Abstract
In addition to proton-pumping complex I, plant mitochondria contain several type II NAD(P)H dehydrogenases in the electron transport chain. The extra enzymes allow the nonenergy-conserving electron transfer from cytoplasmic and matrix NAD(P)H to ubiquinone. We have investigated the type II NAD(P)H dehydrogenase gene families in Arabidopsis. This model plant contains two and four genes closely related to potato (Solanum tuberosum) genes nda1 and ndb1, respectively. A novel homolog, termed ndc1, with a lower but significant similarity to potato nda1 and ndb1, is also present. All genes are expressed in several organs of the plant. Among the nda genes, expression of nda1, but not nda2, is dependent on light and circadian regulation, suggesting separate roles in photosynthesis-associated and other respiratory NADH oxidation. Genes from all three gene families encode proteins exclusively targeted to mitochondria, as revealed by expression of green fluorescent fusion proteins and by western blotting of fractionated cells. Phylogenetic analysis indicates that ndc1 affiliates with cyanobacterial type II NADH dehydrogenase genes, suggesting that this gene entered the eukaryotic cell via the chloroplast progenitor. The ndc1 should then have been transferred to the nucleus and acquired a signal for mitochondrial targeting of the protein product. Although they are of different origin, the nda, ndb, and ndc genes carry an identical intron position.
Plant and fungal mitochondria have highly branched electron transport chains. The proton-pumping respiratory complexes I, III, and IV work to a varying extent in parallel with non-proton-pumping enzymes, i.e. type II NAD(P)H dehydrogenases and alternative oxidase. Thus, the coupling of electron transport to ATP formation varies depending on the electron path (Siedow and Umbach, 1995; Joseph-Horne et al., 2001).
Complex I, the proton-pumping (type I) NAD(P)H dehydrogenase, is a multisubunit enzyme that is inhibited by rotenone in most organisms. It is present in α-proteobacteria and mitochondria of all eukaryotes except fermenting yeasts e.g. Saccharomyces cerevisiae. A homologous complex with unclear enzymatic properties is also present in chloroplasts (Yagi, 1991; Friedrich et al., 1995; Rasmusson et al., 1998).
Type II, or rotenone-insensitive, NAD(P)H dehydrogenases have been found in several bacterial species and in plant and fungal mitochondria. The most studied enzymes, Escherichia coli NDH and S. cerevisiae NDI1, are FAD-containing single-polypeptide enzymes of 45 to 50 kD (Jaworowski et al., 1981; de Vries and Grivell, 1988). The S. cerevisiae NDI1 is located on the inner surface of the inner mitochondrial membrane, where it catalyzes the oxidation of matrix NADH (de Vries et al., 1992). Two homologous S. cerevisiae proteins, NDE1 and NDE2, are located on the external side of the inner membrane, where they oxidize cytoplasmic NADH (Luttik et al., 1998; Small and McAlister-Henn, 1998). An NADPH-specific type II dehydrogenase, NDE1, is present on the external surface of the inner membrane of Neurospora crassa mitochondria (Melo et al., 2001).
Plant mitochondria oxidize NADH and NADPH via type II NAD(P)H dehydrogenases. Biochemical characterization of isolated plant mitochondria have demonstrated the presence of an internal rotenone-insensitive NADH dehydrogenase operating in parallel with complex I (Rasmusson and Møller, 1991; Melo et al., 1996). On the external side of the inner membrane, NADH and NADPH are oxidized by separate calcium-dependent dehydrogenases (Roberts et al., 1995). Also, internal NADPH can be oxidized in a calcium-dependent manner, and internal NADH oxidation is the only calcium-independent activity (Møller and Rasmusson, 1998; Møller, 2001). Therefore, plant mitochondria must contain at least four separate rotenone-insensitive NAD(P)H dehydrogenases.
Previously, we characterized two cDNA clones from potato (Solanum tuberosum), St-nda1 and St-ndb1 (Rasmusson et al., 1999). The encoded proteins are homologous to E. coli and S. cerevisiae type II NADH dehydrogenases. Although NDA1 shows conservation to S. cerevisiae NDI1 throughout the protein sequence, NDB1 contains an insertion carrying an EF hand motif for calcium binding, which is absent in NDA1 and the NADH dehydrogenases in E. coli and S. cerevisiae. However, an insertion with an EF hand motif is found in the same sequence position of the external calcium-dependent NADPH dehydrogenase, NDE1, in N. crassa (Melo et al., 1999, 2001). St-NDA1 and St-NDB1 were localized to the internal and external sides of the inner mitochondrial membrane, respectively (Rasmusson et al., 1999).
It was shown previously that the mRNA level of nda1 in potato leaves was under strict light control and showed a diurnal cycle. Also, immunodetectable NDA1 protein abundance and the internal rotenone-insensitive NADH dehydrogenase activity were affected by light, suggesting that the nda1 gene is involved in photosynthetically associated processes, most likely photorespiration (Svensson and Rasmusson, 2001). For a comprehensive investigation of all potential mitochondrial type II NAD(P)H dehydrogenases in a single species, we have analyzed this protein superfamily in Arabidopsis. We report the presence of three nuclear gene families and their expression in response to light stimulus. Two gene families, nda and ndb, have a common origin with fungal homologs. A novel homolog groups with the respective enzymes in cyanobacteria. We suggest the designation of this gene as ndc1 in line with the denomination in potato. Proteins of all three families are targeted to mitochondria. Transcript analysis indicates that Arabidopsis contains both a light-dependent and -independent nda gene.
RESULTS
Arabidopsis Contains Three Gene Families Encoding Homologs of the Type II NAD(P)H Dehydrogenases
Database screening of the Arabidopsis genome reveals seven nuclear-encoded reading frames (Table I) with highest similarity to type II NAD(P)H dehydrogenases of e.g. S. cerevisiae and E. coli (Young et al., 1981; de Vries et al., 1992) and potato NDA1 and NDB1 (Rasmusson et al., 1999). The deduced protein sequences are conserved throughout the complete reading frames, including the two conserved nucleotide-binding motifs. The Arabidopsis homologs group into three families: NDA (two genes), NDB (four genes), and NDC (one gene). At-NDA1 and At-NDA2 are both 78% identical at the amino acid level to potato NDA1 (Rasmusson et al., 1999). The At-NDB1 and At-NDB2–4 show highest sequence identity, 70% and 59% to 63%, respectively, to potato NDB1. The unique EF hand motif domain of potato NDB1 (Rasmusson et al., 1999) is conserved in all four At-NDB homologs (Fig. 1). A much lower sequence similarity is seen to the EF hand domain in the N. crassa NDE1 homolog and a corresponding domain found in Plasmodium falciparum and Plasmodium yoelli homologs (Fig. 1), although these domains are located in similar places in the reading frame. The seventh Arabidopsis type II NAD(P)H dehydrogenase homolog, At-NDC1, shows only approximately 25% amino acid sequence identity to potato NDA1 and NDB1, demonstrating its separation from the NDA and NDB families. In rice (Oryza sativa), two nda-type genes and one ndc-gene (Fig. 2) were found. In addition, expressed sequence tag (EST) clones (CB657084, CB65708,5 and CA767183) were found, more than 70% identical to St-NDB1 in overlaps of deduced amino acid sequence longer than 200 residues. This indicates that all three gene families are present in dicotyledons and monocotyledons. In general, positions of conserved motifs in Arabidopsis NDA, NDB, and NDC protein sequences are consistent with those in bacterial, yeast, and potato NADH dehydrogenase homologs reported previously (Rasmusson et al., 1999). Apart from this group of seven homologs, two putative genes in Arabidopsis (Munich Information Center for Protein Sequences codes At5g22140 and At3g44190) display some similarity to type II NAD(P)H dehydrogenases. However, the reading frames of At5g22140 and At3g44190 are significantly shorter, 365 and 367 residues, respectively, and unlike the other plant homologs, do not contain any putative domains for organellar targeting.
Table I.
Type II NAD(P)H dehydrogenase genes of Arabidopsis
| Gene | Locus | Accession No. | Protein Lengtha |
|---|---|---|---|
| nda1 | At1g07180 | NM_100592b | 510 |
| nda2 | At2g29990 | AC004680c | 508 |
| ndb1 | At4g28220 | NM_118962b | 571 |
| ndb2 | At4g05020 | AY039856b | 582 |
| ndb3 | At4g21490 | AL161555c | 581 |
| ndb4 | At2g20800 | AC006234c | 582 |
| ndc1 | At5g08740 | NM_120955b | 519 |
a Length of proteins are given in amino acid residues without considering putative posttranslational processing. b cDNA sequences. c Genomic DNA sequences.
Figure 1.
Conservation of the EF hand domain in Arabidopsis NDB proteins. The EF hand-carrying domain of potato NDB1 (Rasmusson et al., 1999) was compared with the corresponding sequences in Arabidopsis NDB proteins, N. crassa NDE1 (Melo et al., 1999), an identically positioned large insert in homologs found in Plasmodium falciparum (CAD51833) and Plasmodium yoelli (EAA22988), and Gallus gallus troponin C (5TNC), which carries two EF hands. The potato NDB1 EF hand motif positions (*) are conserved in all deduced Arabidopsis NDB sequences, with the exception of two positions in NDB4. In comparison, the N. crassa NDE1 EF hand motif (#) is not conserved in any of the plant proteins. The P. falciparum and P. yoelli sequences contain no EF hand motifs. Amino acid position is given at the end of each sequence.
Figure 2.
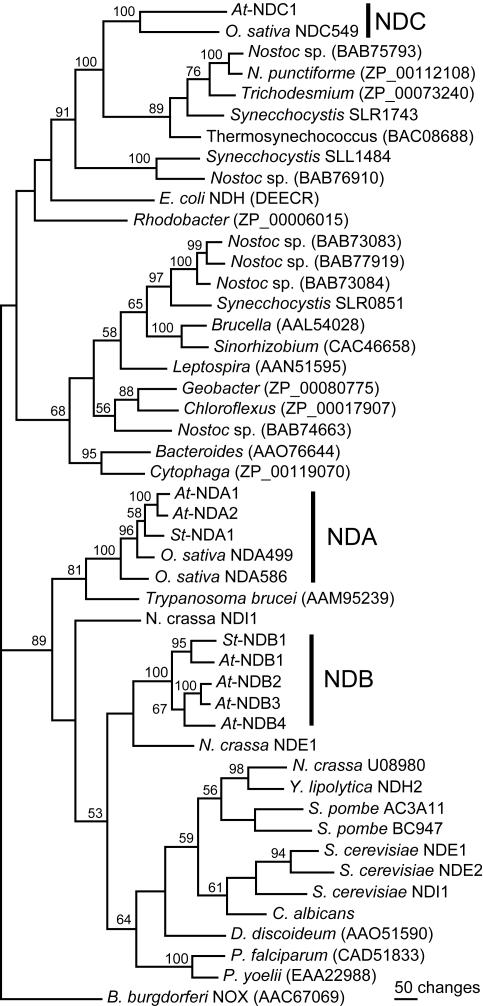
Phylogenetic analysis of bacterial, plant, fungal, and protist NAD(P)H dehydrogenase-like protein sequences. Sequences were aligned using ClustalW in MacVector 7.1 software (Accelrys Inc., San Diego). Alignments were manually inspected and edited, assuring correct matches of conserved regions. The PAUP* 4.0b10 program (Swofford, 2002) was used for phylogenetic analyses of the 648-character-long alignment, excluding the terminal extensions and the EF hand domain of 95 characters. Similar results were obtained when including the latter segment. Maximum parsimony was employed in heuristic searches, resulting in a single tree. Gaps were treated as missing characters, and the tree bisection reconnection option was used as branch-swapping algorithm. One thousand replicates with full heuristic searches were performed in a bootstrap test for the displayed tree. Bootstrap values of 50% or higher are shown on significant nodes. Plant protein families are marked out and include the Arabidopsis proteins in Table I, the potato NDA1 (AJ245861) and NDB1 (AJ245862), two rice cv japonica NDA-type reading frames of 499 and 596 residues (BAB68119 and BAC15811), and a rice cv indica reading frame, NDC549, manually annotated from a genomic clone (AAAA01002183). Other proteins investigated include all homologs from eukaryotes, and the five bacterial homologs giving the highest BLASTP score to St-NDA1, St-NDB1, and At-NDC1, respectively. Remaining homologs in Synechocystis sp. 6803 (Kaneko et al., 1996), Nostoc sp. 7120, and E. coli NDH, an NADH dehydrogenase (Young et al., 1981), were also included. The NOX sequence of Borrelia burgdorferi was included for rooting the tree. GenBank accession numbers are given in the figure for each homolog from bacteria and protists: Nostoc punctiforme, Trichodesmium erythraeum IMS101, Thermosynechococcus elongatus BP_1, Rhodobacter sphaeroides, Brucella melitensis 16M, Sinorhizobium meliloti, Leptospira interrogans serovar lai strain 56601, Geobacter metallireducens, Chloroflexus aurantiacus, Bacteroides thetaiotaomicron VPI_5482, Cytophaga hutchinsonii, P. falciparum 3D7, P. yoelli, Dictyostelium discoideum, and Trypanosoma brucei. Fungal homologs are Schizosaccharomyces pombe AC3A11 (CAB16382) and BC947 (CAA17043); S. cerevisiae NADH dehydrogenases NDI1 (NP_013586), NDE1 (NP_013865), and NDE2 (NP_010198); N. crassa NDE1, NADPH dehydrogenase (CAB41986), NDI1, NADH dehydrogenase (EAA27430), and U08980 (EAA29772); and Yarrowia lipolytica NDH2, NADH dehydrogenase (CAA07265), and Candida albicans (CAB77710).
Type II NAD(P)H Dehydrogenases in Arabidopsis Have Different Evolutionary Origins
The seven identified Arabidopsis protein sequences were compared with homologous sequences from bacteria, fungi, and plants by phylogenetic analysis (Fig. 2). Distance and maximum parsimony phylogenetic approaches were tested, substantially giving identical results. The plant entries group into three distinct subgroups, suggesting that there are three families (nda, ndb, and ndc) of type II NAD(P)H dehydrogenase genes present in plants. While the NDA and NDB sequences of plants are more closely related to the fungal and protist sequences, a clustering of At-NDC1 and the rice NDC homolog with cyanobacterial proteins is supported by 100% bootstrap values (Fig. 2). This indicates that the ndc genes of plants originate from cyanobacteria and most likely entered the plant cell with the chloroplast progenitor. The two At-NDA proteins cluster together, and like the St-NDA1, they are most closely related to one of the rice homologs. The plant NDA family is most similar to a homolog in T. brucei. The N. crassa NDE1 and the P. falciparum and P. yoelli homologs contain a protein segment corresponding to the plant NDB EF hand domain (Fig. 1). However, a closer relationship between these homologs is not supported by the phylogenetic tree, independent of whether the common domain is included in the analysis or not (Fig. 2).
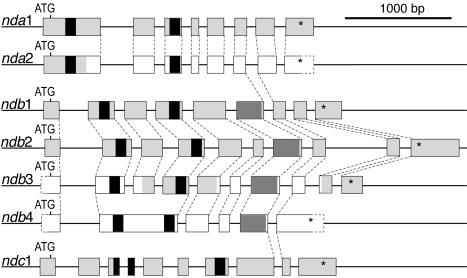
The gene structures were compared for the Arabidopsis type II NAD(P)H dehydrogenase genes (Fig. 3). For nda1, ndb1, and ndb2, complete cDNA sequences were available in the database. For ndc1, a partial cDNA (AY056424) and two ESTs (AU238932 and AV560836) were found that determined the location of all splicing sites. However, the cDNA clone and one EST contain residual introns that in each case are spliced in at least one other clone. Amplification of the complete reading frame by RT-PCR from isolated RNA and restriction of the product with HincII and DraII generated only cDNA fragments consistent with the complete splicing of all introns (results not shown). All intron positions and most intron lengths are conserved between the two nda genes (Fig. 3). Within the ndb family, most intron positions are conserved. The ndb4 gene, however, appears to have lost several introns present in ndb1 and ndb2 and shown or predicted present in ndb3. One intron position is identical in all three families (Fig. 3). This position corresponds to the center of a highly conserved 12 residues protein segment immediately downstream of the EF hand domain of the NDB proteins. All intron positions in the At-nda genes and At-ndc1 are conserved in the rice nda499 and ndc549, respectively, with one additional intron being present in ndc549. The rice nda586 only contains two introns, corresponding to introns 2 and 4 in the At-nda genes. The conservation of gene structures within each gene family provides additional support to the phylogenetic analysis.
Figure 3.
Exon-intron structures of type II NAD(P)H dehydrogenase genes in Arabidopsis. Boxes, exons. For orientation, the segments corresponding to nucleotide-binding motifs are shaded black, and the EF hand domains of NDB proteins are shaded in dark gray. Segments confirmed by isolated cDNA or ESTs are shaded in light gray. Exon segments depicted in white are as annotated in GenBank, with the exceptions for exons 1 and 7 of ndb3. These are conserved in other ndb genes, and their presence in mRNA was confirmed by reverse transcription (RT)-PCR (data not shown). Dashed lines, Identical intron positions. Asterisks, Translational stops.
Mitochondrial Localization of Encoded Proteins
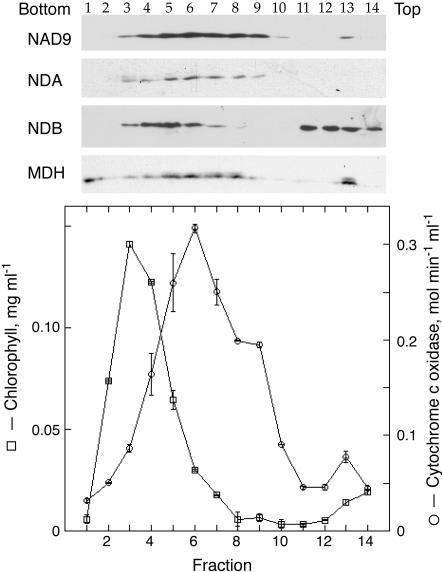
Potato NDA1 was shown previously to be tightly bound to the internal surface of the inner mitochondrial membrane, whereas NDB1 is loosely attached to the external side (Rasmusson et al., 1999; Rasmusson and Agius, 2001). Similar to potato NDA1 and NDB1, all Arabidopsis homologs contain N-terminal domains potentially specifying intracellular targeting (not shown). To investigate the cellular localization of the type II NAD(P)H dehydrogenases in Arabidopsis, mitochondria were purified from leaves by Suc gradient centrifugation. The Suc gradient efficiently separated mitochondria as evaluated by cytochrome c oxidase activity with a peak in fraction 6, from the heavier chloroplasts where chlorophyll peaked in fraction 3 (Fig. 4). Catalase, a marker for peroxisomes, had a wide distribution in the gradient with 4 to 6 μmol min–1 mL–1 in fractions 1 to 9 and less than 2 in fractions 10 to 14 (results not shown). Aliquots of the Suc gradient were subjected to western-blot analyses using antibodies against NDA1 and NDB1 from potato, the NAD9 subunit of wheat complex I, and malate dehydrogenase (Fig. 4). The distribution of malate dehydrogenase deviated from cytochrome c oxidase and NAD9, with malate dehydrogenase being virtually absent in fractions 9 and 10. This indicates that the uppermost part of the cytochrome c oxidase peak contains mitochondria with disrupted inner and outer membranes. NDA1 and NDB1 antibodies specifically detected polypeptides of 48 and 61 kD, respectively, consistent with the apparent molecular masses of St-NDA1 and St-NDB1 (Rasmusson and Agius, 2001). Proteins detected with both antibodies have a different distribution as compared with chlorophyll and catalase on the Suc gradient. The distribution of the immunodetected NDA protein coincided with cytochrome c oxidase activity and the NAD9 protein. The NDB signal peaked in fraction 5, coinciding with the lower part of the mitochondrial peak. This indicates that the protein is present in intact mitochondria but had been released from less dense mitochondria (fractions 7 and 8) with disrupted outer membranes but retaining malate dehydrogenase, i.e. mitoplasts. Protein recognized by the NDB antibodies was to a higher extent than malate dehydrogenase found at the top of the gradient, where small membrane vesicles and soluble proteins are expected, suggesting that more NDB protein had been released from the mitochondria. These results indicate that Arabidopsis homologs recognized by the NDA1 and NDB1 antibodies are localized in mitochondria and imply an external localization of the immunodetected At-NDB proteins, similar to potato NDB1.
Figure 4.
Subcellular localization of NDA and NDB homologs in Arabidopsis. The graph shows distribution of chlorophyll (squares) and cytochrome c oxidase activity (circles) in a Suc gradient analysis of a crude mitochondrial fraction from Arabidopsis leaves. In the upper part, the distribution of mitochondrial inner membrane was detected using antibodies against the NAD9 subunit of wheat (Triticum aestivum) complex I. As a marker for soluble matrix enzymes, malate dehydrogenase (MDH) antibodies were used, detecting distribution of mitochondria with intact inner membranes and enzyme released from disrupted mitochondria. NAD(P)H dehydrogenase homologs were detected using antibodies against potato NDA1 and NDB1. The immunorecognized NDA homologs coincide with NAD9. NDB homologs are detected in the lower part of the mitochondrial peak and at the top of the gradient but not in the chloroplast peak. Malate dehydrogenase has a distribution intermediate to NDB and NAD9 within the mitochondrial peak. The same volume of each fraction was loaded on the gel.
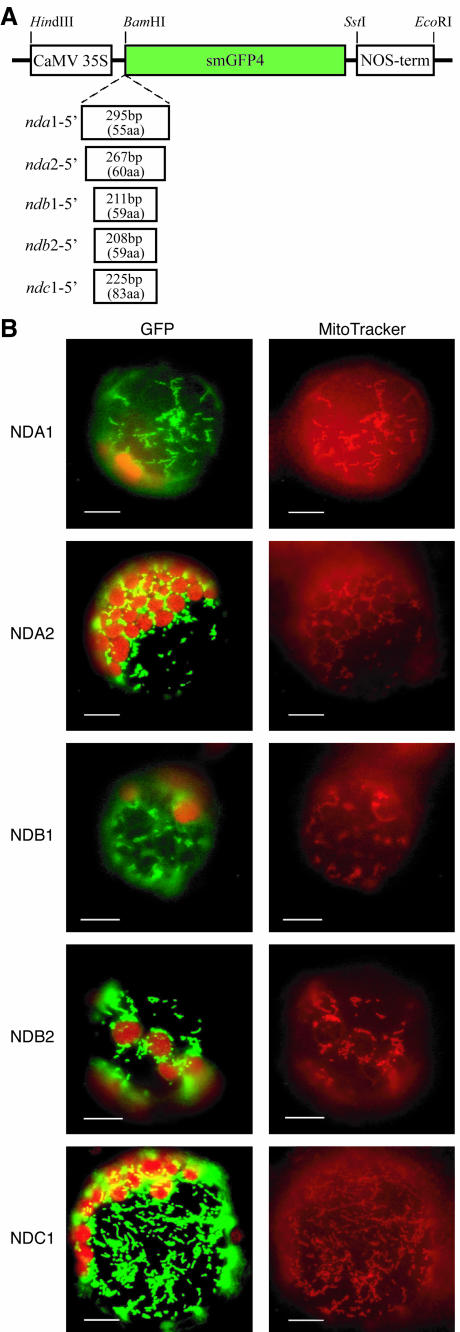
To analyze the subcellular localization of individual proteins, fusions of potential protein targeting domains with green fluorescent protein (GFP) were made for representative genes. Segments of cDNA encoding the N-terminal sequences up to the start of the first conserved nucleotide-binding motif were cloned into psmGFP4 in frame with the smGFP4 (Fig. 5). Tobacco (Nicotiana tabacum) protoplasts were transiently transformed with the different constructs, and expression of the fusion proteins was analyzed by fluorescence microscopy. Transformed protoplasts were simultaneously incubated with MitoTracker Red, a mitochondria-specific dye. All fusion proteins for NDA1, NDA2, NDB1, NDB2, and NDC1 localized to subcellular structures coinciding with MitoTracker Red staining (Fig. 5), and no signal could be detected in chloroplasts. This colocalization suggests that all five analyzed genes encode mitochondrial proteins, consistent with the immunological analyses (Fig. 4).
Figure 5.
Targeting analysis of gene products using GFP fusion proteins. The cDNA encoding the unconserved N-terminal part of each gene product, up to the start of the first nucleotide-binding motif, was fused to the reading frame of the smGFP4 reporter gene (A). The fusion proteins were expressed in transiently transformed protoplasts under the control of cauliflower mosaic virus 35S promoter and the NOS terminator. Location of the fusion proteins was analyzed by fluorescence microscopy (B). Images were taken using filter sets optimal for GFP and chlorophyll autofluorescence (left) and MitoTracker Red (right) and show for each construct the same protoplast. All GFP fusion proteins are observed in small cellular particles, which are also stained with the mitochondria-specific dye MitoTracker Red.
Expression of Arabidopsis NAD(P)H Dehydrogenase Genes
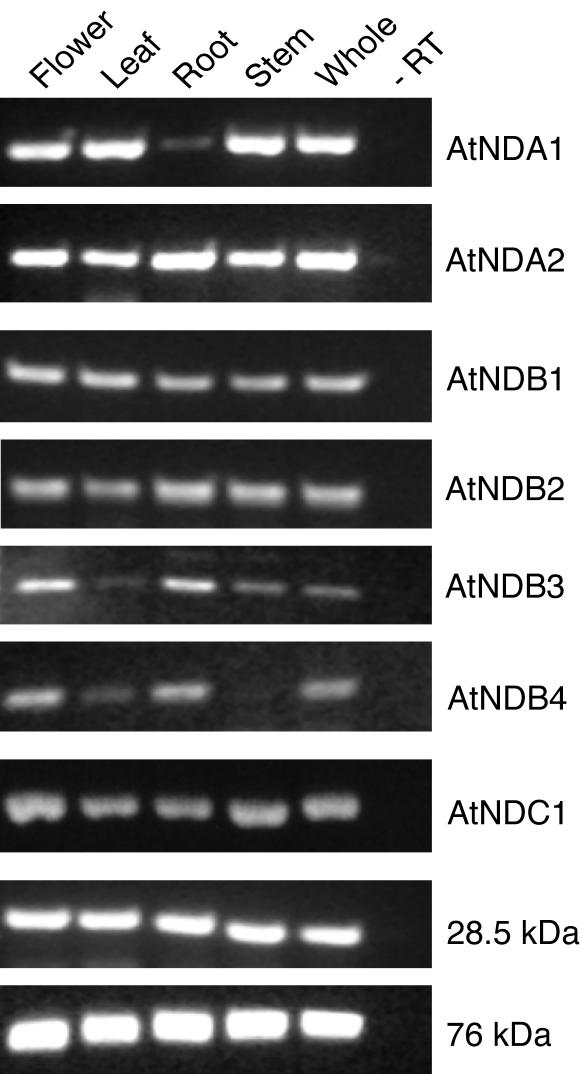
To get a general overview of whether expression is constitutive or confined to certain parts of the plants, nda, ndb, and ndc mRNA of Arabidopsis were analyzed by RT-PCR. Transcripts for the 28.5- and 76-kD complex I subunits (Rasmusson et al., 1998) were used as controls. Figure 6 shows agarose gel analyses of RT-PCR products. The correct identities of all PCR products were controlled by restriction cleavage (results not shown). While nda2 was expressed in all organs investigated, nda1 was preferentially expressed in aboveground tissues with almost no expression in roots. Transcript of ndb1 and ndb2 genes was present in all parts of the plant, whereas ndb3 and ndb4 mRNAs were almost completely absent in leaves, and ndb4 transcript was also absent in stems. The ndc1 gene was expressed at significant levels in all organs. Expression of complex I subunit genes showed little difference between organs, consistent with previous northern-blot analyses (Rasmusson et al., 1998). The observations for nda1, nda2, ndb1, ndc1, and the complex I controls were also supported by real-time RT-PCR (results not shown). The results suggest that all type II NAD(P)H dehydrogenase genes are expressed in several tissues, with potential spatial regulation at the transcript level for nda1, ndb3, and ndb4.
Figure 6.
Expression of type II NAD(P)H dehydrogenase genes of Arabidopsis. Total RNA was prepared from four different organs and whole plants. Transcripts for all seven NAD(P)H dehydrogenase homologs and two complex I subunits were analyzed by RT-PCR (A). To avoid amplification of contaminating genomic DNA, one primer in each pair covers an exon-exon border. Control reactions, where RNA isolated from whole plants were used as a PCR template without previous RT, showed that genomic DNA was not amplified (–RT).
The At-nda1 Transcript Responds to Light
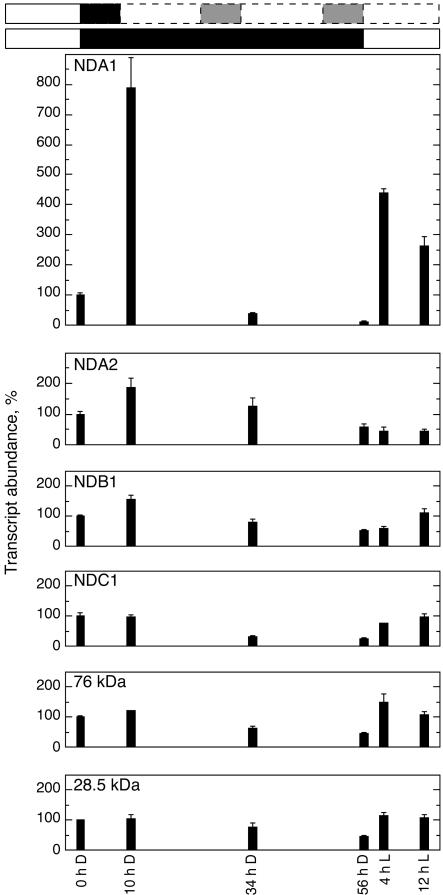
It was found previously that the potato nda1 RNA level is light dependent and has a daily rhythm with a peak in the morning (Svensson and Rasmusson, 2001). The presence of two closely related nda genes in Arabidopsis now raises the question whether any of these are light dependent. We put Arabidopsis plants that had been grown at 16-h-light, 8-h-dark cycles into constant darkness, starting at the end of a light cycle. Leaf samples for RNA isolation were collected after 10 (when the plants normally would have been exposed to light for 2 h), 34, and 56 h, as well as 4 and 12 h after reapplying light and transcript levels determined by real-time RT-PCR (Fig. 7). The NDA1 transcript increased almost 8-fold after 10 h of darkness, an induction that disappeared after 34 and 56 h. Upon reexposure to light, the transcript was again strongly induced. The NDA2 transcript was transiently and less than 2-fold elevated in darkness, with no increase upon reapplication of light. The transcripts for NDB1 and NDC1 showed a profile similar to the 76- and 28.5-kD complex I subunits, displaying an approximately 50% decrease in response to darkness and a return to original level in response to reapplied light. The expression pattern shows that the At-nda1 is specifically controlled by light and may be under circadian regulation, similar to the potato nda1 (Svensson and Rasmusson, 2001). In addition, Arabidopsis contains a light-independent nda2 gene.
Figure 7.
Effects of light and darkness on the expression of type II NAD(P)H dehydrogenase genes. Plants grown with a photoperiod of 16 h of light (7 am–11 pm) of 100 μmol m–2 s–1, were placed in constant darkness at 11 pm for 56 h, followed by light exposure for 12 h. Leaf samples for RNA extraction and concomitant cDNA synthesis were collected at different time points. White and black boxes, Light and dark treatments, respectively. The entrained rhythm is indicated in white and gray boxes with dashed lines. Hours in darkness (D) and light (L) are indicated below for each investigated time point. Error bars = ses for three separate preparation series.
DISCUSSION
Three NAD(P)H Dehydrogenase Protein Families Contain Mitochondrial Proteins
Here, we show the presence of seven nuclear-encoded homologs to type II NAD(P)H dehydrogenases in Arabidopsis that cluster into three gene families. For the nda, ndc1, and two of the four ndb genes, GFP fusion analysis strongly suggests a mitochondrial localization of the respective gene products (Fig. 5). This is consistent with the specific immunodetection of NDA and NDB proteins in the mitochondrial fractions after Suc gradient analysis of a crude mitochondrial fraction, also containing chloroplasts and peroxisomes, from Arabidopsis leaves (Fig. 4). Typically, a crude mitochondrial fraction from any plant tissue contains intact mitochondria and mitochondria with the outer or both membranes disrupted and, thus, depleted in soluble proteins of the enclosed compartments. Broken mitochondria separate on a gradient from intact mitochondria, which are found closer to the bottom. For example, the uppermost part of the mitochondrial peak is depleted in the soluble matrix protein malate dehydrogenase, indicating that the lightest mitochondria have disrupted inner and outer membranes. Immunorecognized NDB protein distributes in the lower one-half of the mitochondrial peak and is also present, more than seen for malate dehydrogenase, at the top of the gradient with small vesicles and soluble proteins. This result suggests that NDB protein but not malate dehydrogenase is released from mitoplast with disrupted outer membranes, indicating an external location of the immunodetectable NDB proteins. The location and release of At-NDB proteins are consistent with observations in potato, where the externally located NDB1 is released from the membrane upon sonication (Rasmusson et al., 1999; Rasmusson and Agius, 2001). Also, external NAD(P)H dehydrogenases have been reported previously to be released in response to osmotic shock and sonication (Douce et al., 1973; Luethy et al., 1995; Menz and Day, 1996). Taken together, the localization results suggest that in Arabidopsis, NDA proteins, NDC1, and at least NDB1 and NDB2 are mitochondrial proteins. It is likely that NDB3 and NDB4 are also mitochondrial, but they appear to be lowly expressed at mRNA level; therefore, the gene products may escape observation by cell fractionation and western blotting.
The nda and ndb gene families are, as expected, most closely related to the NAD(P)H dehydrogenase genes in other mitochondria-containing eukaryotes (Fig. 2). The sequences from yeasts, the plant nda family, and the plant ndb family separate into three different groups. The relationships between these groups are unclear, as is the positioning of N. crassa NDE1 and NDI1 and the protist homologs. An exception is seen in T. brucei, which contains a homolog that partitions with the NDA family at a relatively high bootstrap value. Based on the similar location of the EF hand-carrying domains in potato NDB1 and N. crassa NDE1, it was suggested that these insertions may be descendants of a single evolutionary event, from which the N-terminal EF hand motif is conserved in NDB1 and the C-terminal in NDE1 (Kerscher, 2000). The alignment of the EF hand domains (Fig. 1) reveals a distinct conservation of this protein segment between the plant proteins, indicating that the presence of this domain may be typical for proteins of the NDB family in plants. There is relatively little similarity between plant NDB proteins and N. crassa NDE1, nor to the corresponding segment of P. falciparum and P. yoelli homologs, but in both cases, conservation is seen in the same parts of this domain. Because the phylogenetic tree lends no significant support, it is presently difficult to state a closer relationship between plant ndb genes, N. crassa nde1, and the P. falciparum and P. yoelli homologs.
A Type II NAD(P)H Dehydrogenase Gene of Cyanobacterial Origin
Phylogenetic analyses clearly indicate that the ndc genes in Arabidopsis and rice are most closely related to cyanobacterial genes for type II NAD(P)H dehydrogenases (Fig. 2). This suggests that the ndc genes entered the plant cell via the chloroplast progenitor and later moved to the nucleus (Martin and Herrmann, 1998; Palmer et al., 2000). Genome-wide investigations have revealed several thousands of genes of cyanobacterial origin in the nucleus of Arabidopsis. The majority of them are predicted to encode proteins targeted to cell compartments other than the chloroplast (Martin et al., 2002). It has been reported, for instance, that the mitochondrial RPS13 protein is derived from a chloroplast gene that has been transferred to the nucleus and duplicated. One nuclear gene copy encodes the chloroplast RPS13, and the gene product of the other copy has become exclusively directed to the mitochondrion by acquiring a respective targeting peptide (Adams et al., 2002; Mollier et al., 2002). The exclusive localization of the NDC1 protein in mitochondria (Fig. 5) implies that the gene, after transfer to the nucleus, obtained a targeting sequence specifying mitochondrial import. Based on in silico intracellular prediction, it has been suggested that NDC1, and with lower probability NDA1 and NDA2, may reside in the chloroplast and potentially have a chlororespiratory function (Peltier and Cournac, 2002). However, computer prediction of intracellular localization has been shown previously to be error prone for individual sequences. Of a set of 91 proteins detected in purified Arabidopsis mitochondria, only 50 to 55 were predicted mitochondrial upon analyses with prediction programs (Millar et al., 2001).
Interestingly, ndc1, in addition to encoding a mitochondrial targeting sequence, also shares one common intron position with all but one nda or ndb genes analyzed. It is also possible that the following intron position in At-ndc1 and its rice homolog has an identical counterpart in ndb1 to 3. However, the protein alignment is less clear in this region, with only four conserved residues among the proximal 12, making the exact intron position less certain (results not shown). The identically positioned intron(s) may have been added by gene conversion or independent intron gain. Alternatively, a common intron may be a remnant of an intron present in the common bacterial ancestor, as suggested by the conservation of five intron positions between glyceraldehyde-3-phosphate dehydrogenases of plastid and mitochondrial origin (Kersanach et al., 1994).
It is presently unclear whether cyanobacterial NAD(P)H dehydrogenases have respiratory or photosynthetic functions. Synechocystis sp. strain 6803 contains genes for a complex I-type enzyme and also three reading frames for type II NAD(P)H dehydrogenases, slr0815 (ndbA), slr1743 (ndbB), and sll1484 (ndbC; Kaneko et al., 1996; Howitt et al., 1999). slr1743 especially clusters more closely to the Arabidopsis ndc1 than to the other Synechocystis ndb genes in phylogenetic analysis (Fig. 2). In genome-wide transcript profiling of Synechocystis, slr0815 and slr1743 were light responsive, similar to 25% of the genes in Synechocystis (Gill et al., 2002). All three open reading frames also have been analyzed by mutation in Synechocystis (Howitt et al., 1999). However, the results do not allow clear conclusions about the roles of the individual genes in respiration and photosynthesis. The slr1743 gene was able to complement the MCW232 E. coli NADH dehydrogenase double mutant (Calhoun and Gennis, 1993; Howitt et al., 1999), indicating that the encoded protein can reduce other quinones than plastoquinone, most likely ubiquinone. It is possible that the ndc1 gene serves a respiratory function in Arabidopsis. At-ndc1 is expressed also in heterotrophic tissue and no clear light dependence is seen, indicating that the gene has functions distinct from photosynthesis (Fig. 7).
Consequences of Two nda Genes in Arabidopsis
Analysis of the complete Arabidopsis genome has revealed that a large fraction of genes occur in two or more isoforms and that duplication events followed by extensive reshuffling occurred in the genome (Arabidopsis Initiative, 2000; Blanc et al., 2000). A large section of chromosome 1, containing the nda1 gene, displays synteny to the region of chromosome 2, harboring the nda2 gene (Blanc et al., 2000). The phylogenetic analysis (Fig. 2) and the conserved gene structure, including intron length (Fig. 3), are consistent with a recent duplication being responsible for the presence of two nda genes in Arabidopsis. In such a case, either nda2 has lost the regulation by light, which is a shared characteristic for the nda1 genes in Arabidopsis and potato, or the At-nda1 and St-nda1 acquired light-regulation independently (Fig. 7; Svensson and Rasmusson, 2001).
Based on the high similarity between Arabidopsis NDA1 and NDA2 throughout the reading frame, including the N termini and the recent gene duplication event, it is likely that both have the same enzymatic function and submitochondrial location. The homologous NDA1 in potato is located on the inner surface of the inner mitochondrial membrane, and it was suggested to participate in the oxidation of matrix NADH (Rasmusson et al., 1999). The structure of the second nucleotide-binding motif in potato NDA1 (Rasmusson et al., 1999) is completely conserved in Arabidopsis NDA1 and NDA2 (not shown). The NDA proteins do not contain any segments potentially binding calcium, whereas all investigated NADPH dehydrogenase activities in plant mitochondria are calcium dependent (Møller, 2001). Expression of the potato nda1 gene in response to light and cold is also consistent with a role for the NDA1 protein as an internal NADH dehydrogenase (Svensson and Rasmusson, 2001; Svensson et al., 2002). The difficulties in isolating intact mitochondria from genetic model plants and estimating oxidation of matrix NADH and/or NADPH in isolated mitochondria has, however, so far hampered an absolute assignment of functions to the gene products. This investigation now shows that ndc1 also has to be considered when assigning functions to gene products.
The regulation by light and the diurnal rhythm of the potato nda1 gene expression indicated a role of the encoded enzyme in photosynthetically associated metabolism, most likely in reoxidation of NADH formed by photorespiratory Gly oxidation (Svensson and Rasmusson, 2001). The presence of a light-regulated nda1 in Arabidopsis (Fig. 7) suggests that this is a general phenomenon in plants. The importance of mitochondrial NADH oxidation for optimal photosynthesis has been demonstrated recently in Nicotiana sylvestris mutants, whereby lack of complex I activity disturbed photosynthetic function, most likely via changes in cellular NAD(P)H redox levels (Dutilleul et al., 2003).
The presence of the light-independent nda2 in Arabidopsis indicates that the NDA proteins also may have functions in heterotrophic metabolism, e.g. in darkness or in the root, and that NDA2 has a preferential role in these processes. Analogously, the three alternative oxidase genes in soybean (Glycine max) are independently affected by development, light, and stress (Finnegan et al., 1997; Djajanegara et al., 2002), and a similar differential regulation is likely for the five aox genes in Arabidopsis. Thus, depending on the combinations of alternative NAD(P)H dehydrogenases and oxidases in a cell type, the respiratory chain as a whole may have different properties, adjusting the electron transport to the dominating respiratory processes.
CONCLUSIONS
In the present study, Arabidopsis is shown to contain seven type II NAD(P)H dehydrogenase genes that group into three families, present also in rice. Representative proteins from all three families were shown to be mitochondrial. A family of two genes, similar to potato nda1, was found to contain a light-dependent (nda1) and -independent (nda2) homolog, as determined by real-time RT-PCR. A family closely related to potato ndb1 is present with four expressed homologs (ndb1–4) in Arabidopsis. A homolog of a novel type, ndc1, has a cyanobacterial origin, indicating functional transfer of the protein from the chloroplast to the mitochondria.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia) were grown on soil in climate chambers at 25°C using 100 μmol m–2 s–1 light for 16 h d–1.
Subcellular Fractionation Analysis
Mitochondria were isolated from 100-g leaves of 40-d-old Arabidopsis plants using Suc gradient separation of mitochondria and chloroplasts (Leaver et al., 1983). Two-milliliter fractions were collected from the Suc gradient. Aliquots of identical volume were analyzed by one-dimensional SDS-PAGE (Laemmli, 1970), and proteins were electrotransferred to nitrocellulose membranes (Moos et al., 1988). Immunostaining was performed with antibodies raised against conserved peptides of NDA1 and NDB1 from potato (Solanum tuberosum; Rasmusson and Agius, 2001), the NAD9 subunit of wheat (Triticum aestivum) complex I (Lamattina et al., 1993), and malate dehydrogenase (Gietl et al., 1996) using ECL detection (Amersham, Buck-Inghamshire, UK). Cytochrome c oxidase (EC 1.9.3.1) activity (Rasmusson and Møller, 1990) and catalase (EC 1.11.1.6) activity (Lidén and Møller, 1988) was determined by published methods. Chlorophyll was determined by acetone extraction (Arnon, 1949).
GFP Fusion Construction
Total flower RNA isolation and cDNA synthesis were carried out similarly as described previously (Svensson et al., 2002). Primers adapted for BamHI restriction were designed to amplify sequences coding for the N-terminal domains of each investigated type II NAD(P)H dehydrogenase genes: At-NDA1, 5′-CGG GAT CCT TGG AGT ATT ATT TGC TTC TCT CG-3′ and 5′-CGG GAT CCT CGG CTT CTC TCC CTC TTT CG-3′; At-NDA2, 5′-CGG GAT CCG AAG AAG AAG CGA TGT TCA-3′ and 5′-CGG GAT CCC CGA CCC GAG AAC CAC TAC TC-3′; At-NDB1, 5′-CGG GAT CCT CTG AAG ACT TTT CAC CAA A-3′ and 5′-CGG GAT CCA ACC AGT CCC AAG CAC AAC T-3′; At-NDB2, 5′-CGG GAT CCT CGA ACC AAG AGC GAA AAT G-3′ and 5′-CGG GAT CCT CTT CTT GGT TCC AGT TTC AAC A-3′; and At-NDC1, 5′-CGG GAT CCA GCA ATG GCC GTT CTC TC-3′ and 5′-CGG GAT CCA CAC CCT TGG CCT CTT GTT A-3′. Amplification was made with the Advantage HF2 proof-reading PCR kit (CLON-TECH Laboratories, Palo Alto, CA), using an initial cycle of 60 s at 94°C, 20 s at 63°C. and 60 s at 68°C, and then 29 cycles of 30 s at 94°C, 30 s at 63°C, and 60 s at 68°C. Products were cleaved with BamHI and ligated into psmGFP4, upstream of the smGFP4 reading frame (Davis and Vierstra, 1998). Clones were sequenced for confirmation of the insert using the Thermo Sequenase primer cycle sequencing kit (Amersham).
Protoplast Transformation and Fluorescent Microscopy
Approximately 500,000 protoplasts isolated from tobacco (Nicotiana tabacum cv petit Havana) were transiently transformed with 50 μg of construct DNA using a polyethylene glycol method (Koop et al., 1996). Protoplasts were subsequently incubated with MitoTracker Red according to the manufacturer's protocol (Molecular Probes, Eugene, OR). Fluorescence microscopy was performed using an Axioplan microscope (Carl Zeiss, Oberkochen, Germany). Axiovision software (Carl Zeiss) was used for taking images. Fluorescence filter sets from AHF Analystechnik (Tübingen, Germany) were HQ470/40/HQ500LP for GFP and chlorophyll autofluorescence and HQ545/30/HQ610/75 for MitoTracker Red.
Transcript Analysis
Isolation of total RNA, cDNA synthesis, and real-time PCR were carried out as in Svensson et al. (2002) except for variations in annealing temperatures (see below). Primers were designed so that one primer in each pair spanned an exon border. Control amplifications without RT reaction did not detect genomic signals.
The primer sequences used were (annealing temperatures inside parentheses): At-NDA1 (60°C), 5′-CTC CGT GAG AGC AAG GAA GG-3′ and 5′-GGC GAA GTG GAG GGG ATA TG-3′; At-NDA2 (68°C), 5′-CGA GAG CAA GGA CGC AAA AG-3′ and 5′-CAG TAG GCC GAG ATT GAG AC-3′; At-NDB1 (68°C), 5′-TGC CTG CAA CTG CTC AGG TC-3′ and 5′-GAT GCC CGC CAG TTC TGA AG-3′; At-NDB2 (60°C), 5′-TAC GCC AGT AAG CAA GTG AG-3′ and 5′-TGT GTA TTG GAG CCT TGG AG-3′; At-NDB3 (55°C), 5′-GGT GAG TAG CCA AAG ACG TG and 5′-GAA GAT CGG TAA TGC CAT GC-3′; At-NDB4 (55°C), 5′-CCA ACG CAG AGT CAT GGA AG-3′ and 5′-CAC CAT TTG CAC TCT TGA GC-3′; At-NDC1 (60°C), 5′-CAA TGG CCG TTC TCT CCT C-3′ and 5′-ACA CCC TTG GCC TCT TGT TA-3′; 28.5kDa (58°C), 5′-AAC CGA GAC ACA YGA GGA AC-3′ and 5′-TAG AGR CTC TCC GAC CTC AG-3′; and 76 kD (62°C), 5′-ACA AGG TGT GTA CGA TTT GC-3′ and 5′-TTT GAG GTC AAG GCT CCA AC-3′.
Miscellaneous
General molecular biology methods were performed according to Sambrook et al. (1989). Database searches were mainly performed at the National Center of Biotechnology Information using BLASTP and TBLASTN algorithms (Altschul et al., 1997) for the nr and est Arabidopsis database sets. Pair-wise alignments were performed using LAlign software (Huang and Miller, 1991).
Acknowledgments
We are grateful to Drs. Jean-Michel Grienenberger (IBMP-CNRS, Strasbourg, France) and Christine Gietl (Technische Universität München, Germany) for generous donations of antibodies and Bärbel Weber (Universität Ulm, Germany) for excellent technical assistance.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.024208.
This work was supported by the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (to A.G.R. and U.J.), by Carl Tryggers Stiftelse (to A.G.R.), by Carl Tesdorpfs Stiftelse (to A.G.R.), by Kungliga Fysiografiska Sällskapet i Lund (to A.G.R.), by Erik Philip-Sörensens Stiftelse (to U.J.), and by the Deutsche Forschungsgemeinschaft und Fonds der Chemischen Industrie (to A.B. and S.B.)
References
- Adams KL, Daley DO, Whelan J, Palmer JD (2002) Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14: 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun MW, Gennis RB (1993) Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol 175: 3013–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- de Vries S, Grivell LA (1988) Purification and characterization of a rotenone-insensitive NADH - Q6 oxidoreductase from mitochondria of Saccharomyces cerevisiae. Eur J Biochem 176: 377–384 [DOI] [PubMed] [Google Scholar]
- de Vries S, Van Witzenburg R, Grivell LA, Marres CAM (1992) Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem 203: 587–592 [DOI] [PubMed] [Google Scholar]
- Djajanegara I, Finnegan PM, Mathieu C, McCabe T, Whelan J, Day DA (2002) Regulation of alternative oxidase gene expression in soybean. Plant Mol Biol 50: 735–742 [DOI] [PubMed] [Google Scholar]
- Douce R, Mannella CA, Bonner WD (1973) External NADH dehydrogenases of intact plant mitochondria. Biochim Biophys Acta 292: 105–116 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Whelan J, Millar AH, Zhang QS, Smith MK, Wiskich JT, Day DA (1997) Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol 114: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Steinmuller K, Weiss H (1995) The proton-pumping respiratory complex I of bacteria and mitochondria and its homolog in chloroplasts. FEBS Lett 367: 107–111 [DOI] [PubMed] [Google Scholar]
- Gietl C, Seidel C, Svendsen I (1996) Plant glyoxysomal but not mitochondrial malate dehydrogenase can fold without chaperone assistance. Biochim Biophys Acta 1274: 48–58 [DOI] [PubMed] [Google Scholar]
- Gill RT, Katsoulakis E, Schmitt W, Taroncher-Oldenburg G, Misra J, Stephanopoulos G (2002) Genome-wide dynamic transcriptional profiling of the light-to-dark transition in Synechocystis sp. strain PCC 6803. J Bacteriol 184: 3671–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt CA, Udall PK, Vermaas WF (1999) Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. strain PCC 6803 are involved in regulation rather than respiration. J Bacteriol 181: 3994–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XQ, Miller W (1991) A time-efficient, linear-space local similarity algorithm. Adv Appl Math 12: 337–357 [Google Scholar]
- Jaworowski A, Campbell HD, Poulis MI, Young IG (1981) Genetic identification and purification of the respiratory NADH dehydrogenase of Escherichia coli. Biochemistry 20: 2041–2047 [DOI] [PubMed] [Google Scholar]
- Joseph-Horne T, Hollomon DW, Wood PM (2001) Fungal respiration: a fusion of standard and alternative components. Biochim Biophys Acta 1504: 179–195 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803: II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3: 109–136 [DOI] [PubMed] [Google Scholar]
- Kersanach R, Brinkmann H, Liaud MF, Zhang DX, Martin W, Cerff R (1994) Five identical intron positions in ancient duplicated genes of eubacterial origin. Nature 367: 387–389 [DOI] [PubMed] [Google Scholar]
- Kerscher SJ (2000) Diversity and origin of alternative NADH: ubiquinone oxidoreductases. Biochim Biophys Acta 1459: 274–283 [DOI] [PubMed] [Google Scholar]
- Koop HU, Steinmuller K, Wagner H, Rossler C, Eibl C, Sacher L (1996) Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199: 193–201 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM (1993) Higherplant mitochondria encode an homolog of the nuclear-encoded 30-kda subunit of bovine mitochondrial complex I. Eur J Biochem 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Leaver CJ, Hack E, Forde BG (1983) Protein synthesis by isolated plant mitochondria. Methods Enzymol 97: 476–484 [DOI] [PubMed] [Google Scholar]
- Lidén AC, Møller IM (1988) Purification, characterization and storage of mitochondria from Jerusalem artichoke tubers. Physiol Plant 72: 265–270 [Google Scholar]
- Luethy MH, Thelen JJ, Knudten AF, Elthon TE (1995) Purification, characterization, and submitochondrial localization of a 58-kilodalton NAD(P)H dehydrogenase. Plant Physiol 107: 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MAH, Overkamp KM, Kotter P, de Vries S, van Dijken JP, Pronk JT (1998) The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem 273: 24529–24534 [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol 118: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo AMP, Duarte M, Møller IM, Prokisch H, Dolan PL, Pinto L, Nelson MA, Videira A (2001) The external calcium-dependent NADPH dehydrogenase from Neurospora crassa mitochondria. J Biol Chem 276: 3947–3951 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Duarte M, Videira A (1999) Primary structure and characterisation of a 64 kDa NADH dehydrogenase from the inner membrane of Neurospora crassa mitochondria. Biochim Biophys Acta 1412: 282–287 [DOI] [PubMed] [Google Scholar]
- Melo AMP, Roberts TH, Møller IM (1996) Evidence for the presence of two rotenone-insensitive NAD(P)H dehydrogenases on the inner surface of the inner membrane of potato tuber mitochondria. Biochim Biophys Acta 1276: 133–139 [Google Scholar]
- Menz RI, Day DA (1996) Identification and characterization of an inducible NAD(P)H dehydrogenase from red beetroot mitochondria. Plant Physiol 112: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Mollier P, Hoffmann B, Debast C, Small I (2002) The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr Genet 40: 405–409 [DOI] [PubMed] [Google Scholar]
- Moos M, Nguyen NY, Liu TY (1988) Reproducible high-yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem 263: 6005–6008 [PubMed] [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Møller IM, Rasmusson AG (1998) The role of NADP in the mitochondrial matrix. Trends Plant Sci 3: 21–27 [Google Scholar]
- Palmer JD, Adams KL, Cho YR, Parkinson CL, Qiu YL, Song KM (2000) Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci USA 97: 6960–6966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G, Cournac L (2002) Chlororespiration. Annu Rev Plant Biol 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Agius SC (2001) Rotenone-insensitive NAD(P)H dehydrogenases in plants: immunodetection and distribution of native proteins in mitochondria. Plant Physiol Biochem 39: 1057–1066 [Google Scholar]
- Rasmusson AG, Heiser V, Zabaleta E, Brennicke A, Grohmann L (1998) Physiological, biochemical and molecular aspects of mitochondrial complex I in plants. Biochim Biophys Acta 1364: 101–111 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM (1990) NADP-utilizing enzymes in the matrix of plant mitochondria. Plant Physiol 94: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Møller IM (1991) NAD(P)H dehydrogenases on the inner surface of the inner mitochondrial membrane studied using inside-out submitochondrial particles. Physiol Plant 83: 357–365 [Google Scholar]
- Rasmusson AG, Svensson AS, Knoop V, Grohmann L, Brennicke A (1999) Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J 20: 79–87 [DOI] [PubMed] [Google Scholar]
- Roberts TH, Fredlund KM, Møller IM (1995) Direct evidence for the presence of 2 external NAD(P)H dehydrogenases coupled to the electron transport chain in plant mitochondria. FEBS Lett 373: 307–309 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Siedow JN, Umbach AL (1995) Plant mitochondrial electron transfer and molecular biology. Plant Cell 7: 821–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small WC, McAlister-Henn L (1998) Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J Bacteriol 180: 4051–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson AS, Johansson FI, Møller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517: 79–82 [DOI] [PubMed] [Google Scholar]
- Svensson AS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73–82 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP: Phylogenetic Analysis Using Parsimony, Version 4. Sinauer Associates, Sunderland, MA
- Yagi T (1991) Bacterial NADH-quinone oxidoreductases. J Bioenerg Biomembr 23: 211–225 [DOI] [PubMed] [Google Scholar]
- Young IG, Rogers BL, Campbell HD, Jaworowski A, Shaw DC (1981) Nucleotide sequence coding for the respiratory NADH dehydrogenase of Escherichia coli: UUG initiation codon. Eur J Biochem 116: 165–170 [DOI] [PubMed] [Google Scholar]