgene expression level was higher in SS lines than in LS lines. Expression of AtWBC11, the closest homolog of GhWBC1 in Arabidopsis, was not altered in either SS or LS transgenic plants examined. These data suggest that GhWBC1 interferes with substance translocation that is required for Arabidopsis seed and silique development. Characterization of Arabidopsis WBC members, particularly AtWBC11, will help to dissect the role of GhWBC1 in cotton fiber development and elongation.
The ATP-binding cassette (ABC) transporters, known for their function of translocating a broad range of substances across biological membranes powered by ATP hydrolysis, are ubiquitously present in bacteria, fungi, plants, and animals, and they constitute one of the largest protein families (Higgins, 1992; Henikoff et al., 1997; Theodoulou, 2000).
Functional ABC transporters typically consist of two transmembrane domains (TMDs) and two nucleotide-binding folds (NBFs). In eukaryotic organisms, these four domains are often arranged into a single polypeptide, forming a full-sized transporter. However, a number of eukaryotic genes also encode half-sized transporters, with each peptide containing one NBF and one TMD (Higgins, 1992). These half-sized transporters fulfill their functions through formation of homodimers or heterodimers with other partners.
In bacteria, many ABC transporters are involved in nutrient acquisition, such as His and maltose (Higgins, 1992). For eukaryotic ABC transporters, much attention has been paid to their roles in multidrug resistance. For example, overexpression of STS1 in yeast (Saccharomyces cerevisiae) and BCRP/MXR/ABCG2 in cancer cells increased cellular resistance to cytotoxic agents (Bissinger and Kuchler, 1994; Doyle et al., 1998; Litman et al., 2000). In addition, a number of eukaryotic ABC transporters have been shown to participate in other distinct biological processes, such as translocation of lipids, excretion of yeast mating factors, and regulation of ion channel activities (Higgins, 1995; Ketchum et al., 2001; Oram, 2002).
Plants, with a sessile life-style, have a particularly large number of ABC transporters. The genome of Arabidopsis contains 129 open reading frames (ORFs) capable of encoding ABC proteins (Sánchez-Fernández et al., 2001). Based on their size, orientation, domain organization, and resemblance to ABC proteins from other organisms, these proteins can be classified into 12 subfamilies: multidrug resistance proteins, multidrug resistance-associated proteins (MRPs), pleiotropic drug resistance proteins, and ABC1 homolog are full-sized transporters; the peroxisomal membrane proteins (PMPs), white/brown complex proteins (WBCs), ABC2 homologs, ABC transporters of the mitochondrion (ATMs), and transporters associated with antigen processing are half-sized transporters. In addition, three groups of soluble proteins are also categorized within this superfamily (Sánchez-Fernández et al., 2001).
Plant ABC transporters have been found to be involved in detoxification by means of vacuolar sequestration of various metabolites or excretion of toxic compounds out of the cell (Rea et al., 1998; Theodoulou, 2000; Jasinski et al., 2001; Martinoia et al., 2002). Members of the MRP subfamily (AtMRP1, AtMRP2, AtMRP3, and AtMRP5) exhibit glutathione S-conjugates transport activity (Lu et al., 1997, 1998; Tommasini et al., 1998; Gaedeke et al., 2001). Two pleiotropic drug resistance proteins, NpABC1 from Nicotiana plumbaginifolia and SpTUR2 from Spirodela polyrrhiza, are able to excrete diterpenoid antifungal agent, the sclareolide derivatives (Jasinski et al., 2001; van den Brûle et al., 2002). However, function of plant ABC proteins is not restricted to detoxification because genetic analysis of several loss-of-function mutants has revealed their diverse roles in plant growth and development, including metabolite translocation, far-red light response, stomata movement, auxin transport, maturation of cytosolic Fe/S proteins, and seed germination (Sidler et al., 1998; Debeaujon et al., 2001; Gaedeke et al., 2001; Kushnir et al., 2001; Møller et al., 2001; Noh et al., 2001; Footitt et al., 2002). Despite progresses made in the last several years, functions of most plant ABC transporters remain unknown.
Cotton (Gossypium hirsutum) fibers are single-celled seed hairs developed from the outmost layer of the integument. After anthesis, differentiated fiber cells begin to expand and undergo rapid elongation, followed by overlapping stages of secondary cell wall synthesis and dehydration (Applequist et al., 2001; Kim and Triplett, 2001). Fiber quality is of paramount importance for industry, and quality characters are mostly controlled by genetic factors (Paterson et al., 2003). Several spontaneous mutants defective in fiber development were found during cotton breeding (Kim and Triplett, 2001); however, none of the mutated genes has been cloned, partly because of the large genome size of cotton. Quite a number of cDNAs, on the other hand, have been isolated from developing cotton fibers with expression patterns characterized (John and Crow, 1992; Orford and Timmis, 1998; Li et al., 2002; Cedroni et al., 2003; Ji et al., 2003). Although precise mechanisms controlling fiber initiation and growth are still elusive, the popular assumption for fiber cell elongation is, to date, turgor-based cell expansion (Smart et al., 1998). Recent investigation showed the importance of plasmodesmatal gating, substance transport (Suc and K+), and carbon partitioning in cotton fiber development and expanding (Haigler et al., 2001; Ruan et al., 2001, 2003). Therefore, the cell-cell communication and substance exchange between fibers and adjacent cells of ovules are critical to fiber elongation.
Here, we report cDNA cloning and expression analysis of a half-sized ABC transporter, GhWBC1, from cotton. GhWBC1 is predominantly expressed in developing fibers and exhibited a temporal expression pattern correlating with fiber elongation. enhanced green fluorescence protein (EGFP)-GhWBC1 fusion protein was localized to plasma membrane. Interestingly, overexpression of GhWBC1 in Arabidopsis plants resulted in impaired seed development and short siliques (SSs), suggesting that this transporter plays an important role in plant development.
RESULTS
Isolation of GhWBC1
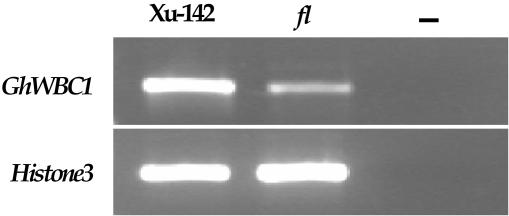
Upland cotton is a tetraploid species widely cultivated in the world. An fl (fuzzless-lintless) cotton mutant, which was isolated from cotton cv Xu-142, has seeds that are nearly glabrous, and the mutant plants do not show other phenotypic changes (Zhang and Pan, 1991; Yu et al., 2000). To isolate mRNAs that are enriched in fiber cells, we performed comparative reverse transcription (RT)-PCR analysis of ovules of the wild-type cotton cultivar and the fl mutant. By analyzing transcripts corresponding to randomly selected clones from a cDNA library of cotton ovules (–3–15 DPA), we found one of the cDNAs, ZHU93, that showed a much higher level of transcripts in wild type than in fl mutant ovules at 5 DPA (Fig. 1).
Figure 1.
RT-PCR analysis of GhWBC1 (Zhu93) gene expression in developing ovules of cotton cv Xu-142 and fl mutant, collected at 5 DPA. The cotton Histone-3 (GhHIS3) was amplified as a control; in negative control (–), no template DNA was added. The PCR was performed with primers GH1 and GH2.
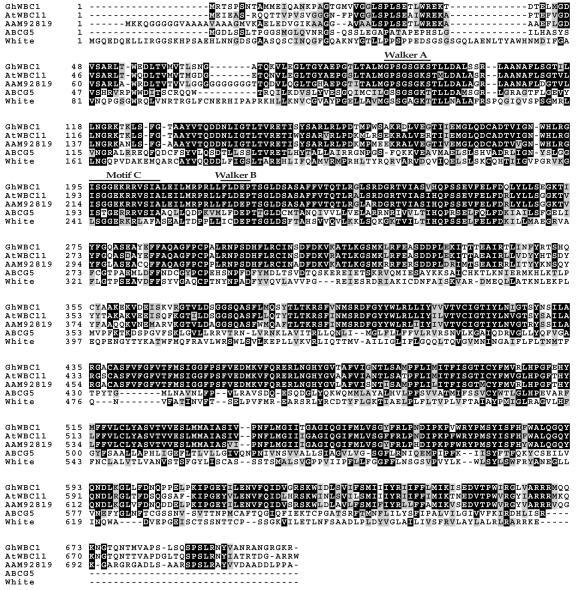
The ZHU93 cDNA is 2,372 bp in length (Gene Bank accession no. AY255521). An in-frame stop codon was present upstream to the translation start codon ATG, suggesting a complete ORF. This ORF encodes a protein of 705 amino acid residues with a predicted molecular mass of 77 kD. BLAST analysis indicated that the deduced protein was highly similar to ABC transporter proteins, and the highest sequence identities were found with a group of WBC proteins of the ABC transporter superfamily, including AtWBC11 (At1g17840) of Arabidopsis, with an identity of 82%, and a putative transporter of rice (Oryza sativa; AAM92819), with an identity of 79%. In this cotton protein, and in AtWBC11 and the putative WBC transporter of rice, an NBF is located near the N-terminal, where there are three conserved motifs: the Walker motif A and B and motif C (Fig. 2). Motif C is situated between two Walker motifs, and the sequence here (ISGGEKRRVSIA) is consistent with the consensus of ABC signature (Bairoch, 1992). Hydrophobicity analysis showed that the C-terminal one-half of this cotton protein is highly hydrophobic, and six putative transmembrane segments are predicted to be present (data not shown). As revealed by protein sequence alignment (Fig. 2), this reverse orientation (NBF-TMD) is shared by all the WBC proteins (Sánchez-Fernández et al., 2001), including fruitfly (Drosophila melanogaster) WBC proteins (Dreesen et al., 1988), human (Homo sapiens) BCRP (Doyle et al., 1998), ABCG5, and ABCG8 (Berge et al., 2000; Klucken et al., 2000). Taken together, sequence information indicates that this cotton cDNA encodes a half-sized ABC transporter of the WBC subfamily, and it is then named GhWBC1.
Figure 2.
Alignment of amino acid sequences of GhWBC1, Arabidopsis AtWBC11 (At1g17840), a rice putative ABC transporter (AAM92819), human ABCG5 (AAG40003), and fruitfly White protein (P10090). All the proteins aligned here are WBC members of the ABC transporter superfamily. The sequences were aligned with ClustalW (Thompson et al., 1994) using default parameters through EMBnet (http://www.ch.embnet.org/software/ClustalW.html), and the box shade was created by BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Southern blot showed that the diploid genome of Gossypium arboreum contains a small family of two to three genes encoding GhWBC1 homologous proteins (data not shown).
Spatial and Temporal Expression Pattern
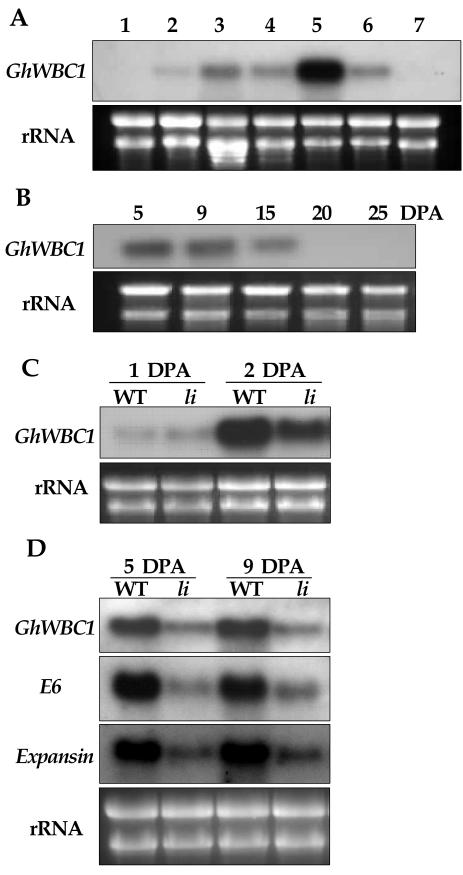
As shown in Figure 1, RT-PCR analysis of ZHU93 revealed a significantly higher level of GhWBC1 gene expression in wild-type ovules containing fibers than in fiberless fl ovules, both at 5 DPA. We then examined GhWBC1 expression in other tissues. RNA blot showed that, in contrast to a high level of GhWBC1 transcripts in cotton fibers of 9 DPA, only very low levels were present in leaves, cotyledons, and hypocotyls of the 10-d-old seedlings. The transcripts were undetectable in either roots or fl ovules (Fig. 3A). These results indicate that GhWBC1 is regulated in a fiber-preferential manner.
Figure 3.
Northern-blot analysis of spatial and temporal pattern of GhWBC1 expression. A, Expression in seedlings (10 d old) and developing ovules (9 DPA); total RNA was isolated from roots (1), hypocotyls (2), cotyledons (3), young true leaves (4), fiber cells (5), wild-type intact ovules (6), and fl ovules (7); 20 μg of total RNA was loaded each lane. B, Expression of GhWBC1 in developing fibers of 5, 9, 15, 20, and 25 DPA, respectively; 1.25 μg of total RNA was loaded each lane. C and D, expression of GhWBC1, E6, and Expansin genes in li mutant and wild-type cotton cv Xu-142 ovules. C, GhWBC1 expression in 1- and 2-DPA ovules; D, GhWBC1, E6, and Expansin expression in 5- and 9-DPA ovules. Ten micrograms of total RNA was loaded each lane.
Accumulation of GhWBC1 mRNA in fibers at different developmental stages was also analyzed. In rapidly elongating fibers (5 and 9 DPA), the GhWBC1 transcript level was high. The transcript abundance decreased significantly at 15 DPA and diminished at 20 and 25 DPA (Fig. 3B), a stage when fibers entered the secondary cell wall formation. The results indicate that GhWBC1 gene expression is up-regulated in rapidly elongating fibers, with the mRNA level peaking around 5 to 9 DPA, the stage in which the fastest elongation of fibers occurs (Basra and Malik, 1984; Smart et al., 1998).
Then, we investigated GhWBC1 expression in fiber cells of an li (ligon-lintless) mutant of cotton, which has fibers that are not longer than 6 mm (Karaca et al., 2002), significantly shorter than the fibers of wild-type cotton cultivars (approximately 30 mm long). For both the wild type and li mutant, expression level of GhWBC1 was low in 1-DPA ovules (Fig. 3C). Immediately afterward (at 2 DPA), the transcript abundance increased dramatically in wild-type cotton ovules and also to a lesser extent in li ovules. However, a more profound difference of transcript abundance appeared in 5- and 9-DPA ovules, with a much lower level in the li mutant (Fig. 3D). At the same time, two fiber-enriched genes, E6 and Expansin, were also down-regulated in li mutant fibers at 5 and 9 DPA (Fig. 3D). E6 is the first identified fiber-specific gene (John and Crow, 1992), and Expansin protein has been proposed to function in cell wall loosening and cell expansion (Orford and Timmis, 1998; Cosgrove, 2000). These results further support the notion that GhWBC1 expression correlates with fiber elongation.
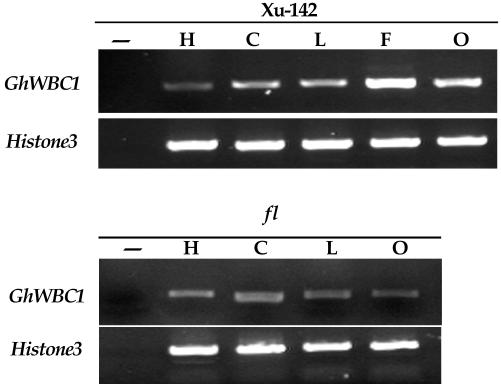
In northern analysis described above, the probe used covered part of the coding region, and it may have hybridized with transcripts of other members of the family. We then conducted the more specific RT-PCR with primers GH3 and GH4. The latter was designed according to the sequences of the 3′-untranslated region of the GhWBC1 cDNA. As shown in Figure 4, RT-PCR again showed the highest level of transcripts in developing fibers and a lower level in cotyledons, hypocotyls, and leaves. In addition, RT-PCR also revealed a weak expression of GhWBC1 in aerial organs of the fl mutant (Fig. 4). This indicates that, except for the fiberless ovule in which the GhWBC1 transcript abundance is significantly reduced, the GhWBC1 gene expression in the fl mutant is not altered.
Figure 4.
RT-PCR analysis of GhWBC1 expression in different aerial organs of cotton cv Xu-142 and fl mutant. Total RNAs were isolated from hypocotyls (H), cotyledons (C), and young true leaves (L) of the 10-d-old seedlings and from 9-DPA ovules (O) or fibers (F). The PCR was conducted by 27 cycles of PCR amplification with primers GH3 and GH4.
Subcellular Localization
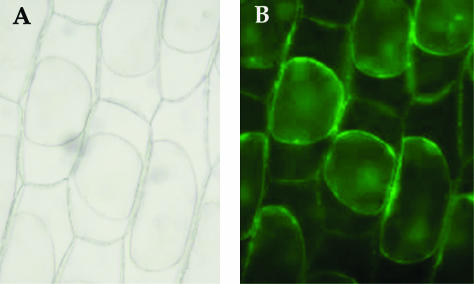
Cellular and subcellular localization of a transporter is often related to its function. To examine in which cellular compartment GhWBC1 is present, a 35S:EGFP-GhWBC1 expression cassette was transferred into onion (Allium cepa) epidermal cells by particle bombardment for transient expression of the fusion protein. After an overnight incubation, the green fluorescence signal of EGFP-GhWBC1 was observed around the cell surface, whereas the signal of EGFP alone was ubiquitously spread in the cell (data not shown). To determine whether GhWBC1 is localized to the cell wall or to the plasma membrane, a plasmolysis experiment was performed. As shown in Figure 5, GhWBC1-directed green fluorescence signal was internalized and associated with plasma membrane after plasmolysis, whereas no strong signal remained present around the cell wall. This observation suggests that GhWBC1 is likely a plasma membrane protein.
Figure 5.
Subcellular localization of EGFP-GhWBC1 fusion protein in onion epidermal cells. A, Cell structure under light microscopy. B, Green fluorescence signal of EGFP-GhWBC1 after plasmolysis with 20% (w/v) Suc for 5 min. Plasma membrane and GFP signal shrank together from cell wall.
Phenotypic Changes of Transgenic Arabidopsis
To get further clues to its function, GhAWC1 cDNA driven by a 35S promoter was introduced into Arabidopsis. About 400 primary transformants were obtained on the basis of kanamycin resistance and PCR identification. In the early stage of vegetative growth, no phenotypic changes were observed. In the reproductive stage, however, 53 of 400 (13.3%) T1 transgenic plants produced SSs, whereas other lines had siliques with similar length to the wild type (long siliques [LSs]; Fig. 6, A and B). The SS phenotype was also observed in T2 and T3 generations, although with a percentage lower than expected (data not shown).
Figure 6.
Phenotype of transgenic Arabidopsis plants overexpressing GhWBC1 under a 35S promoter. A, Inflorescence of mature plants of WT (left) and SS-29 transgenic line (right); B, siliques of WT (left) and SS-29 line (right); C, seeds in the 3-d-old siliques of WT (left) and of SS-29 line (right).
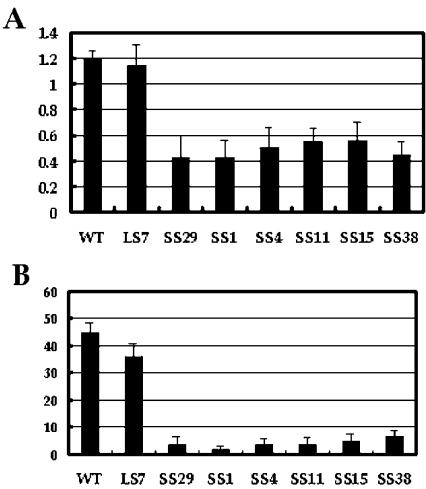
Several transgenic plant lines were used for further measurements. In our greenhouse conditions, the average silique length of SS lines was around 0.5 cm, significantly shorter than that of LS lines (e.g. LS-7, 1.14 ± 0.16 cm) and wild-type plants (1.20 ± 0.06 cm; Fig. 7A). The SS plants produced fewer seeds per silique (Fig. 7B). In SS-29 plants, for example, there were three to four seeds per silique, whereas the numbers for WT and LS-7 plants were 44.9 ± 3.38, and 35.87 ± 4.9, respectively. In developing siliques of the wild type (about 3 DPA), the embryos were regularly lined. In siliques of SS lines, most embryos were severely shriveled (Fig. 6C).
Figure 7.
Silique length and seed number per silique of WT and GhWBC1-transgenic lines of Arabidopsis. A, Length of siliques; B, seed number per silique. LS and SS represent the long (normal) silique lines and the SS lines of transgenic Arabidopsis, respectively. For each line, around 10 siliques were measured.
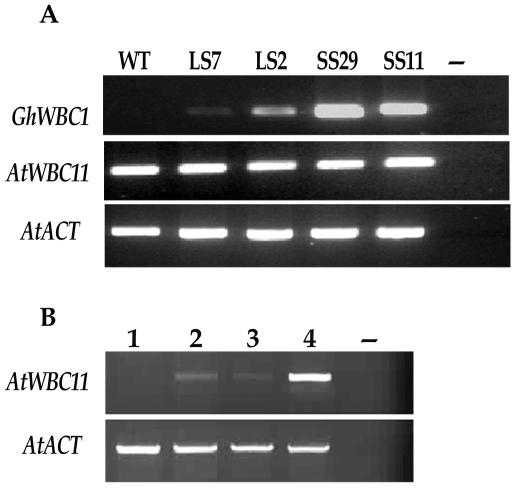
The SS-29 and SS-11 lines (T3) then were selected for further molecular examination. RT-PCR analysis of transgenic plants was performed to see if the phenotypic changes were related to transgene expression. Higher levels of GhWBC1 transcripts were found in both SS-29 and SS-11 lines, whereas in LS lines (LS-7 and LS-2) that showed a wild-type phenotype, the GhWBC1 expression level was lower (Fig. 8A). We further investigated whether the SS phenotype was caused by cosuppression of an Arabidopsis homolog. The closest homolog of GhWBC1 in the Arabidopsis genome is AtWBC11, which is predominantly expressed in inflorescences and siliques according to our preliminary analysis (Fig. 8). RT-PCR showed that AtWBC11 transcript levels in siliques of LS and SS transgenic lines and of the wild-type Arabidopsis were comparable (Fig. 8A). These data suggest that a high-level (and probably ectopic) expression of GhWBC1 in transgenic Arabidopsis plants may have impeded seed and silique development.
Figure 8.
A, RT-PCR analysis of transcripts of GhWBC1 and AtWBC11 in siliques of GhWBC1 transgenic lines of Arabidopsis; higher level of GhWBC1 gene expression was detected in siliques of SS lines. B, RT-PCR analysis of AtWBC11 expression in WT Arabidopsis plants. 1, Roots; 2, leaves; 3, stem; 4, inflorescence; –, negative control without template DNA. The PCR was performed by 30 cycles of amplification. Arabidopsis ACTIN2 (AtACT) was amplified as a control.
DISCUSSION
GhWBC1 encodes a half-sized ABC transporter of the WBC subfamily. In Arabidopsis genome, the full- and half-sized ABC transporters are equally represented. The largest group of Arabidopsis ABC proteins is the WBC subfamily, with 29 genes annotated (Sánchez-Fernández et al., 2001), but to date, none of them has a known function reported. In fruitfly, three WBC transporter genes are expressed in the eye pigmentary cells, and they form two types of heterodimers that mediate transport of guanine (WBC) and Trp (White/Scarlet complex), respectively (Dreesen et al., 1988). In the human genome, there are four WBC proteins that have been identified. ABCG1 (ABC8) regulates the transport of macrophage cholesterol and phospholipids (Klucken et al., 2000), ABCG5 and ABCG8 form a functional complex that mediates efflux of cholesterol and phospholipids (Berge et al., 2000), and BCRP forms homodimer to extrude drugs (Kage et al., 2002).
GhWBC1 transcripts are enriched in developing fibers during the elongation stage. This expression pattern suggests that GhWBC1 protein may be involved in the transport of a substance during fiber growth. If GhWBC1 protein is present in plasma membrane, as revealed by the EGFP-GhWBC1 fusion protein, it is unlikely that this transporter is directly involved in vacuolar sequestration that contributes to maintaining turgor pressure of expanding fiber cells. The GhWBC1 protein may not transport cell wall building blocks, either, because during the stage of secondary cell wall deposition, GhWBC1 transcripts in fibers were hardly detectable. Several genes whose products are involved in lipid biosynthesis and translocation are actively transcribed in rapidly expanding fibers, including an acyl carrier protein gene, ACP (Song and Allen, 1997); a lipid transfer protein gene, GH3 (Ma et al. 1995); and a putative β-ketoacyl-CoA synthase (Fiddlehead-like protein) gene, GhFDH (Li et al., 2002). All three genes show similar expression pattern to GhWBC1, with the highest level of expression taking place in fast-expanding fiber cells. Three of the four human WBCs are found to mediate lipid efflux (Berge et al., 2000; Klucken et al., 2000). Whether GhWBC1 also participates in lipid translocation is worthy of further investigation. Clearly, our results presented here do not naturally lead to an assumption that it is the mutation of GhWBC1 that has caused the mutant phenotype of either fl or li plants. However, based on its expression pattern, GhWBC1 may play an important role in cotton fiber development.
It is interesting that a high-level expression of GhWBC1 in Arabidopsis driven by the 35S promoter resulted in phenotypic changes: Seed development was largely impaired, and the siliques were significantly shorter. Severity of the phenotype was related to the transgene expression level, and low levels of expression did not cause visible phenotypic changes. In addition, expression of the closest homolog AtWBC11 was not altered in SS lines; thus, cosuppression is unlikely the cause of developmental abnormalities, although possibilities of suppressing other genes cannot be fully excluded. It is possible that the impeded seed and silique development of transgenic SS lines was caused by excessive and/or ectopic activities of the introduced transporter GhWBC1, which resulted in disrupted homeostasis of related component(s), such as membrane or extracellular lipids. Alternatively, GhWBC1 might form heterodimers with Arabidopsis half-sized ABC protein(s), such as AtWBC11, thus reducing activities of endogenous transporter(s) or acquiring novel activities. As mentioned above, different heterodimer complexes of WBCs in fruitfly mediate intercellular transport of different substances (Dreesen et al., 1988). Knockout lines of several Arabidopsis WBC members are now available. Detailed investigation of these knockout lines and further analysis of transgenic Arabidopsis plants will be greatly helpful in elucidation of GhWBC1 function in cotton fiber development.
MATERIALS AND METHODS
Plant Materials
Cotton (Gossypium hirsutum L. cv Xu-142) plants, the fl mutant (Zhang and Pan, 1991), and an li mutant of cotton (Karaca et al., 2002) were grown in the field. Cotton bolls were tagged on the day of anthesis. Ovules were harvested at various developing stages and frozen in liquid nitrogen. Plants of Arabidopsis ecotype Columbia were grown in soil under continuous light at 22°C.
DNA Analysis
Genomic DNA of the diploid cotton G. arboreum was extracted from etiolated seedlings (10 d old) as described by Tan et al., 2000). After an overnight digestion with Hind III, EcoR V, EcoR I, and XbaI, respectively, 40 μg of DNA in each lane was separated by electrophoresis and blotted onto nylon Hybond-XL (Amersham-Pharmacia Biotech, Uppsala). PCR performed on GhWBC1 cDNA with primers GH1056 (5′-TCCCCCGGGCCAAAGTTAGAGGAACCGTGC-3′) and GHR1850 (5′-CGGGATCCTCCATTTCGACCGTCCG-3′), and the amplified product was 32P labeled by using the Prime-a-Gene labeling system (Promega, Madison, WI) and used as a probe. Hybridization and washing were performed according to a standard protocol (Sambrook et al., 2001). The membrane was exposed to x-ray film for 2 d.
cDNA Library Construction and RNA Analysis
Arabidopsis total RNA was isolated by using the TRIzol method (Invitrogen, Carlsbad, CA). Cotton Total RNA was isolated from different organs using a cold phenol method (Groppe and Morse, 1993). For cDNA library construction, total RNA was isolated from ovules of cotton cv Xuzhou-142, which were collected at –3, –1, 0, 1, 3, 5, 7, 9, 11, 13, and 15 DPA, respectively. The RNA samples were then pooled with equal quantity for mRNA extraction. The library was constructed by using a ZAP Express cDNA Synthesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's manual.
For RNA blots, total RNA was separated by denaturing agarose gel and transferred onto Hybond-XL membranes (Amersham-Pharmacia Biotech). The blots were hybridized with 32P-labeled DNA probes of GhWBC1, E6, and Expansin, respectively. Hybridization and washing were performed according to a standard protocol (Sambrook et al., 2001). The blots were finally exposed to x-ray film for 2 d.
The DNA probe of GhWBC1 was obtained as described for Southern blot. For E6 and Expansin, PCR was performed on corresponding cDNA clones. The E6 fragment was amplified with primers E60 (5′-CCATGGCTTCCTCACCAA-3′) and E63 (5′-GGCATTCTTGTTGTAGTAG-3′), and the Expansin fragment was amplified with primers EXP1 (5′-CGAACCATAACCGTGACAGC-3′) and EXP2 (5′-AATTTCTGGAC-ATAGGTAG-3′).
For RT-PCR analysis, the first strand cDNA was synthesized with 1 μg of total RNA in a volume of 20 μL using the RNA PCR kit (TaKaRa, Dalian, China). Reaction mixture of 1 μL was used as template in PCR amplification with gene-specific primers. The primers used were: GH1 (5′-CTTATTAACTTCTACCGTACATCAC-3′), GH2 (5-′AACACGAAGAA CATATAATGTTCA-3′), GH3 (5-′TTCCTGGCGAATACATCCTT-3′), and GH4 (5-′AACTTCAAACGCACAATACAGAT-3′) for GhWBC1 (Zhu93); HIS1 (5-′GAAGCCTCATCGATACCGTC-3′) and HIS2 (5-′CTACCACTACCATCATGGC-3′) for GhHIS3 (AF024716); AT1 (5′-TTGGTTGACTATTACCACACTTCTG-3′) and AT2 (5′-AGAACAAAGAAGAGGTAGTGAGTG-3′) for AtWBC11; and ACT3 (5′-GACCTGCCTCATCATACTCG-3′) and ACT5 (5′-TTCCTCAATCTCATCTTCTTCC-3′) for AtACT (ACTIN2, At3g18820).
Subcellular Localization
The coding sequence of EGFP was amplified from the plasmid pEGFP-1 (CLONTECH, Palo Alto, CA) and in-frame fused to the 5′-terminal of GhWBC1. An intervening unstructured region (GPGGGG; Peck et al., 2001) was introduced between EGFP and GhWBC1. The resultant chimerical fusion gene GFP-GhWBC1 and the EGFP coding region alone were inserted into pBI121 vectors (CLONTECH) behind the 35S promoter, respectively. For transient expression, inner onion (Allium cepa) peels (2 × 2 cm2) were placed on a filter soaked with liquid Murashige and Skoog medium and bombarded with 2 μg of each plasmid under a vacuum of 27 inches Hg and a helium pressure of 1,100 psi. After incubation on Murashige and Skoog medium for about 16 h, green fluorescence was examined under a microscope (BX51, Olympus, Tokyo).
Arabidopsis Transformation
PCR was performed with Pyrobest DNA polymerase (TaKaRa) on GhWBC1 cDNA to introduce a SacI site after the stop codon; then, the ORF of GhWBC1 was inserted into a pBI121 vector (CLONTECH) between the SmaI and SacI restriction sites, replacing the GUS reporter gene. The 35S: GhWBC1-NOS expression cassette was also cloned into pCAMBIA2301. The resultant binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 via electroporation. Transformation of Arabidopsis was performed by a floral dip method (Clough and Bent, 1998). T1 transgenic plants were selected on one-half-strength Murashige and Skoog agar plates containing kanamycin (50 μg mL–1). One-week-old resistant seedlings were transferred to soil.
Acknowledgments
We thank Professors Tian-Zhen Zhang, Bao-Liang Zhou, and Song Chen for providing cotton cultivars and for their various help.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027052.
This work was supported by the National HighTech Program of China (grant no. 2001AA222121) and by the National Key Basic Research Program of China (grant no. 2002CB111301).
References
- Applequist WL, Cronn R, Wendel JF (2001) Comparative development of fiber in wild and cultivated cotton. Evol Dev 3: 3–17 [DOI] [PubMed] [Google Scholar]
- Bairoch A (1992) Prosite: a dictionary of sites and patterns in proteins. Nucleic Acids Research Suppl 20: 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basra AS, Malik CP (1984) Development of cotton fiber. Int Rev Cytol 89: 65–113 [Google Scholar]
- Berge K, Tian H, Graf GA, Yu L, Grishin NV, Schultze J, Kwiterovich P, Shan B, Barnes R, Hobbs HH (2000) Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290: 1771–1775 [DOI] [PubMed] [Google Scholar]
- Bissinger PH, Kuchler K (1994) Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product: a yeast ABC transporter conferring mycotoxin resistance. J Biol Chem 269: 4180–4186 [PubMed] [Google Scholar]
- Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wendel JF (2003) Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol Biol 51: 313–325 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJ, Leon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD (1998)A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA 95: 15665–15670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen TD, Johnson DH, Henikoff S (1988) The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol Cell Biol 8: 5206–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Muller A, Ansorge M, Becker D, Mamnun Y, Kuchler K, Schulz B et al. (2001) The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J 17: 1875–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe JC, Morse DE (1993) Isolation of full-length RNA templates for reverse transcription from tissues rich in RNase and proteoglycans. Anal Biochem 210: 337–343 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov VV, Hwang S, Martin K, Delmer DP (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47: 29–51 [PubMed] [Google Scholar]
- Henikoff S, Greene EA, Pietrokovski S, Bork P, Attwood TK, Hood L (1997) Gene families: the taxonomy of protein paralogs and chimeras. Science 278: 609–614 [DOI] [PubMed] [Google Scholar]
- Higgins CF (1992) ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8: 67–113 [DOI] [PubMed] [Google Scholar]
- Higgins CF (1995) The ABC of channel regulation. Cell 82: 693–696 [DOI] [PubMed] [Google Scholar]
- Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M (2001) A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13: 1095–1107 [PMC free article] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX (2003) Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res 31: 2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Crow LJ (1992)Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc Natl Acad Sci USA 89: 5769–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage K, Tsukahara S, Sugiyama T, Asada S, Ishikawa E, Tsuruo T, Sugimoto Y (2002) Dominant-negative inhibition of breast cancer resistance protein as drug efflux pump through the inhibition of S-S dependent homodimerization. Int J Cancer 97: 626–630 [DOI] [PubMed] [Google Scholar]
- Karaca M, Saha S, Jenkins JN, Zipf A, Kohel R, Stelly DM (2002) Simple sequence repeat (SSR) markers linked to the Ligon Lintless (Li) mutant in cotton. J Hered 93: 221–224 [DOI] [PubMed] [Google Scholar]
- Ketchum CJ, Schmidt WK, Rajendrakumar GV, Michaelis S, Maloney PC (2001) The yeast a-factor transporter Ste6p, a member of the ABC superfamily, couples ATP hydrolysis to pheromone export. J Biol Chem 276: 29007–29011 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Klucken J, Buchler C, Orso E, Kaminski WE, Porsch-Ozcurumez M, Liebisch G, Kapinsky M, Diederich W, Drobnik W, Dean M et al. (2000) ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci USA 97: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, Rycke RD, Engler G, Stephan UW, Lange H, Kispal G et al. (2001) A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Zhu YQ, Meng YL, Wang JW, Xu KX, Zhang TZ, Chen XY (2002) Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR. Plant Sci 163: 1113–1120 [Google Scholar]
- Litman T, Brangi M, Hudson E, Fetsch P, Abati A, Ross DD, Miyake K, Resau JH, Bates SE (2000) The multidrug-resistant phenotype associated with overexpression of the new ABC half-transporter, MXR (ABCG2). J Cell Sci 113: 2011–2021 [DOI] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Drozdowicz YM, Hoertensteiner S, Martinoia E, Rea PA (1998) AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparison with AtMRP1. Plant Cell 10: 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Rea PA (1997) AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci USA 94: 8243–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DP, Tan H, Si Y, Creech RG, Jenkins JN (1995) Differential expression of a lipid transfer protein gene in cotton fiber. Biochim Biophys Acta 257: 81–84 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Muller-Rober B, Schulz B (2002) Multifunctionality of plant ABC transporters: more than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- Møller SG, Kunke T, Chua NH (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like Genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram JF (2002) Molecular basis of cholesterol homeostasis: lessons from Tangier disease and ABCA1. Trends Mol Med 8: 168–173 [DOI] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN (1998) Specific expression of an expansin gene during elongation of cotton fibers. Biochim Biophys Acta 1398: 342–346 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Saranga Y, Menz M, Jiang CX, Wright RJ (2003) QTL analysis of genotype-environment interactions affecting cotton fiber quality. Theor Appl Genet 106: 384–396 [DOI] [PubMed] [Google Scholar]
- Peck SC, Nühse TS, Hess D, Iglesias A, Meins F, Boller T (2001) Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13: 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA, Li ZS, Lu YP, Drozdowicz YM, Martinoiz E (1998) From vacuolar GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49: 727–760 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Liewellyn DJ, Furbank RT (2001) The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- Sánchez-Fernández R, Davies TGE, Coleman JOD, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276: 30231–30244 [DOI] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 10: 1623–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Allen D (1997) Identification of a cotton fiber-specific acyl carrier protein cDNA by differential display. Biochim Biophys Acta 1351: 305–312 [DOI] [PubMed] [Google Scholar]
- Tan XP, Liang WQ, Liu CJ, Luo P, Heinstein P, Chen XY (2000) Expression pattern of (+)-δ-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta 210: 644–651 [DOI] [PubMed] [Google Scholar]
- Theodoulou FL (2000) Plant ABC transporters. Biochim Biophys Acta 1465: 79–103 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Fromentau M, Hörtensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J 13: 773–780 [DOI] [PubMed] [Google Scholar]
- van den Brûle S, Müller A, Fleming AJ, Smart CC (2002) The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J 30: 649–662 [DOI] [PubMed] [Google Scholar]
- Yu XH, Zhu YQ, Lu S, Zhang TZ, Chen XY, Xu ZH (2000) A comparative analysis of a fuzzless-lintless mutant of Gossypium hirsutum L. cv. Xu-142. Sci China Ser C 43: 623–630 [DOI] [PubMed] [Google Scholar]
- Zhang TZ, Pan JJ (1991) Genetic analysis of fuzzless-lintless mutant in upland cotton. Jiangsu J Agric Sci 7: 14–16 [Google Scholar]