Abstract
Gene expression during the potato (Solanum tuberosum) tuber lifecycle was monitored by cDNA-amplified fragment-length polymorphism, and several differentially expressed transcript-derived fragments were isolated. One fragment, named TDFL431, showed high homology to a copper (Cu) chaperone for Cu/zinc superoxide dismutase (CCS). The Ccs protein is responsible for the delivery of Cu to the Cu/zinc superoxide dismutase enzyme. The potato CCS (StCCS) full-length gene was isolated, and its sequence was compared with CCSs from other species. The promoter region of this gene was isolated, fused to the firefly luciferase coding sequence, and used for transformation of potato plants. The highest level of StCCS-luciferase expression was detected in the cortex of stem (like) tissues, such as stem nodes, stolons, and tubers; lower levels were detected in roots and flowers. The StCCS promoter contains regions highly homologous to several plant cis-acting elements. Three of them are related to auxin response, whereas four others are related to response to various stresses. Induction of the StCCS promoter was analyzed on 18 media, differing in hormone, sugar, and Cu content. StCCS expression was induced by auxin, gibberellins (GA4 + 7), fructose, sucrose, and glucose and was inhibited by relatively high concentrations of Cu.
Potato (Solanum tuberosum) plants undergo several developmental stages during their tuber lifecycle involving stolon formation, tuberization, tuber filling, dormancy, and tuber sprouting (Ewing and Struik, 1992). According to the classical hypothesis (Gregory, 1956), a substance is synthesized in the leaves under short-day conditions and transported to the stolons, where it triggers tuber formation (Struik et al., 1987). Tuberization is a complex developmental process resulting in the differentiation of an underground stolon into a specialized storage organ, the tuber (Taylor et al., 1992a; Visser et al., 1994). The process of tuberization comprises inhibition of longitudinal growth of the stolon followed by the initiation and growth of the tuber (Cutter, 1978). Tuberization is a continuous process, meaning that different developmental stages, from stolon induction to tuber growth, can occur in one plant simultaneously (Vreugdenhil and Struik, 1989). From a physiological point of view, dormancy can be considered to begin at tuber initiation and ends when buds are capable of extension growth (Wiltshire and Cobb, 1996). The starting moment of tuber formation is of importance for the length of the sprouting period (Claassens and Vreugdenhil, 2000). Thus not only tuberization but also the other phases of potato tuber life cycle, such as dormancy and sprouting, are asynchronous processes in nature and are therefore difficult to analyze individually.
To study gene expression during potato tuber life cycle, the screening of differentially expressed genes was performed in tuberization-related tissues obtained with a well-defined synchronous in vitro tuberization and dormancy system (Hendriks et al., 1991; Bachem et al., 2000). Using an RNA fingerprinting technique called cDNA-amplified fragment-length polymorphism (AFLP; Bachem et al., 1998), several hundred transcript-derived fragments (TDFs) were identified (Bachem et al., 2000; Trindade, 2003). One of them, TDFL431, was homologous to Cu chaperones for Cu/Zn superoxide dismutases (CCS) of different species.
Cu chaperones are small proteins that bind Cu in the cytosol and mediate its delivery to Cu-dependent target proteins (Harrison et al., 1999). In Brewer's yeast (Saccharomyces cerevisiae), three members of the Cu chaperones protein family have been described: Cox17p (cytochrome oxidase; Glerum et al., 1996), Atx1p (anti-oxidant; Lin and Culotta, 1995; Lin et al., 1997), and Lys7p (Lys biosynthetic pathway; Gamonet and Lauquin, 1998). Cox17 protein is involved in Cu delivery to the mitochondrial cytochrome oxidase, and Atx1 protein is responsible for targeting Cu into the Golgi vesicles of the secretory pathway. Lys7p inserts Cu into Cu/Zn superoxide dismutase (Sod1p) enzyme and is therefore called Ccsp (Cu chaperone for Sod) in humans (Homo sapiens; Culotta et al., 1997). The Ccs protein is the largest Cu metallochaperone identified to date, and it folds into three functionally distinct protein domains, whereas Atx1p and Cox17p represent single-domain proteins in yeast and human (Schmidt et al., 1999).
As in other species, plant Ccs proteins are responsible for the delivery of Cu to the Cu/Zn Sodp (Zhu et al., 2000; Wintz and Vulpe, 2002). Plant Sodps are known to play a role in defense against toxic reduced oxygen species and are consequently important for plant stress tolerance (Bowler et al., 1994). The Cu/Zn superoxide dismutase is a homodimeric zinc and Cu-containing enzyme that catalyzes the disproportionation of superoxide (O–2) to hydrogen peroxide (H2O2) and oxygen (O2) in a reaction mediated through the cyclic reduction and oxidation of the bound Cu ion (McCord and Fridovich, 1969).
In plants, the different members of the Cu chaperone family have been identified and characterized on different levels. The Arabidopsis CCH gene, highly homologous to the yeast ATX1 (Himelblau et al., 1998), has been the most extensively studied of the three Cu chaperones in plants, both at the mRNA (Mira et al., 2001b) and protein level (Mira et al., 2001a). The second type of Cu chaperones, the AtCOX17 gene, was recently isolated in Arabidopsis (Balandin and Castresana, 2002) and is a functional homolog of the yeast COX17. The plant homolog of the yeast LYS7 gene has been identified in tomato (Lycopersicon esculentum; LeCCS) as an expressed sequence tag (Zhu et al., 2000). Furthermore, analysis of the Arabidopsis genome resulted in the identification of one CCS gene (Wintz and Vulpe, 2002).
Although the three members of the Cu chaperone family have been identified in plants and proven to complement their homologs in yeast, besides the CCH gene (Mira et al., 2001b), no information is available about the expression of the COX17 and CCS genes in the different plant organs and tissues and/or during plant development. In this article, we describe the isolation and characterization of the Cu chaperone gene StCCS, coding for a protein responsible for delivering Cu to the Cu/Zn superoxide dismutase (Sodp), and the analysis of its expression through out the potato plant and during the potato tuber lifecycle.
RESULTS
Isolation of TDFL431
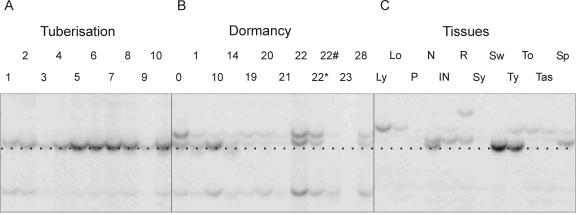
Gene expression during the potato tuber lifecycle was studied using the cDNA-AFLP technique, and a number of differentially expressed TDFs were identified. The fragment named TDFL431 showed increased expression during tuberization, whereas during dormancy, its expression decreased to a non-detectable level 10 weeks after harvest (Fig. 1). In addition, L431 transcript was detected in stem nodes, swelling stolons, and young tubers.
Figure 1.
cDNA AFLP analysis of TDFL431 expression during the tuber life cycle. The expression profile during the 10 d of the in vitro tuberization system (A) and in the 12 time points of dormancy template (B). The numbers indicate days in A and weeks in B; *, 1 week after sprouting; #, 3 weeks after sprouting. C, The expression in 12 different plant tissues: young leaves (Ly), old leaves (Lo), petioles (P), stem nodes (N), stem internodes (IN), roots (R), young stolons (Sy), swelling stolons (Sw), young tubers (Ty), dormant tubers (To), tubers after sprouting (Tas), and sprouts (Sp). The black dotted line indicates the band corresponding to the TDFL31.
The 175-bp-long TDFL431 is highly homologous to a Cu chaperone of the Cu/Zn superoxide dismutase (CCS) derived from different species.
Molecular Analysis
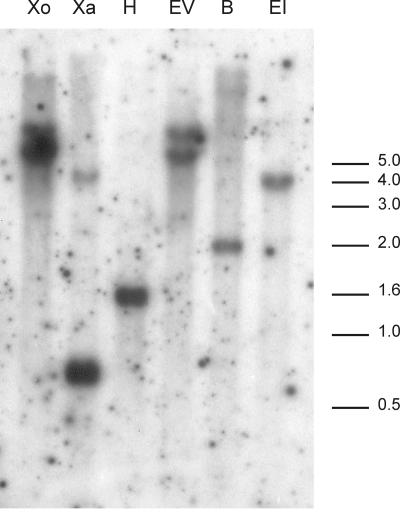
The results of the Southern-blot analysis using six different restriction enzymes are depicted in Figure 2. Hybridizations with the 175-bp-long TDFL431 resulted in two bands with the enzymes EcoRV, XbaI, and XhoI and one band with EcoRI, BamHI, and HindIII. These results suggested the presence of two copies or allelic versions of TDFL431 in the tetraploid potato genome of cv Bintje.
Figure 2.
Genomic organization of potato CCS gene(s) revealed by Southern-blot analysis. Genomic DNA from potato cv Bintje was digested with the following restriction enzymes: XhoI (Xo), XbaI (Xa), HindIII (H), EcoRV (EV), BamHI (B), and EcoRI (EI). The immobilized fragments were hybridized with the TDFL431 fragment.
Isolation of the Potato Cu Chaperone for Cu/Zn Superoxide Dismutase (StCCS) Gene and Its Promoter
Because StCCS showed a transient expression pattern during the tuber lifecycle, was only expressed in stem-like tissues (stems, stolons, and tubers), and had a low copy number, it was selected for promoter isolation. In total, 2,933 bp were identified, from which 954 bp were upstream of the start codon (ATG). Both Southern-blot analysis and the sequence identification revealed the presence of at least two StCCS copies or alleles.
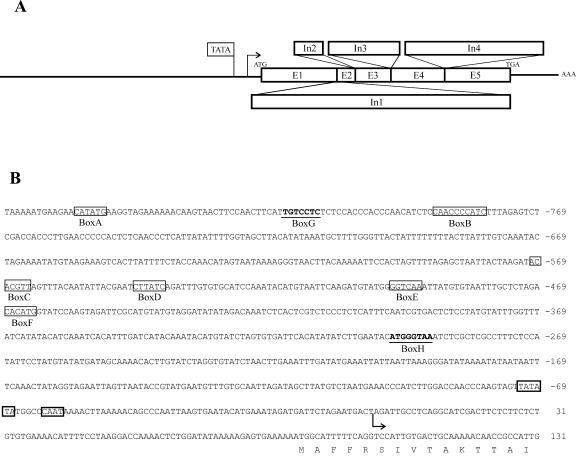
To identify the sequence of the whole gene and to determine the exon/intron borders, a genomic DNA fragment and a cDNA fragment downstream the TDFL431 were isolated. Combining all of the sequencing data, we concluded that the StCCS gene is 2,802 nucleotides long from the ATG till the stop codon (TGA) and encodes a protein of 311 amino acids (Figs. 3A and 4). Comparing the sequence of StCCS gene with the potato tentative consensus (TC 49957) in The Institute for Genomic Research database (http://www.tigr.org), six exons have been identified, interrupted by five introns as depicted in Figure 3A. The 3′-untranslated region is 179 bp long ending with a poly(A) tail. The genomic sequence of the StCCS gene is available in the National Center for Biotechnology Information (NCBI) databases (accession no. AY196210).
Figure 3.
StCCS gene and its promoter. A, Organization of the five introns (In1, 290 bp; In2, 679 bp; In3, 113 bp; In4, 266 bp; and In5, 521 bp) and six exons (E1, 255 bp; E2, 27 bp; E3, 73 bp; E4, 135 bp; E5, 200 bp; and E6, 243 bp) in the StCCS genomic fragment of 2,802 bp encoding a protein of 311 amino acids. The TIS is represented with an arrow and the poly(A) tail is represented with AAA. B, StCCS promoter sequence. Putative promoter cis-elements are labeled with boxes (Box A–H). Both the TATA box and CAAT box are built-in bold lines. The TIS is labeled with an arrow, and the translation start site with M.
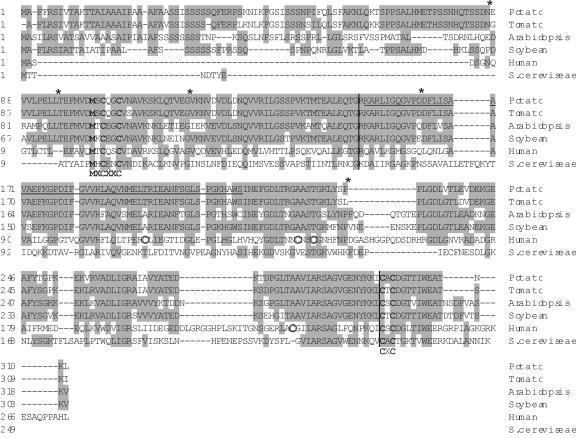
Figure 4.
The amino acid sequence of the potato StCcsp (accession no. AY196210) is shown in alignment with the sequences from other species available in GenBank. These include tomato (AF117707.2), Arabidopsis (AF179371.1), soybean (AF329816.1), human (AF002210.1), and yeast Lys7p (AAC49068.1). The amino acids different from the consensus are shaded. Vertical bars separate the three protein domains identified in human and mouse. The asterisks represent the localization of the introns in potato CCS, and the black horizontal line represents the TDFL431. The conserved motifs are in bold lettering, and the consensus sequence is indicated underneath. The circles indicate the Cys residues conserved in human and mouse.
Alignment of StCCS with CCS Genes from Other Species
The StCCS coding region was compared with sequences available from other species, such as tomato (Zhu et al., 2000), soybean (Glycine max), Arabidopsis, human (Culotta et al., 1997), and yeast (Gamonet and Lauquin, 1998). The result of this analysis is depicted in Figure 4. The potato CCS gene (StCCS) showed high similarity at the amino acid level to the CCS gene from tomato (LeCCS, 91.3% identical), soybean (GmCCS, 66.6%), and Arabidopsis (AtCCS, 60.5%).
Ccs proteins from human, mouse, and yeast are organized in three domains as shown in Figure 4 by vertical bars. StCcsp domain I is 151 amino acids long, as in the other plant Ccs proteins. However, the human and yeast domain I are much smaller, 70 and 66 amino acids, respectively. Domain I is homologous to Atx1p Cu chaperone, and it contains a highly conserved region, MXCXXC, which is thought to be a Cu-binding site (Zhu et al., 2000; Fig. 4). StCcsp domain II has some similarity to the superoxide dismutase protein, but not as similar as the human and mouse Ccs proteins. This domain contains 144 amino acids in potato, approximately the same as in the other plants, whereas in yeast and human, this domain is 162 and 174 amino acids long, respectively. The human and mouse proteins contain four extra Cys residues (marked with circles in Fig. 4) in domain II, which are present neither in plants nor in yeast. The third domain is specific for CCS genes and is the smallest with only 15 residues in potato and tomato, 21 in Arabidopsis, soybean, and Brewer's yeast, and 31 residues in human. In this domain, there is another highly conserved Cys motif, which is a putative binding site for Cu, CXC (Zhu et al., 2000).
Analysis of the 5′ Regulatory Region
The transcription initiation site (TIS) was determined experimentally with the GeneRacer Kit (Invitrogen, Carlsbad, CA), and it was localized 86 bp upstream of the first ATG. A putative TATA box was found 72 bp upstream of the TIS followed by a CAAT motif 11 bp downstream (Fig. 3B).
The StCCS 5′ region showed no overall sequence homology to any other promoter in the NCBI databases (Altschul et al., 1997). For the identification of putative cis-elements, the CCS promoter region was analyzed using the PLACE database (Higo et al., 1999). Six boxes (box A–F) that are 100% identical to other plant cis-acting elements (Fig. 3B) and two (box G and H) containing one nucleotide difference were found.
Box A is located 856 bp upstream of the TIS and has the same sequence as the auxin-responsive element small auxin-up RNA in soybean (Xu et al., 1997). Box B, located 790 bp upstream of the TIS, encloses a sequence that confers circadian mRNA oscillations in tomato (Piechulla et al., 1998). At position –570, there is a putative cis-element, box C, with the reverse complementary sequence of the quantitative activator region of the extensin gene (EXTA) in canola (Brassica napus; Elliott and Shirsat, 1998). Box D contains a sequence conserved among the light-regulated genes in Arabidopsis and tomato (Giuliano et al., 1988), and it is located 545 bp upstream of the TIS. Forty-eight nucleotides downstream of this box, another putative cis-element was identified (box E), with a sequence similar to the elicitor responsive element in the promoter of parsley PR1 genes (Rushton et al., 1996). Finally, at position –400 (box F), a putative recognition site for MYC transcription factor previously identified in the dehydration-responsive gene (RD22) in Arabidopsis (Abe et al., 1997) was found. At positions –817 (box G) and –299 (box H), there are two boxes (Fig. 3B) that resemble two auxin-responsive promoter elements, TGTCTC in the soybean GH3 promoter (Liu et al., 1994; Hagen and Guilfoyle, 2002) and the (G/T) GTCCCAT motif of the pea PS-IAA 4/5 promoter (Ballas et al., 1993; Guilfoyle et al., 1998), respectively.
Analysis of StCCS Expression in Different Tissues
The 954-bp fragment upstream of the translation initiation site from the StCCS gene was fused to the firefly luciferase+ reporter gene in the pBRH1 plasmid (Hulzink et al., 2002). After regeneration of transformants, 25 independent lines were further analyzed.
In vitro plantlets were assayed for luciferase activity, including roots, stems, and leaves. Luciferase expression was detected in roots and stems, but no signal was identified in leaves. The highest expression was observed in root tips and in the stem nodes (data not shown).
The in vitro plantlets were transferred to soil and analyzed for LUC+ gene expression. Several different tissues were assayed, such as leaves (young and fully grown), petioles, stems, roots, stolons (young, swelling, primary, and secondary), tubers (growing, mature, and dormant), flowers, and tuber sprouts. Potato fruits were not obtained under the greenhouse conditions used. The Lucp activity detected in the different tissues is presented in Figure 5. The different tissues were measured during different periods of time, different distances to the detector, and with different modes. To correct for all of these differences and to give a better idea of the relative expression in each organ, a relative time was added to each picture. The higher the relative time, the lower the expression in a particular organ.
Figure 5.
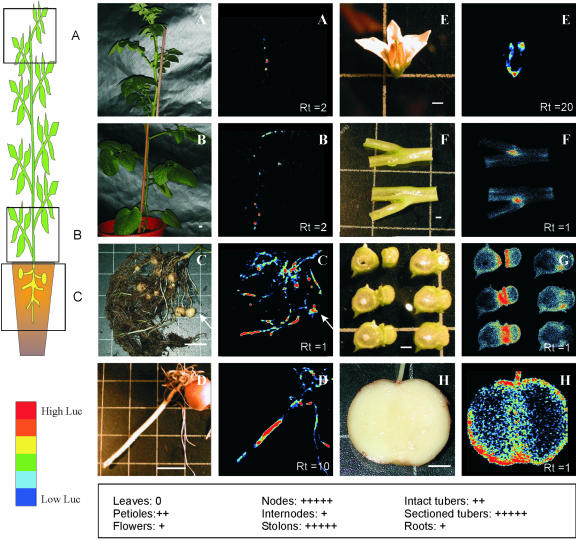
Expression of the firefly luciferase gene under the control of the potato StCCS promoter. On the left side of this figure, there is a schematic drawing illustrating the organs where luciferase expression has been measured. The second panel includes 16 pictures of the organ(s) measured in the luminometer (left side) and the results of the luciferase measurements (right side). Both pictures of the luciferase activity and corresponding plant organs are displayed with the same amplification. The colors resulting from the luciferase activity range from blue (low activity) to red (high activity) as represented in the lower left corner of the figure. A and B illustrate LUC+ expression in the aerial part, whereas C shows the underground part of a 2.5-month-old potato plant. The part of the plant where measurements A, B, and C were performed is indicated in the drawing next to these pictures. D shows expression in a tuber sprout, E in a flower, F and G in a sectioned stem node, and H in a sectioned tuber. The white bars in A through D correspond to 3.5 cm and in E through H to 0.35 cm. The white arrow in C points to the tuber. Rt, Relative time of luciferase activity measurements. Underneath D and H, an overview of the relative expression in different organs is given (0 corresponds to no activity and +++++ to the highest activity).
Concerning the aboveground organs of the potato plant, the highest luciferase gene expression was detected in stems (Fig. 5, A, B, F, and G). The stronger signal was identified in the stem nodes, mostly in the apical part of the plant (Fig. 5A) and just above the ground (Fig. 5B). Sectioning of stem nodes revealed that the highest LUC+ gene expression was found in the axillary buds and in the cortex of the stem (Fig. 5, F and G). As shown in Figure 5B, luciferase activity was also detected in some petioles, although at a lower level than in the stem nodes. A much weaker signal was measured in flowers, more specifically in the stamens and in the calyx (Fig. 5E). No LUC+ expression was found in leaves.
Regarding the underground tissues, the highest luciferase signal was identified in stolons, both primary and secondary (Fig. 5C). Some young (growing) intact tubers showed a medium LUC+ expression (Fig. 5C, white arrow), although after sectioning, a high expression was detected in the cortical region and in the pith (Fig. 5H).
Analysis of the potato tuber sprouts revealed that StCCS is expressed in this organ although at a lower level (Fig. 5D). Also in sprouts, LUC+ expression was found in the cortex and in the internodes (data not shown).
Northern-Blot Analysis
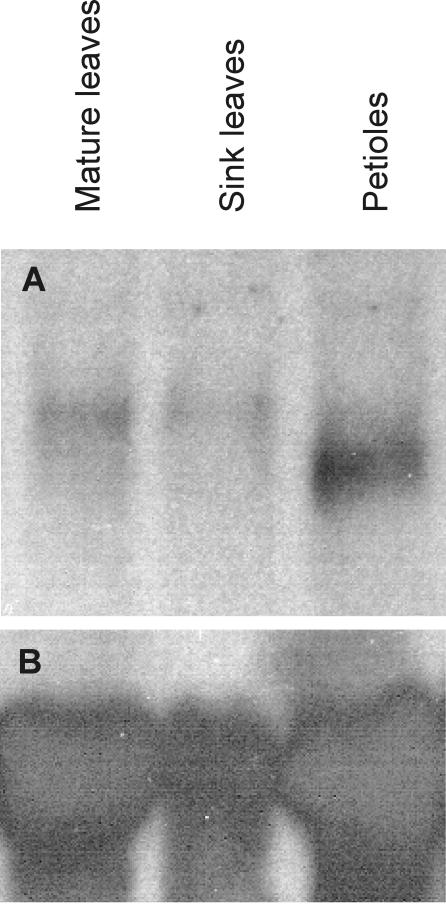
Rojas-Beltran et al. (2000) have shown that both cytosolic and plastidic isoforms of the Cu/Zn SOD are expressed in leaves. Thus, one would expect that StCCS would be expressed in leaves as well. However, no StCCS expression was detected in leaves either with the StCCS promoter/LUC+ fusions or by cDNA-AFLP analysis. A northern-blot analysis was carried out to investigate whether another StCCS copy or allele is expressed in leaves. Analysis of the northern-blot results revealed the presence of at least two transcripts hybridizing to the StCCS probe. The shorter transcript was only present in the petioles (Fig. 6, Petiole), whereas the longer transcript was detected in both young and mature leaves (Fig. 6, Sleaves and Mleaves) and was not present in petioles.
Figure 6.
Northern-blot analysis of StCCS transcript. A indicates the results of the hybridization with the StCCS gene and B with the 25S ribosomal probe.
Expression of CCS in Different Tissues after Spraying with CuSO4
Given that StCCS is a Cu chaperone, it was expected that its expression would change in the presence of high (or low) concentrations of Cu. Therefore, greenhouse-grown potato plants were sprayed with 50 mm CuSO4, and luciferase signal was measured 30 min, 1 h, and 2 h after treatment. No significant changes in luciferase activity were detected in any of the tissues at the different time points.
Because no changes were detected in greenhouse-grown plants after they were sprayed with CuSO4, the effect of Cu was tested in in vitro-grown plantlets. In these plantlets, high concentrations of Cu (10 mm of CuSO4) showed a negative effect on StCCS gene expression, bringing Luc+ activity down to less than one-third (Fig. 7) relative to the expression found in plantlets grown in MS10.
Figure 7.
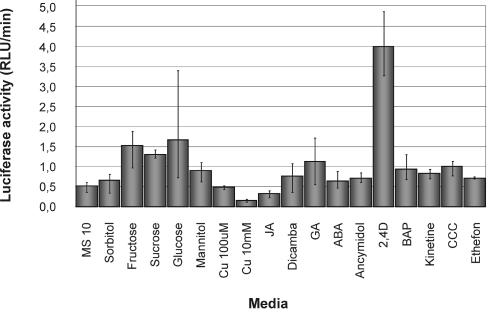
Activity of StCCS promoter driving luciferase gene in transgenic plants grown for 8 d on media supplemented with 18 different sugars, hormones, and Cu. Cu, Cu sulfate; JA, jasmonic acid; GA, gibberellins (GA4 + 7); ABA, abscisic acid; 2,4-D is a synthetic auxin; BAP, benzylaminopurine; CCC, 2-chloroethyltrimethylammonium chloride. In each sample, the luciferase expression was measured for 1 h, and the results are shown in relative light units per minute (RLU/min).
Regulation of CCS by Several Hormones and Sugars
To identify factors influencing StCCS gene expression, transgenic plants containing the StCCS promoter fused to the luciferase gene were grown in vitro in media supplemented with several hormones and sugars (Fig. 7).
With regard to the sugars, plantlets grown in media containing Fru, Suc, or Glc showed a higher LUC+ gene expression relative to the plants grown in MS10, whereas the non-metabolizable sugar alcohols, sorbitol and mannitol, did not show any significant effect. Concerning the hormones, the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) showed a more than 5-fold increase in StCCS expression, whereas gibberellins (GA4 + 7) led to a 2-fold increase in luciferase activity.
Although the different sugars and hormones seem to influence StCCS expression, the t test indicates, with 99% confidence, that the synthetic auxin 2,4-D has an inducing effect and the 10 mm CuSO4 an inhibitory affect on StCCS expression.
DISCUSSION
Gene expression during the potato tuber life cycle was monitored using the cDNA-AFLP technique, and several differentially expressed genes were identified. One of them, named TDFL431 was expressed during tuber formation and early stages of tuber development. Its expression was uniquely detected in stem-like tissues, such as stems, stolons, and tubers (Fig. 1). In addition, it showed the highest level of expression in newly developing organs, as swelling stolons and young tubers. Similar results were obtained with the StCCS promoter/LUC+ fusion, where the highest signal was identified in stolons, sectioned tubers, and nodes. A lower level of expression was detected in sprouts and even lower in flowers and roots. No expression was observed in leaves either with cDNA-AFLP or with promoter LUC+ fusions. Interestingly, high levels of expression of other Cu chaperones, such as CCH, have been also found in stems of Arabidopsis plants (Mira et al., 2001b).
Until now, no reports have been published on the presence of Ccsp in leaves of any plant species. Because both isoforms of Cu/Zn Sodp, cytosolic and plastidic, are expressed in potato leaves (Rojas-Beltran et al., 2000), one would expect that also the Cu chaperone for these enzymes would be present in the same tissues. The difference in the localization of Cu/Zn Sodp and StCCS expression could have different explanations. First, if Cu is present at high concentrations in potato leaves, the presence of a chaperone would not be required for the incorporation of Cu in the Sodp. As shown in the case of yeast, the Sod has the ability to incorporate Cu when this is present at high concentrations (O'Halloran and Culotta, 2000). Another explanation could be that the other copy (or allele) of StCCS, detected by Southern hybridization, is expressed in leaves. This hypothesis is supported by the northern hybridization where two different StCCS transcripts were detected, one in petioles, which is likely to be derived from StCCS, and the other in leaves.
Analysis of the 954-bp sequence corresponding to the StCCS promoter resulted in the identification of several potential binding sites for transcription factors. Three of them were highly similar to auxin-responsive elements. The possible function of these controlling sequences is supported by the fact that StCCS expression was increased 5-fold in the presence of the synthetic auxin 2,4-D (50 μm).
It is known that the expression of the Cu/Zn SOD is induced by stress, such as light (Rao et al., 1996), wounding (van Camp et al., 1997), and dehydration (Yu and Rengel, 1999; Borsane et al., 2001; Martinez et al., 2001). Plants overexpressing Cu/Zn SOD are protected from photo-oxidative damage caused by high light intensity (Allen et al., 1997). Considering that the activity of Sodp is dependent on its chaperone, the chaperone genes are likely to be induced by the different forms of stress as well. Four putative cis-elements related to stress induction were identified in the StCCS promoter. Two boxes, B and D (Fig. 3B), are related with circadian mRNA oscillations and light response. Box E is an elicitor-responsive element, which is active in response to a pathogen. Finally, box F is most likely a water stress-responsive element. Greenhouse-grown plants were measured for LUC+ expression before and after incubation under light and water stress conditions for 24 h. Preliminary results showed no significant differences in Luc activity between the two conditions compared with the control plants (data not shown), although further research is needed.
Alignment of the sequence of the potato CCS gene with other species resulted in the identification of differences in the protein structure between plant, human, and yeast, although the conserved motifs MXCXXC in domain I and CXC in domain III were present in the Ccsp of all of the species. The differences found at the nucleotide and amino acid level between the Ccsp of plants and other eukaryotic species supported the hypothesis that Cu chaperones function in a slightly different manner in plants and other species (Wintz and Vulpe, 2002).
Promoter-luciferase fusions showed that CCS expression was confined to the cortical region of several stem (-like) tissues. The cortex of the stem contains parenchyma cells usually with high abundance of plastids (Esau, 1977). This result, together with the presence of a putative plastidic transit-peptide in the N terminus of the Ccs protein (Wintz and Vulpe, 2002), suggests that the StCcs protein might be active in plastids of stem (-like) tissues.
Pekker et al. (2002) reported an induction in the cytosolic ascorbate peroxidase (cAPX) at the mRNA and protein level in response to iron overload. cApxp, like Sodp, is a detoxifying enzyme of reactive oxygen intermediates (Asada, 1992). Therefore we decided to test the effect of Cu overload on StCCS gene expression. Plants sprayed with CuSO4 did not respond with a significant change in StCCS expression. It is known Cu/Zn Sodp itself can bind Cu when this is present at high concentrations, and in this case, the Cu/Zn Sodp would not require the chaperone to bind Cu (O'Halloran and Culotta, 2000). This is consistent with the inhibition of StCCS gene expression when in vitro potato plants were grown in media supplemented with 10 mm CuSO4. On the other hand, one cannot exclude that the inefficient penetration of CuSO4 through the epidermis of greenhouse-grown plants, in the time tested, could be the cause of the non-detectable changes in StCCS expression.
Although Cu can be toxic, the Cu chaperones are not detoxification proteins; they clearly function in a “chaperone” like manner, guiding and protecting the metal ion while facilitating appropriated partnerships (O'Halloran and Culotta, 2000). In fact, studies in yeast showed that metallochaperones became critical for cell function only under Cu-limiting conditions (O'Halloran and Culotta, 2000).
StCCS expression is induced by auxin, which is known to play a role in different stages of potato development. Auxins have a promoting effect on cell elongation/expansion, and depending on the time point at which they are applied, they can have an inhibitory (Kumar and Wareing, 1974) or stimulating effect (Xu et al., 1998) on tuberization.
The results described herein will contribute to a better understanding of CCS genes in plants, which might function in a slightly different manner than in other eukaryotic organisms. Additionally, because the StCCS promoter confers expression mostly in stem (-like) tissues, it could be used for the unraveling of cis-elements responsible for specific expression in these organs.
MATERIALS AND METHODS
Plant Material
Two different tetraploid varieties of potato (Solanum tuberosum) used were potato cvs Karnico and Bintje. Potato cv Bintje was used to isolate genomic DNA and total RNA, whereas potato cv Karnico was used for transformation experiments because of its outstanding transformation efficiency (Heeres et al., 2002).
For the identification of genes involved in tuber lifecycle, a well-defined in vitro synchronous tuberization and dormancy system, optimized for potato cv Bintje, was applied (Hendriks et al., 1991; Bachem et al., 2000).
Nucleic Acid Manipulation
DNA manipulations were conducted using standard procedures as described by Sambrook et al. (1989), unless specified otherwise. DNA was extracted from potato leaves according to Rogers and Bendich (1988). For Southern-blot analysis, DNA digested with restriction enzymes was electrophoresed through 1% (w/v) agarose gel (4 μg lane–1) and transferred onto a nylon membrane (Hybond N+, Amersham). The membrane was prehybridized for 3 h at 65°C in modified Church buffer (Church and Gilbert, 1984). Hybridization was carried out for 16 h at 65°C in modified Church buffer with a 175-bp fragment, corresponding to TDFL431 (described in “Results”), labeled with [α-32P]dCTP by random priming. Membranes were washed at 65°C for 15 min each time, twice in 2× SSC and 0.1% (w/v) SDS and finally in 1× SSC and 0.1% (w/v) SDS.
Total RNA used for the northern-blot analysis was isolated from the different tissues according to Bachem et al. (1998). Escherichia coli strain DH5α served as a host for plasmid amplification.
cDNA-AFLP Reactions and Isolation of TDFs
Total RNA was isolated from axillary buds harvested on each of the 10 d of in vitro tuber formation, as described by Bachem et al. (1998). The templates used to analyze gene expression during dormancy and sprouting were derived from samples collected at 12 time points starting at the moment the tubers were harvested until 28 weeks after storage at 10°C in the dark. The first sample was collected before storage, sample 2 after 1 week of storage, sample 3 after 10 weeks, sample 4 corresponded to 14 weeks of storage, sample 5 to 19 weeks, sample 6 to 20 weeks, sample 7 to 21 weeks, sample 8 to 22 weeks, and sample 11 to 23 weeks. All of these samples were collected from tubers where no tuber sprouts were visible. Samples 9 and 10 were collected from tubers incubated for 22 weeks in dark where the first sprouts had appeared 1 and 3 weeks before, respectively. Sample 12 was collected from sprouts after 28 weeks of storage. Other tissues used for RNA isolation and template preparations included potato young and mature leaves, nodes, internodes, roots, young stolons (not swollen), swelling stolons, small tubers (<1 cm), mature tubers, dormant tubers, and tuber sprouts. All of the tissue samples were collected from potato cv Bintje.
The templates used for cDNA-AFLP were prepared according to Bachem et al. (1996) using AseI and TaqI restriction enzymes. AFLP reactions and isolation of the TDFs from the polyacrylamide gel were performed according to Bachem et al. (1998). PCR products were controlled on a 1.5% (w/v) agarose gel and were subsequently sequenced using an anchor-related primer (CTCGTAGACTGCGTACCTAAT or/and GACGATGAGTCCTGACCGA).
Nucleotide and translated sequences were compared with nucleotide and amino acid sequences of the GenBank nonredundant databases by using the Blastn and Blastx sequence alignment programs (Altschul et al., 1997).
Isolation of StCCS Promoter Region
Isolation of the promoter region was carried out with the help of the Universal GenomeWalker Kit (BD Biosciences Clontech, Palo Alto, CA) according to manufacturer's instructions (Siebert et al., 1995). Two rounds of PCR were performed using the Advantage Genomic Polymerase mix (BD Biosciences Clontech) in a thermocycler (2400, PerkinElmer Life Sciences, Boston). The primary PCR was performed using an adaptor-related primer (AP1) and a gene-specific primer (GSPx, where x is odd number), followed by a second PCR with a nested adaptor-related primer (AP2) and a nested gene-specific primer (GSPy, where y is even number). In total, three genome walks were performed. The DNA sequences of the primers used in these three walks are shown in Table I. The parameters used during the primary PCR reactions were as follows: seven cycles [94°C, 2s; 72°C, 3 min] and 37 cycles [94°C, 2s; 67°C, 3 min]; within the last cycle, the PCR samples were incubated for an additional 7 min at 67°C. For the nested PCR, the same profiles were used, however, the number of cycles was different: five cycles with the first profile and 25 cycles with the second profile. The PCR fragments were analyzed on a 1% (w/v) agarose gel and cloned into pGEM-T Easy Vector System I (Promega, Madison, WI) according to manufacturer's recommendations.
Table I.
Primers used for the isolation of the promoter region and the 3′ end of the copper chaperone for a Cu/Zn Sodp (StCCS) gene and for the determination of the TIS
The first part of the table indicates the primers used in the three genome walks to isolate the promoter region of StCCS. AP1 stands for primary and AP2 for nested adaptor-related primer, GSP with odd numbers for primary gene-specific primer, and GSP with even numbers for nested gene-specific primer. In the second part of the table, the primers used in the GeneRacer kit to isolate the StCCS TIS and the 3′ region are depicted. GR3′/5′ stands for primary adaptor-related primers, GR3′/5′N for nested adaptor-related primers, GSP with odd numbers for primary gene-specific primer, and GSP with even numbers for nested gene-specific primer.
| Experiment | Primer Name | Primer Sequence (5′→3′) |
|---|---|---|
| Walk No. | ||
| 1, 2, and 3 | AP1 | GTAATACGACTCACTATAGGGC |
| 1, 2, and 3 | AP2 | ACTATAGGGCACGCGTGGT |
| 1 | GSP1 | AATATCTGGTCCTTTGAATTCGGCAAC |
| 1 | GSP2 | GCAGCAGATATAAGGAAATCTTCGGGT |
| 2 | GSP3 | CCCCCAAACTAAACCTTCAATAACCAA |
| 2 | GSP4 | AAATGGGCTACATTAAGCCATCAAGAT |
| 3 | GSP5 | CGGAGGAAGTTTGGTGATTGGAAGAAG |
| 3 | GSP6 | CGATGGACGTTCAAACTGCGAAGAAGA |
| Isolation of 3″ or TIS | ||
| 3′ and TIS | GR5′ | CGACTGGAGCACGAGGACACTGA |
| 3′ and TIS | GR5′N | GGACACTGACATGGACTGAAGGAGTA |
| 3′ and TIS | GR3 | GCTGTCAACGATACGCTACGTAACG |
| 3′ and TIS | GR3′N | CGCTACGTAACGGCATGACAGTG |
| TIS | GSP7 | GAAGGCGTTTCCATATGAAGAGCTGAA |
| TIS | GSP8 | GAAGCAGCAAAAGCAGCAGCTGGAATA |
| 3′ | GSP9 | AGCCCGTTTGATTGGACAAGGAGTACC |
| 3′ | GSP10 | TATCTGCTGCTGTTGCCGAATTCAAAG |
Three walks were performed from the TDFL431 toward and into the promoter region. As a result of the first walk, two PCR fragments were obtained from the DraI digested library, with a length of 815 and 549 bp. They were almost identical except for the 266-bp addition, which corresponded to an intron. The PCR fragment obtained in the second walk was amplified from the PvuII library and was 1,058 bp long, and the third one was a 1,074-bp product from the DraI library.
Analysis of the StCCS Promoter and Gene Sequence
The StCCS promoter sequence was compared with the GenBank eukaryotic promoter database (epd) using the BLASTn sequence alignment program (Altschul et al., 1997) and with the PLACE database (Higo et al., 1999). For the localization of the putative TATA box and CAAT box site, two different programs were used: PC gene and Gene Runner (v3.05; Hastings Software, Hastings on Hudson, NY).
The sequence of StCCS gene was compared with nonredundant NCBI database with the BLASTx protein alignment program (Altschul et al., 1997). The sequences obtained in each walk were analyzed using the DNA-Star software package. The alignment between StCCS and CCS genes from other species was performed according to the Clustal method (Higgins and Sharp, 1989).
Identification of StCCS TIS
Total RNA was isolated from potato stolons from potato cv Bintje with the GenElute Mammalian total RNA miniprep Kit (Sigma-Aldrich) according to Fossati (2002). mRNA was extracted from 5 μg of total RNA with paramagnetic streptavidin coated beads as described by Bachem et al. (1998). About 200 ng of mRNA was used to proceed with the GeneRacer Kit (Invitrogen), and the identification of the TIS was performed as previously described by Trindade et al. (2003). The sequences of the primers used are indicated in Table I: GR5′ and GSP7 for the primary PCR and for the nested PCR GR5′N and GSP8.
Isolation of the StCCS 3′ Region
The StCCS 3′ region was isolated using two different approaches. First, the transcribed region between amino acid 209 and the poly(A) tail was isolated with the GeneRacer Kit (Invitrogen). Second, to identify possible introns in the same region, a PCR was performed with a primer starting at amino acid 189 (TGGCTCAAGTCAATATGG) and another primer from amino acid 308 (GTTGCTTCCCAGATAGTT). The GeneRacer kit reactions were performed as described in the previous section and the StCCS gene-specific primers used were: GSP9 and GR3′ for the primary PCR and GSP10 and GR3′N for nested PCR (Table I).
Transformation and Regeneration of Transgenic Plants
To be able to clone the regulatory region of the StCCS gene, a 954-bp fragment upstream of the ATG was isolated by PCR using the primers L431luc2 (–863AAGCTT GAAGAACATATGAAGGTAG) and L431luc3 (60CTCGAGTTTTCACTCTTTTTA TATCC). This fragment was fused to a modified firefly luciferase gene, LUC+ (Sherf and Wood, 1994) in the binary vector pBRH1. The 35S303 5′/303 3′ vector contains the 35S promoter followed by a ntp303 5′-untranslated region, LUC+ gene, and the ntp303 3′-untranslated region (Hulzink et al., 2002). The pBRH1 vector is derived from the 35S303 5′/303 3′ vector (Hulzink et al., 2002) after removal of the 35S promoter. Potato transformation with Agrobacterium tumefaciens was carried out essentially as described by Visser (1991). Untransformed plants were used as negative control, whereas plants transformed with a plasmid containing luc+ gene driven by the cauliflower mosaic virus 35S promoter (35S303 5′/303 3′; Hulzink et al., 2002) served as positive control. Twenty-five independent transformed lines were obtained containing the StCCS promoter/luc+ construct.
To determine the luciferase activity in the transformants, eight clonal plants were produced from each of the primary transformants and grown on Murashige and Skoog medium supplemented with 10 g of Suc and 50 mg mL–1 kanamycin at 25°C with 16 h of light. When the plantlets had six to seven leaves, they were transferred to soil.
Several tissues of the in vivo and in vitro plants were analyzed for luciferase gene expression including roots, stolons (young and swelling stolons), tubers (young <1 cm and mature), stems (nodes and internodes), leaves (young and full-grown), petioles, flowers, and tuber sprouts. During these experiments, no fruits were obtained.
Potato Plants Sprayed with CuSO4
To analyze the effects of Cu on the Cu chaperone for the Cu/Zn Sodp, the greenhouse-grown transgenic plants containing the StCCS promoter region fused with the luciferase gene were sprayed with 50 mm CuSO4. Luciferase activity was measured 30 min, 1 h, and 2 h after treatment.
In Vitro Plants Grown on Different Media
Three randomly chosen transgenic clones containing the StCCS promoter/LUC+ and one untransformed potato cv Karnico were propagated so that 150 plantlets from each clone were available. They were grown in vitro for 20 d on Murashige and Skoog medium supplemented with 3% (w/v) Suc. Eight plants from each clone were grown on 18 different media for 8 d at 25°C with 16 h of light. Sixteen media were supplemented with different hormone and sugar conditions, whereas two other media were supplemented with 100 μm and 10 mm CuSO4. The hormones were added to Murashige and Skoog medium supplemented with 1% (w/v) Suc in the following concentrations: 25 mg L–1 benzylaminopurine, 50 μm dicamba, 50 μm 2,4-D, 7 mm ethephon, 50 μm kinetine, 100 μm abscisic acid, 200 μm methyl jasmonate, 5 mg L–1 ancymidol, 1.75 mg mL–1 2-chloroethyltrimethylammonium chloride, and 50 μm GA4 + 7. The hormones and sugars were chosen based on their influence (inducing or inhibiting) in gene expression during the potato life cycle (Li, 1985; Vreugdenhil and van Dijk, 1989; Müller-Röber et al., 1990; Perl et al., 1991; Jackson and Willmitzer, 1994). Both dicamba and 2,4-D are synthetic auxins, ethefon is a substance that releases ethylene, whereas ancymidol and 2-chloroethyltrimethylammonium chloride are anti-gibberellins. The sugars sorbitol, Fru, Glc, mannitol, and Suc were added in a concentration of 300 mm, and as a control, MS10 was used. All plantlets were grown at 25°C with 16 h light.
Luc Protein Extraction and Quantification
Protein extraction was carried out essentially as described by Leeuwen et al. (2000). Eight in vitro plants of each of the four different clones grown on the 18 media were frozen in liquid nitrogen and subsequently ground with a pestle. Eight plants from five different clones transformed with the 35S promoter/LUC+ were grown in MS10 and used as positive control.
As expected, the plantlets incubated in some media showed yellow leaves and did not grow as much. To make the luciferase measurements comparable, only green tissue was harvested.
Determination of protein concentration in these extracts was performed with the Protein Assay ESL (Roche Diagnostics, Mannheim, Germany) according to manufacturer's recommendations. To standardize further measurements, all of the samples were diluted to the same protein concentration. For the luciferase measurements, 10 μL of each protein extract was used mixed with 40 μL of luciferase extraction buffer.
In Vitro Luciferase Assay
For the measurement of luciferase activity, the protein extracts were thawed on ice, and 5 μL of each sample was mixed with 100 μL of flash-assay buffer (20 mm Tricine, 2.67 mm MgSO4, 0.1 mm EDTA, 2 mm dithiothreitol, 470 μm d-luciferin, and 5 mm ATP, pH 7.8). The luciferase activity was measured in a luminometer (Luminoskan LS, Labsystem, Helsinki) at 25°C. Luciferase signal was quantified for 2 s, 10 s, and 1 min. Several dilutions of luciferase (Roche Diagnostics) in luciferase extraction buffer were used for calibration (0.1–200 units mL–1). To determine whether the differences in luciferase expression, under the influence of the different hormones and sugars, were statistically significant, a Student's t test was performed for each sample.
Statement on Materials Availability
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are very grateful to Irma Straatman-Engelen for the northern-blot analysis and to Dirk-Jan Huigen for helping with the greenhouse-grown plants. We also thank Dr. Anne-Marie Wolters, Dr. Niek Appeldoorn, and Dr. Krit Raemakers for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025320.
References
- Abe H, Yamaguchi-shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD, Webb RP, Schake SA (1997) Use of transgenic plants to study antioxidant defenses. Free Radic Biol Med 23: 473–479 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K (1992) Ascorbate peroxidase: a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85: 235–241 [Google Scholar]
- Bachem C, van der Hoeven R, Lucker J, Oomen R, Casarini E, Jacobsen E, Visser R (2000) Functional genomic analysis of potato tuber life-cycle. Potato Res 43: 297–312 [Google Scholar]
- Bachem CWB, Oomen RJFJ, Visser RGF (1998) Transcript imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol Biol Rep 16: 157–173 [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF (1996) Visualisation of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J 9: 745–753 [DOI] [PubMed] [Google Scholar]
- Balandin T, Castresana C (2002) AtCOX17, an Arabidopsis homolog of the yeast copper chaperone COX17. Plant Physiol 129: 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Theologis A (1993) Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-induced gene, PS-IAA4/5, of pea (Pisum sativum). J Mol Biol 233: 580–596 [DOI] [PubMed] [Google Scholar]
- Borsane O, Díaz P, Agius MF, Valpuesta V, Monza J (2001) Water generates an oxidative stress through the induction of a specific Cu/Zn superoxide dismutase in Lotus corniculatus leaves. Plant Sci 161: 757–763 [Google Scholar]
- Bowler C, van Camp W, van Montagu M, Inzé D (1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13: 199–218 [Google Scholar]
- Church MG, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassens MMJ, Vreugdenhil D (2000) Is dormancy of potato tubers the reverse of tuber initiation? Potato Res 43: 347–369 [Google Scholar]
- Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD (1997) The copper chaperone for superoxide dismutase. J Biol Chem 272: 23469–23472 [DOI] [PubMed] [Google Scholar]
- Cutter EG (1978) Structure and development of the potato plant. In PM Harris, ed, The Potato Crop. Chapman and Hall, London, pp 70–152
- Elliott KA, Shirsat AH (1998) Promoter region of the extA extensine gene from Brassica napus control activation in response to wounding and tensile stress. Plant Mol Biol 37: 675–687 [DOI] [PubMed] [Google Scholar]
- Esau K (1977) Membranous modifications in sieve element plastids of spinach affected by the aster yellows disease. J Ultrastruct Res 59: 87–100 [DOI] [PubMed] [Google Scholar]
- Ewing EE, Struik PC (1992) Tuber formation in potato: induction, initiation, and growth. In Horticultural Review, Vol 14. John Wiley & Sons, New York, pp 89–198 [Google Scholar]
- Fossati T (2002) Plant RNA extraction using GenElute mammalian total RNA miniprep kit. Origins Nr 6: 12–13 [Google Scholar]
- Gamonet F, Lauquin GJM (1998) The Saccharomyces cerevisiae LYS7 gene is involved in oxidative stress protection. Eur J Biochem 251: 716–723 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionary conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glerum DM, Shtanko A, Tzagoloff A (1996) Characterisation of COX17, a yeast gene in copper metabolism and assembly of cytochrome oxidase. J Biol Chem 271: 14504–14509 [DOI] [PubMed] [Google Scholar]
- Gregory LE (1956) Some factors for tuberisation in the potato plant. Am J Bot 43: 281–288 [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998) How does auxin turn on genes? Plant Physiol 118: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Harrison MD, Jones CE, Dameron CT (1999) Copper chaperones: function, structure and copper-binding properties. J Biol Inorg Chem 4: 145–153 [DOI] [PubMed] [Google Scholar]
- Heeres P, Schipers-Rozenboom M, Jacobsen E, Visser RGF (2002) Transformation of a large number of potato varieties: genotype-dependent variation in efficiency and somaclonal variability. Euphytica 124: 13–22 [Google Scholar]
- Hendriks T, Vreugdenhil D, Stiekema WJ (1991) Patatin and four serine proteinase inhibitor genes are differentially expressed during potato tuber development. Plant Mol Bol 17: 385–394 [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM (1989) Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS 5: 151–153 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Mira H, Lin SJ, Culotta VC, Peñarrubia L, Amasino RM (1998) Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol 117: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulzink RJM, de Groot PFM, Croes AF, Quaedvlieg W, Twell D, Wullems GJ, van Herpen MMA (2002) The 5′-untranslated region of the ntp303 gene strongly enhances translation during pollen tube growth, but not during pollen maturation. Plant Physiol 129: 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, Willmitzer L (1994) Jasmonic acid spraying does not induce tuberisation in short-day-requiring potato species kept in non-inducing conditions. Planta 194: 155–159 [Google Scholar]
- Kumar D, Wareing PF (1974) Studies on tuberisation of Solanum andigena: II. Growth hormones and tuberisation. New Phytol 73: 833–840 [Google Scholar]
- Leeuwen W, Hagendoorn MJM, Ruttink T, Poecke R, van der Plas LHW, van der Krol AR (2000) The use of luciferase reporter system for in planta gene expression studies. Plant Mol Biol Rep 18: 143a–143t [Google Scholar]
- Li PH (1985) Potato Physiology. Academic Press, Harcourt Brace Jovanovich Publishers, Philadelphia, PA, pp 1–586
- Lin S, Culotta VC (1995) The ATX1 Gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc Natl Acad Sci USA 92: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Pufahl RA, Dancis A, O'Holloran TV, Culotta VC (1997) A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem 272: 9215–9220 [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) The soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez CA, Loureiro ME, Oliva MA, Maestri M (2001) Differential responses of superoxide dismutases in freezing resistant Solanum curbilobum and freezing sensitive Solanum tuberosum subjected to oxidative and water stress. Plant Sci 160: 505–515 [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I (1969) Superoxide dismutase: an enzymatic function for erythrocuprein (hemocuprein). J Biol Chem 244: 6049–6055 [PubMed] [Google Scholar]
- Mira H, Martinez-Garcia F, Peñarrubia L (2001b) Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J 25: 521–528 [DOI] [PubMed] [Google Scholar]
- Mira H, Vilar M, Péres-Payá E, Peñarrubia L (2001a) Functional and conformational properties of the exclusive C-domain from the Arabidopsis copper chaperone (CCH). Biochem J 357: 545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Röber BT, Koβmann J, Hannah C, Willmitzer L, Sonnewald U (1990) One of two different ADP-glucose pyrophosphorylase genes from potato responds strongly to elevated levels of sucrose. Mol Gen Genet 224: 136–146 [DOI] [PubMed] [Google Scholar]
- O'Halloran TV, Culotta VC (2000) Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem 275: 25057–25060 [DOI] [PubMed] [Google Scholar]
- Pekker I, Tel-Or E, Mittler R (2002) Reactive oxygen intermediates and glutathione regulate the expression of cytosolic ascorbate peroxidase during iron-mediated oxidative stress in bean. Plant Mol Biol 49: 429–438 [DOI] [PubMed] [Google Scholar]
- Perl A, Aviv D, Willmitzer L, Galun E (1991) In vitro tuberization potatoes harboring β-glucuronidase linked to a patatin promoter: effects of sucrose levels and photoperiods. Plant Sci 73: 87–95 [Google Scholar]
- Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter region necessary for circadian expression. Plant Mol Biol 38: 655–662 [DOI] [PubMed] [Google Scholar]
- Rao MV, Paliyath G, Ormrod DP (1996) Ultraviolet-B and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol 110: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1988) Extraction of DNA from plant tissues. Plant Mol Biol Manual A6: 1–10 [DOI] [PubMed] [Google Scholar]
- Rojas-Beltran JA, Dejaeghere F, Abdallakotb M, duJardin P (2000) Expression and activity of antioxidant enzymes during potato dormancy. Potato Res 43: 383–393 [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding protein with elicitor response elements in the promoter of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmidt PJ, Rae TD, Pufahl RA, Hamma T, Strain J, O'Halloran TV, Culotta VC (1999) Multiple protein domains contribute to the action of the copper chaperone foe superoxide dismutase. J Biol Chem 274: 23719–23725 [DOI] [PubMed] [Google Scholar]
- Sherf BA, Wood KV (1994) Firefly luciferase engineered for improved genetic reporting. Promega Notes 49: 14–21 [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23: 1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struik PC, Boon EJ, Vreugdenhil D (1987) Effects of extracellular extracts from leaves on the tuberization of cuttings of potato (Solanum tuberosum L.). Plant Physiol 84: 214–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, Mad Arif SA, Kumar A, Davies HV, Srobie LA, Pearce S, Flavell AJ (1992) Expression and sequence analysis of cDNAs induced during early stages of tuberization in different organs of the potato plant (Solanum tuberosum L.). Plant Mol Biol 20: 641–651 [DOI] [PubMed] [Google Scholar]
- Trindade LM (2003) Potato life-cycle: from genes to promoters to cis-acting elements. PhD thesis. Wageningen University, The Netherlands
- Trindade LM, Horvath B, Bachem C, Jacobsen E, Visser RGF (2003) Isolation and functional characterisation of a stolon specific promoter from potato (Solanum tuberosum L.). Gene 303: 77–87 [DOI] [PubMed] [Google Scholar]
- van Camp W, Inzé D, van Montagu M (1997) The regulation and function of tobacco superoxide dismutases. Free Radic Biol Med 23: 515–520 [DOI] [PubMed] [Google Scholar]
- Visser RGF (1991) Regeneration and transformation of potato by Agrobacterium tumefaciens. In K Lindsey, ed, Plant Tissue Culture Manual B5. Dordrecht, The Netherlands, pp 1–9
- Visser RGF, Vreugdenhill D, Hendriks T, Jacobsen E (1994) Gene expression and carbohydrate content during stolon to tuber transition in potatoes (Solanum tuberosum). Physiol Planta 90: 285–292 [Google Scholar]
- Vreugdenhil D, van Dijk W (1989) Effects of ethylene on the tuberization of potato (Solanum tuberosum) cuttings. Plant Growth Regul 8: 31–39 [Google Scholar]
- Vreugdenhil D, Struik PC (1989) An integrated view of the hormonal regulation of tuber formation in potato. Physiol Plant 75: 525–531 [Google Scholar]
- Wiltshire JJJ, Cobb AH (1996) A review of the physiology of potato tuber dormancy. Ann Appl Biol 129: 553–569 [Google Scholar]
- Wintz H, Vulpe C (2002) Plant copper chaperones. Biochem Soc Trans 30: 732–735 [DOI] [PubMed] [Google Scholar]
- Xu N, Hagen G, Guilfoyle T (1997) Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Sci 126: 193–201 [Google Scholar]
- Xu X, van Lammeren AAM, Vermeer E, Vreugdenhil D (1998) The role of gibberellins, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol 117: 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Rengel Z (1999) Drought and salinity differentially influence activities of superoxide dismutase in narrow leafed lupins. Plant Sci 142: 1–11 [Google Scholar]
- Zhu H, Shipp E, Sanchez RJ, Liba A, Stine JE, Hart J, Gralla EB, Nersissian AM, Valentine JS (2000) Cobalt(2+) binding to human and tomato copper chaperone for superoxide dismutase: implications for the metal ion transfer mechanism. Biochemistry 39: 5413–5421 [DOI] [PubMed] [Google Scholar]