Abstract
We utilized a proteomic approach to investigate seed development in Medicago truncatula, cv Jemalong, line J5 at specific stages of seed filling corresponding to the acquisition of germination capacity and protein deposition. One hundred twenty proteins differing in kinetics of appearance were subjected to matrix-assisted laser desorption ionization time of flight mass spectrometry. These analyses provided peptide mass fingerprint data that identified 84 of them. Some of these proteins had previously been shown to accumulate during seed development in legumes (e.g. legumins, vicilins, convicilins, and lipoxygenases), confirming the validity of M. truncatula as a model for analysis of legume seed filling. The study also revealed proteins presumably involved in cell division during embryogenesis (β-tubulin and annexin). Their abundance decreased before the accumulation of the major storage protein families, which itself occurs in a specific temporal order: vicilins (14 d after pollination [DAP]), legumins (16 DAP), and convicilins (18 DAP). Furthermore, the study showed an accumulation of enzymes of carbon metabolism (e.g. sucrose synthase, starch synthase) and of proteins involved in embryonic photosynthesis (e.g. chlorophyll a/b binding), which may play a role in providing cofactors for protein/lipid synthesis or for CO2 refixation during seed filling. Correlated with the reserve deposition phase was the accumulation of proteins associated with cell expansion (actin 7 and reversibly glycosylated polypeptide) and of components of the precursor accumulating vesicles, which give rise to a trypsin inhibitor on maturation. Finally, we revealed a differential accumulation of enzymes involved in methionine metabolism (S-adenosyl-methionine synthetase and S-adenosylhomo-cysteine hydrolase) and propose a role for these enzymes in the transition from a highly active to a quiescent state during seed development.
The development of the angiosperm seed proceeds through histodifferentiation and seed filling and terminates with a desiccation phase after which the embryo enters into a quiescent state, thereby permitting its storage and survival in various environmental conditions (Bewley and Black, 1994). The storage compounds found in most mature seeds accumulate during seed filling. They are principally storage proteins, oil (often triacylglycerols) and carbohydrates (often starch; Baud et al., 2002). These reserves are of major importance for two reasons: (a) They support early seedling growth when degraded upon germination and, therefore, participate in crop establishment; and (b) they are widely used for human and animal nutrition. Seeds of legume species, such as soybean (Glycine max), pea (Pisum sativum), and fava bean (Vicia faba), are an important protein source, with 20% to as much as 40% protein content, depending on species, genotype, and environment. In contrast, seeds of graminaceous species, such as maize (Zea mays) and wheat (Triticum aestivum), are a major source of starch and contain less than 16% protein content. However, the major proteins stored in legume seeds are poor in sulfur containing amino acids and the presence of nutritionally undesirable compounds, such as protease inhibitors, remain limiting factors. Because of its nutritional and economic importance, much effort by plant breeders is directed toward the improvement of seed quality, and both plant breeding and molecular technologies can be used to produce plants carrying the desired traits (Mazur et al., 1999). Therefore, there is strong interest in identifying the processes occurring during seed filling and the proteins involved.
The use of the most agriculturally important legume crops to study legume biology is limited by the large size of their genome and the complex ploidy. Unlike the major crop legumes, Medicago truncatula is diploid, has a small genome size (approximately 500 Mb), and is currently the subject of major genomic initiatives. To date, more than 180,000 M. truncatula expressed sequence tag (EST) sequences are available in public databases, and a sequencing project for the entire genome is underway (Bell et al., 2001). This annual relative of alfalfa (Medicago sativa) produces seeds rich in proteins (35%–45%) and oil (approximately 12%) but low in starch (<1%) at maturity (G. Duc, personal communication). Moreover, because M. truncatula is phylogenetically related to the major legume crops, its use provides the potential to transfer information into crop improvement.
To take advantage of the available genomic resources of M. truncatula, we have characterized seed development in this species at the level of its protein complement. Proteomics offers the opportunity to examine simultaneous changes in, and to classify temporal patterns of, protein accumulation occurring in complex developmental processes such as seed filling (Bove et al., 2002). This approach was recently applied to study the changes in proteins that occur during development of starch-rich grains (Finnie et al., 2002). More recently, Watson et al. (2003) performed a survey of the organ-/tissue-specific proteomes of M. truncatula. However, to date, there has been no proteomic or transcriptomic project to study protein or gene expression profiles during seed filling in a legume species.
Here, we provide a framework of physiological data relevant for M. truncatula seed development and report the identification by mass spectrometry (MS) of many seed proteins. This study has not only cataloged proteins but has also described their accumulation patterns at specific stages during seed development, before and during protein deposition. These findings contribute to our understanding of how metabolic networks are regulated at the protein level during reserve deposition in seeds of a legume species. This knowledge will support our attempts to engineer legume seed composition for added end user value.
RESULTS
Physiology of M. truncatula Seed Development
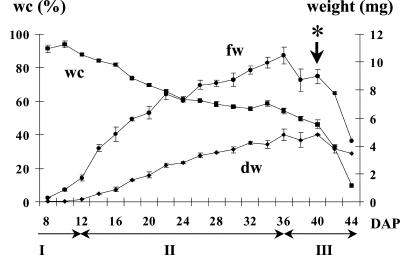
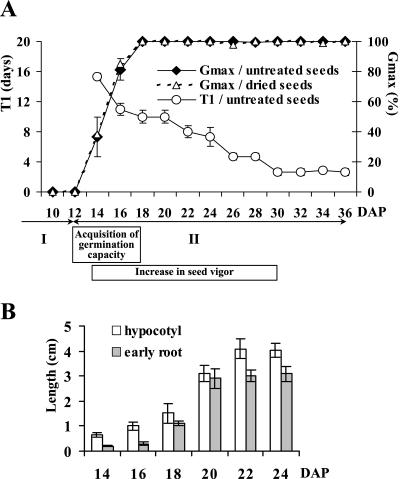
To provide a framework for the proteomic study of seed filling, a series of stages of seed development, from embryogenesis to seed dispersal, were defined. Three phases were characterized by distinct physiological events and the associated changes in seed dry weight and moisture status. The first phase, corresponding to stages preceding 12 d after pollination (DAP), was characterized by a water content of about 90% of the seed fresh weight (Fig. 1). During this phase, which corresponds to histodifferentiation or embryogenesis (Bewley and Black, 1994), the whole seeds removed from pods were not able to germinate on water (Fig. 2A), indicating that internal seed features permitting germination are not yet developed, or that constraints imposed by the tissue surrounding the embryo prevent germination.
Figure 1.
Characterization of the different phases of M. truncatula (line J5) seed development. This graph represents changes in whole-seed fresh weight (fw), dry weight (dw), and water content (wc) from 8 to 44 DAP. The three following phases are indicated: I, histodifferentiation; II, seed filling; and III, desiccation. Data are the mean ± sd of three replicates of 15 seeds. Asterisk, Pod abscission. The accumulation kinetics of the storage compounds during M. truncatula seed development were very similar to those previously described for Arabidopsis seeds (Baud et al., 2002). Early stages of seed development were characterized by a transient accumulation of starch; the synthesis of both proteins and triacylglycerols occurred during seed filling (to a final concentration of about 40% and 10%, respectively), and soluble sugars belonging to the raffinose family accumulated to a final concentration of about 10% during the last phase of seed development (J.-P. Boutin, personal communication, unpublished data).
Figure 2.
Viability, vigor, and dehydration tolerance of M. truncatula seeds collected during development. A, Seed germination. T1, Start of germination (time to reach 1% of germination ± sd) for seeds freshly harvested; Gmax, final percentage of germination obtained for fresh and dried seeds. The arrows indicate histodifferentiation (I) and seed filling (II). B, Hypocotyl elongation (length between cotyledons and cotyledonary node) and early root development (length between cotyledonary node and root apex) of seedlings after germination of untreated seeds collected at defined developmental stages and grown for the same time period (20 d) on water. Data are the mean ± sd of three replicates of 15 seeds.
The second phase was associated with a large increase in the seed dry matter from 12 to 36 DAP (Fig. 1) and characterized by the acquisition of the ability to germinate (Fig. 2A). The time from imbibition to germination (T1 in Fig. 2A) declined gradually from about 16 d at 14 DAP to about 3 d at 30 DAP, indicating that seed vigor clearly increased during this phase. Similarly, seedling vigor, expressed as hypocotyl elongation and early root development, increased from 14 to 22 DAP (Fig. 2B). These data indicate that physiological and biochemical features of M. truncatula seeds, which allow vigorous germination and subsequent growth, are established during seed filling. Germination occurred without removing the seed coat, indicating that in M. truncatula the surrounding testa and endosperm are not constraints on germination in contrast to many species where these structures must be removed to allow the immature embryo to germinate (Bewley and Black, 1994).
The terminal phase of M. truncatula seed development was characterized by a decrease in seed fresh weight and a drastic loss of water as the seed undergoes drying (Fig. 1). This loss of water may play a role in the switch in cellular activities from a seed formation-oriented program to an exclusively germination/growth-oriented program (Kermode et al., 1986). Figure 2A shows that the onset of desiccation tolerance is earlier than the drying phase. This is consistent with previous studies showing a similar rapid acquisition of desiccation tolerance in developing seeds of several species, such as castor bean (Ricinus communis) or rapeseed (Brassica napus; Bewley and Black, 1994).
Strategy Adopted for the Proteomic Analysis of Seed Filling
We characterized the proteome of seeds harvested at five stages during seed filling: seeds with low dry weight and unable to germinate (12 DAP) and seeds increasing in dry weight and developing the ability to germinate (14, 16, 18, and 20 DAP; Figs. 1 and 2A). The protein samples were analyzed by two-dimensional (2-D) gel electrophoresis. Seed filling was accompanied by an increase in the number of protein spots detected in Coomassie Blue gels up to 18 DAP (172 ± 1 spots at 12 DAP, 252 ± 3 at 18 DAP, and 245 ± 3 at 20 DAP). In the 2-D gels, the abundance of the protein spots corresponded to their volumes, which were determined by the ImageMaster 2-D Elite software as described in “Materials and Methods.” The volume varies as a function of both the area and the densitometry of the detected protein spot. Therefore, spot volume is the total intensity of a defined spot and corresponds to the amount of proteins in that spot. We conducted some experiments consisting in loading five different amounts of the total seed protein extracts (from 2–45 μL) in 2-D gels. The results indicated that for approximately 85% of the studied spots, there was a strong linear relationship (0.90 ≤ R2 ≤ 0.99) between spot volume and the total amount of proteins loaded into the Coomassie Blue gels. For the remaining 15% of spots, the relationship was slightly less linear (R2 approximately 0.85). To discard experimental variations in 2-D gels between the different stages, the volume of each spot was normalized to the total volume of 20 spots, which were chosen as references because they did not show any qualitative variation in silver- and Coomassie Blue-stained 2-D gels during all developmental stages analyzed.
The ANOVA of the relative abundance of the 274 different spots detected over the five stages permitted a classification according to their accumulation patterns (Table I). Among these polypeptides, 90 belong to class 0 proteins, that is, spots whose level did not significantly vary from 12 to 20 DAP. Class 1 and 2 proteins were represented by spots whose abundance significantly increased or decreased, respectively, during the 12- to 20-DAP period. Class 3 and 4 proteins corresponded to spots showing a transient increase or decrease, respectively, in their abundance, and class 5 proteins were represented by spots showing varying changes in their levels during this period. The protein spots were further separated in two categories: the highly abundant polypeptides (normalized volumes ranging from 10,000–150,000), and the less abundant polypeptides (normalized volumes ranging from 21–10,000).
Table I.
Types of variation in the level of expression of the 274 individual spots reproducibly detected in 2-D gels between 12 and 20 DAP
| Proteins with NV > 10,000
|
Proteins with NV < 10,000
|
||||
|---|---|---|---|---|---|
| Class | Characteristics | No. of proteins detected | No. of proteins identified | No. of proteins detected | No. of proteins identified |
| 0 | Constant level | 0 | - | 90 | 0 |
| 1 | Increased level | 32 | 11 | 61 | 32 |
| 2 | Decreased level | 0 | - | 25 | 13 |
| 3 | Transiently increased level | 0 | - | 59 | 23 |
| 4 | Transiently decreased level | 0 | - | 6 | 4 |
| 5 | Varying level | 0 | - | 1 | 1 |
| Total | 32 | 11 | 242 | 73 | |
NV, Normalized spot volume obtained from densitometric analysis of individual spots.
Kinetics of Storage Protein Accumulation
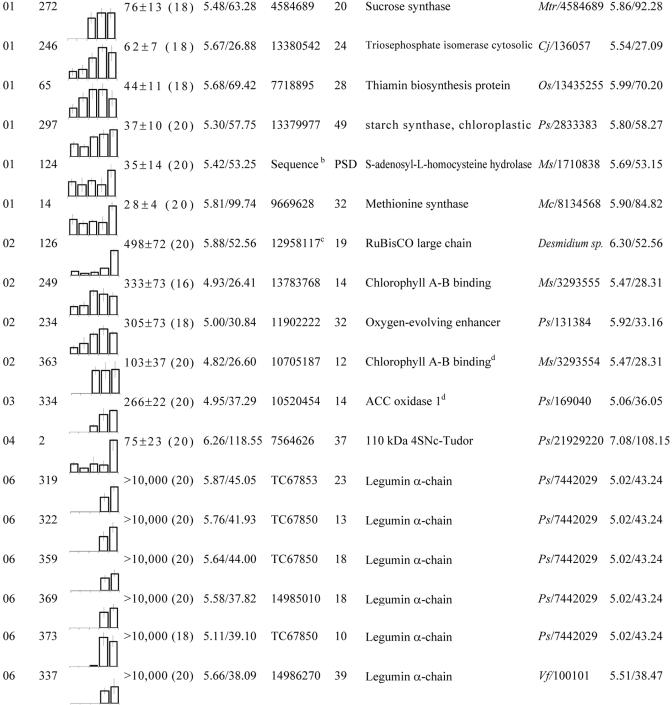
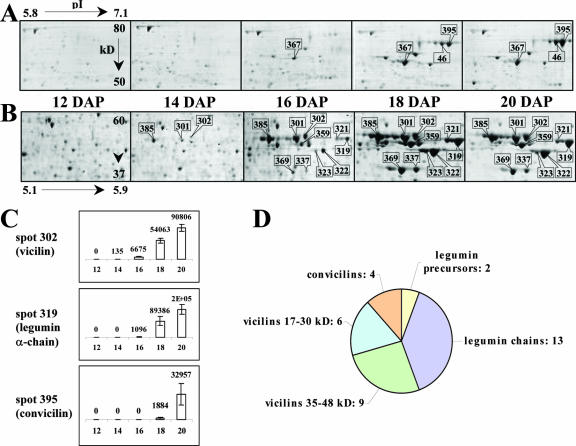
All of the highly abundant polypeptides belong to class 1 of proteins, whose abundance increased during the 12- to 20-DAP period (Table I). Only one abundant protein spot detected in extracts from dry mature seeds (stage 44 DAP in Fig. 1) accumulated after the 20-DAP stage (data not shown), indicating that storage protein deposition is an early event in M. truncatula seed formation. Eleven well-resolved spots were analyzed by matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) MS, revealing their identity as members of the major storage protein families: the legumins, vicilins, and convicilins (group 06 in Table II; species terminology medicagins, alfins, and conalfins not used further here for ease of comparison). Each of these protein families appeared at different stages in seed development. Although the vicilin spots were detected in 2-D gels at 14 DAP, the legumin spots first appeared at 16 DAP, and the convicilin spots were not detected until 18 DAP (Fig. 3). In pea, vicilins have also been shown to appear earlier than the other globulins, but convicilins started to accumulate before legumins (Wenzel et al., 1993). Our experimental conditions did not reveal abundant proteins corresponding to the 2S albumin family found in pea and alfalfa seeds (Higgins et al., 1986; Tabe et al., 1995).
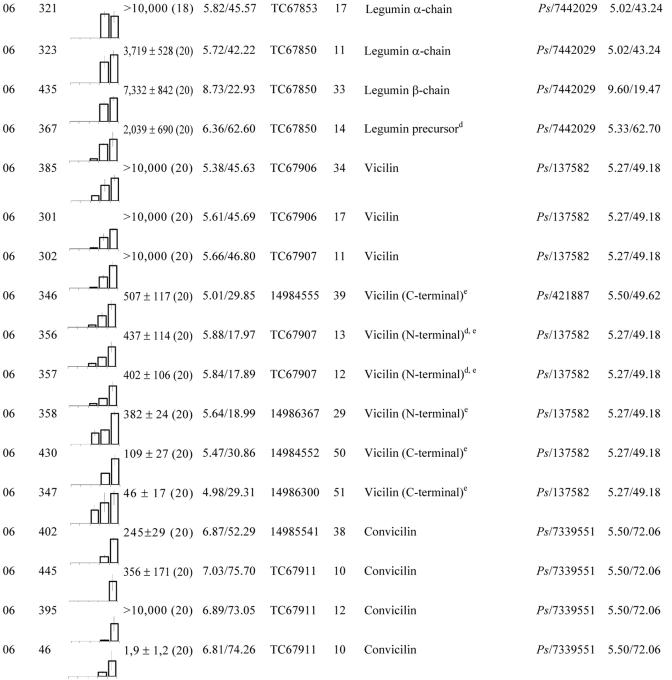
Table II.
M. truncatula polypeptides whose levels increased (class 1 proteins in Table I) between 12 and 20 DAP
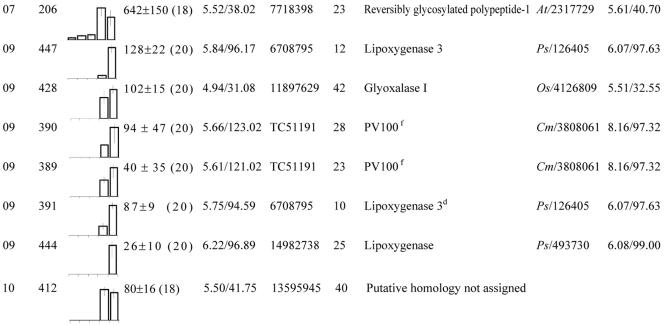
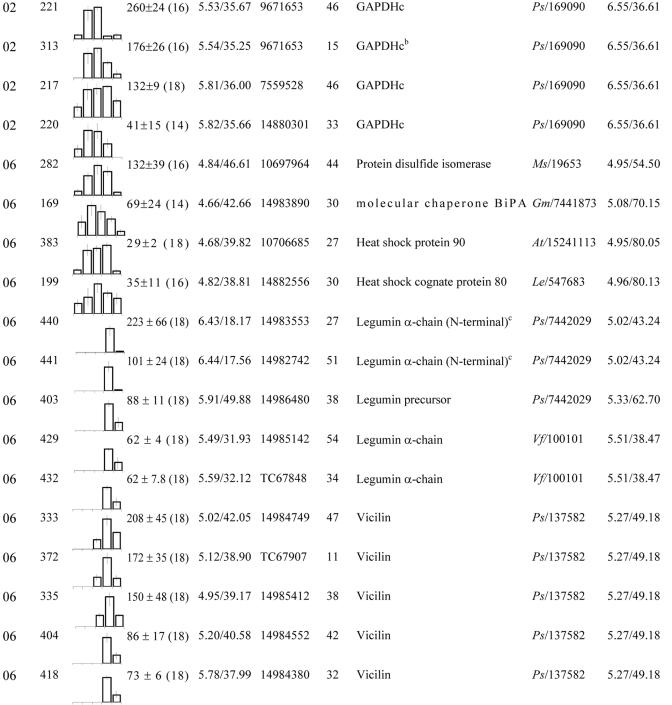
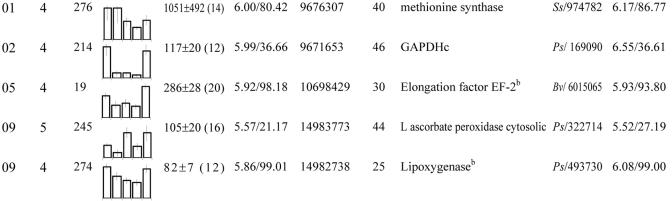
These proteins were grouped according to their biological function (see Fig. 4C). Within each group, spots were classified according to their maximum level detected in 2-D gels between 12 and 20 DAP. No., Spot no. gi, GenBank accession no. TC, Identifier of consensus sequences retrieved from The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/tgi/mtgi/). Cov, Coverage. At, Arabidopsis. Cj, Coptis japonica. Cm, Pumpkin. Mc, Mesembryanthemum crystallinum. Ms, Alfalfa. Mtr, M. truncatula. Os, Rice (Oryza sativa). Ps, Pea. Vf, Fava bean.
Figure 3.
Kinetics of the accumulation of some storage proteins in seeds harvested 12, 14, 16, 18, and 20 DAP. A and B, Two portions of Coomassie Blue gels show protein spots belonging to class 1 (Table II) and identified by mass spectrometry as being legumin precursors (367 in A), α-subunits of legumins (319, 321, 322, 323, 337, 359, and 369 in B), vicilins (301, 302, and 385 in B), and convicilins (46 and 395 in A). Range in pI and Mr are indicated in the panels for 12-DAP seeds. C, Example of changes in the normalized volume of some spots (302, 319, and 395) between 12 and 20 DAP. D, Proportions of the storage protein families identified by mass spectrometry.
Identification of Less Abundant Proteins
Many of the 242 individual less abundant proteins (normalized volumes between 21 and 10,000) showed constant level, increased, or transiently increased between 12 and 20 DAP (class 0, class 1, and class 3 proteins in Table I). Some proteins showed decreased levels (class 2 proteins in Table I), and a few spots transiently decreased (class 4 proteins in Table I) or showed varying changes in their levels (class 5 proteins in Table I). Because the main objective of this study was to reveal molecular and metabolic processes, which could play a role specifically during the phase of protein deposition, we identified by MS, in parallel to abundant seed storage proteins, those less abundant proteins whose abundance varied between 12 (stage preceding storage protein accumulation) and 20 (stage after protein deposition) DAP. One hundred nine polypeptides of low abundance, taken from the protein classes 1 to 5 and well resolved, were analyzed by MALDI-TOF MS, and 73 proteins were successfully identified (Tables II, III, IV, V). Among these proteins, 23 corresponded to minor legumin chains or vicilins, many of which accumulated at 16 to 18 DAP and disappeared after this stage (Table IV). Some of these may correspond to unstable forms of storage proteins not yet fully processed or to proteins that were either misdirected or misfolded and degraded. Some could also correspond to fragments released by specific proteolysis of the storage protein, as observed before Arabidopsis seed germination (Gallardo et al., 2001).
Table III.
M. truncatula polypeptides whose levels decreased (class 2 proteins in Table I) between 12 and 20 DAP
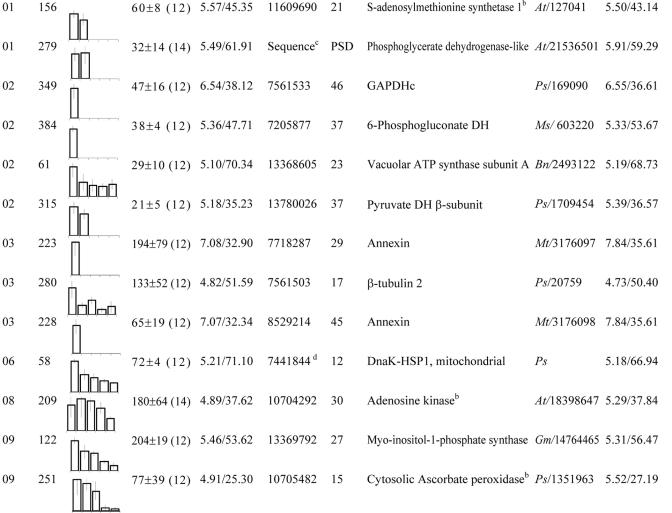
These proteins were grouped according to their biological function (see Fig. 4C). Within each group, spots were classified according to their maximum level detected in 2-D gels between 12 and 20 DAP. No., Spot no. DH, Dehydrogenase. gi, GenBank accession no. Cov, Coverage. At, Arabidopsis. Bn, rapeseed. Gm, soybean. Mtr, M. truncatula. Ms, Alfalfa. Os, Rice. Ps, Pea.
Table IV.
M. truncatula polypeptides whose levels transiently increased (class 3 proteins in Table I) between 12 and 20 DAP
These protein spots were grouped according to their biological function (see Fig. 4C). Within each group, spots were classified according to their maximum level detected in 2-D gels between 12 and 20 DAP. No., Spot no. gi, GenBank accession no. TC, Identifier of consensus sequences retrieved in TIGR (http://www.tigr.org/tdb/tgi/mtgi/). Cov, Coverage. At, Arabidopsis. Le, Lycopersicon esculentum. Md, Malus domestica. Ms, M. sativa. Nt, Tobacco. Ps, Pea. Vf, Fava bean.
Table V.
M. truncatula polypeptides whose levels transiently decreased (class 4 proteins in Table II) or showed various changes (class 5 proteins in Table II) between 12 and 20 DAP
These protein spots were grouped according to their biological function (see Fig. 4C). Within each group, spots were classified according to their maximum level detected in 2-D gels between 12 and 20 DAP. No., Spot no. gi, GenBank accession no. Cov, Coverage. Bv, Beta vulgaris. Ps, Pea. Ss, Solenostemon scutellarioides.
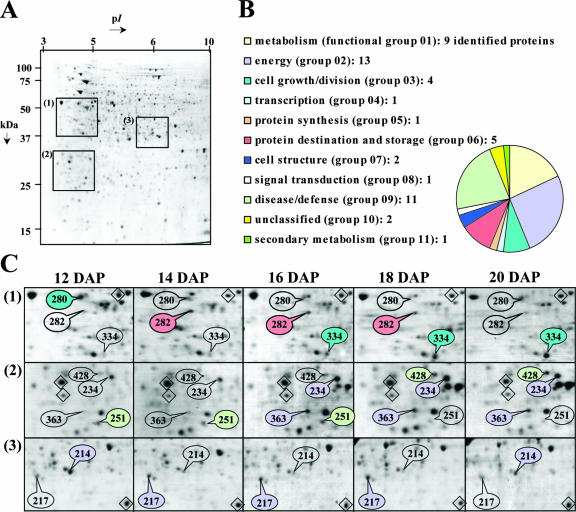
The 50 other less abundant proteins identified by MS were classified in different groups corresponding to their presumed biological function. Figure 4 shows that many of them are presumed to be involved in energy, disease/defense, metabolism, protein destination and storage, cell growth/division, and cell structure. Some of these proteins are nutritionally undesirable in legume seeds, such as lipoxygenases (spots 391, 444, and 447 and group 09 in Table II and 274 in Table V) known to produce hydroperoxides that attack nutritionally essential components (e.g. proteins, vitamins, and polyunsaturated fatty acids) and to release off-flavors compounds (Robinson et al., 1995). Despite the low number of spots identified for certain functional groups, they may also play an essential role during seed development. For example, protein spot 65 (group 01 in Table II) is involved in the biosynthesis of thiamine that is absolutely required for seed formation as shown using mutants defective in this biosynthetic pathway (Li and Redei, 1969; Komeda et al., 1988).
Figure 4.
Characterization of less abundant M. truncatula seed proteins whose levels vary between 12 and 20 DAP. A, Example of Coomassie Blue 2-D gel from 12-DAP seeds. Sections 1 to 3 are zoomed in C. B, Distribution of the minor proteins in the functional groups reported for Arabidopsis genes and used to classify M. truncatula proteins (Bevan et al., 1998; Watson et al., 2003). C, Variations of protein profiles in the three different sections shown in A between 12 and 20 DAP. Colored labels indicate some polypeptides identified within some functional classes, and uncolored labels show the location of these spots when they are undetectable or present at low levels. The spots 251 and 280 decreased in their relative abundance between 12 and 18 DAP (class 2 proteins), spot 214 transiently decreased (class 4 protein), protein spots 234, 334, 363, and 428 increased (class 1), and protein spots 217, 282, 383 transiently increased (class 3 proteins). These spots were identified by mass spectrometry as being cytosolic ascorbate peroxidase (spot 251), β-tubulin (280), GAPDHc (214 and 217), oxygen-evolving enhancer protein (234), 1-aminocyclopropane-1-carboxylate (ACC) oxidase (334), chlorophyll a/b-binding protein (363), glyoxalase I (428), and protein disulfide isomerase (PDI; 282). Protein spot quantitation and ANOVA were carried out from four gels for each seed sample. Some spots, chosen as references to discard experimental variations, are shown as diamonds.
Correlation of Protein Abundance with mRNA Abundance
To get a first indication of the extent of the correlations between mRNA and protein levels during seed development, the frequency of occurrence of the transcripts corresponding to the proteins identified was determined in the EST data sets of the cDNA libraries from early and late developing M. truncatula seeds. The Medicago EST Navigation System, previously used to characterize the sets of genes expressed in roots of M. truncatula during symbiosis (Journet et al., 2002), was used to count the number of sequences from the same mRNA in each EST data set. In this program, the ESTs are clustered based on sequence similarity and assembled into contigs reflecting the transcripts. From the proteins identified by MS, 35 different clusters were identified, and 29 corresponded to transcripts expressed in developing seeds. The data were then grouped according to the expression profiles of the mRNAs during seed development (Table VI).
Table VI.
Relative expression level of M. truncatula transcripts based on the repetitive occurrence of sequences in the EST data sets from immature seeds
The cluster accession nos. of the M. truncatula ESTs identified by MS were extracted from the Medicago EST Navigation System (http://medicago.toulouse.inra.fr/) release of January 2003. The cluster accession nos. were used to search the no. of sequences from the same mRNA in two different data sets: a cDNA library of immature seeds ranging in age from 11 to 19 DAP (MtGESD, 4,525 ESTs) and a cDNA library of immature seeds ranging in age from 25 to 35 DAP (MtGLSD, 4,866 ESTs). The M. truncatula transcripts whose expression was found to be significantly variable in the two different data sets (χ2 test, P < 0.050) were grouped according to their expression profiles during seed development. In each class of proteins (see Table I), the corresponding M. truncatula transcripts were sorted in the descending order based on their EST counts in both data sets.
| Protein Class | Strongest BLAST Hit | EST gi | Cluster Accession No. | MtGESD (11-19 DAP) | MtGLSD (25-35 DAP) |
|---|---|---|---|---|---|
| Transcripts preferentially expressed during early stages of seed development | |||||
| 1 | Vicilin | 14986367 | MtC60032.3_GC | 282 | 55 |
| 1 | Vicilin | 14984555 | MtC60159_GC | 41 | 21 |
| 1 | Chlorophyll a/b binding | 13783768 | MtC00119_GC | 10 | 0 |
| 2 | Vicilin | 14984749 | MtC60032.4_GC | 280 | 55 |
| 2 | Annexin | 7718287 | MtC20316_GC | 14 | 1 |
| 2 | S-adenosylmethionine synthetase | 11609660 | MtC00046_GC | 4 | 0 |
| 3 | Flavonone 3-hydroxylase | 13596069 | MtC93321_GC | 12 | 0 |
| 3 | Protein disulfide isomerase | 10697964 | MtC10403_GC | 10 | 0 |
| 3 | GAPDHc | 9671653 | MtC00021.1_GC | 4 | 0 |
| Transcripts preferentially expressed during late stages of seed development | |||||
| 1 | Legumin | 14985010 | MtC60042_GC | 119 | 451 |
| 1 | Legumin | 14986270 | MtC60076_GC | 67 | 311 |
| 1 | Convicilin | 14985541 | MtC60090_GC | 71 | 209 |
| Transcripts whose relative levels did not vary significantly during seed development | |||||
| 1 | Lipoxygenase | 14982738 | MtC60669_GC | 11 | 15 |
| 1 | ACC oxidase | 10520454 | MtC10108.2_GC | 9 | 6 |
| 1 | Met synthase | 9669628 | MtC00018_GC | 4 | 6 |
| 1 | Homology not assigned | 13595945 | MtD00176_GC | 1 | 2 |
| 1 | Glyoxalase 1 | 11897629 | MtC60084_GC | 3 | 0 |
| 1 | Oxygen-evolving enhancer | 11902222 | MtC60015_GC | 2 | 1 |
| 1 | Starch synthase | 13379977 | MtC90709_GC | 2 | 1 |
| 1 | Reversibly glycosylated polypeptide-1 | 7718398 | MtC10969_GC | 2 | 0 |
| 1 | 110-kD 4SnNc-Tudor | 7564626 | MtC50269.1_GC | 1 | 1 |
| 1 | Triosephosphate isomerase | 13380542 | MtC00059_GC | 1 | 0 |
| 1 | Thiamine biosynthesis protein | 7718895 | MtD00077_GC | 1 | 0 |
| 1 | Lipoxygenase | 6708795 | MtC60306_GC | 0 | 0 |
| 1 | S-adenosyl-l-homo-Cys hydrolase | 1710838 | MtC30011_GC | 0 | 0 |
| 2 | β-Tubulin | 7561503 | MtC00356.1_GC | 2 | 1 |
| 2 | Adenosine kinase | 10704292 | MtC10064_GC | 1 | 0 |
| 2 | Pyruvate DH β-subunit | 13780026 | MtC30080_GC | 0 | 0 |
| 2 | Myo-inositol-1-P synthase | 13369792 | MtC60071_GC | 0 | 0 |
| 3 | Heat shock cognate protein 80 | 14882556 | MtD00014_GC | 5 | 1 |
| 3 | Molecular chaperone BiP A | 14983890 | MtC00550.1_GC | 3 | 0 |
| 3 | Heat shock protein 90 | 10706885 | MtC00079_GC | 3 | 0 |
| 3 | Putative homology not assigned | 9678016 | MtC30333_GC | 0 | 0 |
| 3 | Resistance gene analog protein | 13779916 | MtD02701_GC | 0 | 0 |
| 4 | Elongation factor EF-2 | 10698429 | MtC00166.1_GC | 2 | 0 |
The comparison of the results showing significant variations between early and late developing seeds with the proteomic data revealed some correlations between transcript and protein levels. For example, the results suggest that the levels of mRNAs encoding proteins specifically associated with the early stages of seed filling (class 2 and class 3 in Table I), such as Ado-Met synthetase and PDI (Table VI), were higher in early developing seeds, whereas the transcripts encoding proteins that accumulated during the later stages (18–20 DAP), such as legumins and convicilins (Table VI), were preferentially expressed in the late developing seeds. The results also suggest that the levels of transcripts encoding some proteins of class 1 (Table I) decreased during the later stages of seed development. For those proteins whose abundance is maintained up to the desiccation stage, such as vicilins, the absence or decrease of the transcript at late stages suggests a high stability for these proteins throughout seed development (data not shown). Although this approach is not sufficient to establish a rigorous validation of gene expression, the results provide a basis to elucidate the mechanisms of regulation of protein accumulation and stability.
DISCUSSION
The seed occupies a central position in the life cycle of higher plants. In addition to its role in dispersal, the seed determines the success with which germination and early seedling growth occur. Moreover, seeds such as those of legumes are major food sources whose importance lies in the proteins stored during development. Our aim was to identify seed proteins characteristic of specific stages during reserve deposition in M. truncatula. These data will help elucidate the biochemical and molecular processes underlying seed filling in a legume species. Eighty-four proteins whose abundance varied during reserve deposition were identified by MS. As expected, the most abundant proteins corresponded to storage proteins. Many of the weakly abundant proteins could play a role in cell division during embryogenesis, in protein or starch deposition, in defense against herbivores, in cell expansion during reserve deposition, or in the transition from a highly active to a quiescent state during seed development. These results are discussed in the following sections.
The Abundance of Cell Division-Associated Proteins Decreases in Transition from Embryogenesis to Reserve Deposition
Embryogenesis starts with a morphogenesis phase during which the embryo differentiates through several distinct stages (globular, heart, and torpedo) and ends at the cotyledon stage when all embryo structures have been formed. Figure 2A shows that acquisition of ability to germinate does not occur before 14 DAP, presumably because the young embryo is not fully formed before this stage. At the end of embryogenesis, cell division arrests and the seed accumulates the storage components (Raz et al., 2001). Our results showed that the 12-DAP stage corresponded to the end of the embryogenesis phase (Fig. 1) and that the abundance of several proteins decreased in the transition from embryogenesis to seed filling (Table III). This was the case for β-tubulin (spot 280 in Table III) and annexin (spots 223 and 228 in Table III) that are associated with cell cycle events. Tubulins are associated with cell division and cell enlargement aspects of the cell cycle. During cell division, they play an important role in separation of the organelles and daughter chromosomes (mitosis). The accumulation of β-tubulin has been observed during seed germination, in relation to reactivation of cell cycle activity (De Castro et al., 2000). In contrast, the presence of annexin in seeds has not been reported previously. However, there is strong evidence that annexins are involved in cell division. For example, they accumulate during the cell cycle and peak at the end of mitosis in tobacco (Nicotiana tabacum) cells (Proust et al., 1999). Because they are localized at cell junctions and are known to bind secretory vesicles during exocytosis, annexins could play a role in cell wall maturation during cell division (Proust et al., 1999). Because the abundance of annexin and β-tubulin decreased significantly 14 DAP and remained low at the protein and mRNA levels (Fig. 4; Table VI), it is likely that these proteins participate in the stages preceding reserve deposition, presumably in the embryonic divisions. Their turnover reflects the developmental switch from embryogenesis to seed filling, i.e. a switch from cell division to reserve deposition (Figs. 3 and 4).
Synthesis and Maturation of the Storage Protein Families
After the cessation of cell division, the seed storage compounds are synthesized. In M. truncatula, the reserve deposition phase is characterized by a large increase in the protein content of up to 45% (G. Duc, personal communication). In our study, the most abundant protein spots, which are mainly responsible for this increase in the protein content and, thus, for the nutritional value of legume seeds, were identified as being the 7S (vicilin and convicilin) and 11S (legumin) globulins (Table II). These protein families accumulated in a specific temporal order during seed filling: vicilins (14 DAP), legumins (16 DAP), and convicilins (18 DAP; Fig. 3). The transcripts encoding the storage protein families were expressed in a similar time course when compared with protein abundance, with mRNAs encoding vicilins being preferentially expressed in the early stages, and mRNAs encoding the legumin and convicilin families being preferentially expressed in the later stages (Table VI) as observed in our proteome analysis. This suggests that the temporal accumulation of the storage protein families is likely to be transcriptionally controlled during seed development. As expected for proteins encoded by multigene families (Casey et al., 2001), several spots with similar masses but differing in charge were identified as being the same storage protein. Because these polypeptides are broken down during germination and used by the germinating seedling as an initial food source (Bewley and Black, 1994), it was not surprising to observe that their accumulation during seed filling is concomitant with the acquisition of both seed vigor and seedling vigor (Figs. 2 and 3).
The characterization of the globulin families in various dicotyledonous plants has shown that the precursor forms of these proteins are transported from the endoplasmic reticulum lumen to the protein storage vacuoles, where they are processed into specific subunits. These chains are then assembled within the protein bodies, yielding the mature forms, typically trimeric for the 7S globulins and hexameric for the 11S globulins (Gruis et al., 2002). In 2-D gels, two spots were identified as precursor forms of legumin (Tables II and IV) and nine as acidic α-chains (Mr of approximately 40,000) or basic β-chains (Mr of approximately 20,000; Table II). These polypeptides accumulated with the same time course during seed filling, indicating that synthesis and maturation of the 11S globulins are not developmentally separated in M. truncatula. Our results showed that the convicilin family was represented by polypeptides of 50 and 74 kD (Table II). Also, three protein spots with Mr of approximately 47,000 were identified as vicilin and six spots as corresponding to the carboxyl-terminal domain (three spots with Mr of approximately 30,000) or the amino-terminal domain (three spots with Mr of approximately 17,000) of the vicilin precursor forms (Table II), suggesting that the vicilin precursor forms are processed to give products ranging in Mr from approximately 17,000 to 47,000.
Most of the mature products of the M. truncatula 7S and 11S globulins possessed similar masses to those found in pea and soybean seeds (Croy et al., 1980; Bewley and Black, 1994; Jung et al., 1998; Casey, 1999), suggesting that storage protein processing may be conserved within these species. Moreover, the BLASTP analyses of the tag consensus (TC) translated sequences of these storage protein families (Table II) showed higher homology (70%–85%) with those from agriculturally important legume crops (pea, soybean, and fava bean). Together, these data highlight the importance of using M. truncatula as a model to study the cellular and molecular mechanisms related to protein deposition in seeds of legume crops.
Endoplasmic reticulum resident proteins known as molecular chaperones play important roles in the formation and assembly of the seed storage proteins (Li and Larkins, 1996; Hatano et al., 1997; Takemoto et al., 2002). Our study revealed several luminal proteins involved in protein folding, including a PDI and a putative chaperonin of the binding protein (BiP) class (spots 169 and 282 and group 06 in Table IV; Fig. 4C). PDI catalyzes the formation and rearrangement of disulfide bonds of the newly synthesized proteins, whereas BiP, a molecular chaperone related to the heat shock proteins, is involved in the assembling of the nascent proteins by preventing their denaturation or aggregation and in the recognition and disposal of misfolded polypeptides (Kainuma et al., 1995; Zhang et al., 1997). The temporal induction of the proteins identified as BiP and PDI coincided with the onset of storage protein accumulation (see Figs. 3 and 4). Therefore, these molecular chaperones are good candidates to study the folding and assembly of the storage proteins in legume seeds.
Two 100-kD Components of the Precursor-Accumulating Vesicles (PV100) Are Synthesized during M. truncatula Seed Filling
Interestingly, in addition to vicilin polypeptides of the expected 30- to 40-kD class, two protein spots of Mr approximately 120,000 (389 and 390 and group 09 in Table II) that accumulated during seed filling also possessed vicilin-related sequences. In their respective MALDI-TOF spectrums, at least eight peptide masses matched with those from vicilins. Despite their high Mrs, they were identified provisionally, therefore, as vicilins. A similar protein D of approximately 100,000 containing a vicilin-like domain was described in pumpkin (Cucurbita maxima) seeds. This protein, called PV100, is synthesized on rough endoplasmic reticulum as a precursor form consisting of a single protein chain with three domains: a vicilin-like domain, a Cys-rich domain (91 amino acids), and an Arg-/Glu-rich domain (267 amino acids; Yamada et al., 1999). To investigate whether the M. truncatula protein spots with Mr of approximately 120,000 could be the homologs of PV100, the unmatched peptide masses obtained by MALDI-TOF were compared with those predicted from the Cys-rich and Arg-/Glu-rich domains of PV100 from pumpkin. The results showed that several peptide masses for spots 389 and 390 matched well with those predicted from these domains (Table II). Yamada et al. (1999) reported that the precursor form of PV100 is transported by vesicles to protein storage vacuoles, where it is processed to a mature vicilin, a trypsin inhibitor, and a basic cytotoxin-related peptide. Because these latter two peptides might play a herbivore-deterrent role, their elimination could improve the nutritional quality of legume seeds for human and animal use. Such a protein has not been characterized yet in legume seeds. Assuming a functional homology to PV100, it will be of interest, therefore, to characterize the mature peptide products derived from the M. truncatula 120-kD proteins.
Role of Embryonic Photosynthesis during Seed Filling
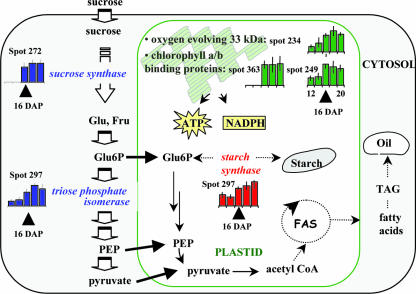
In some legumes, such as soybean, starch can be present early during seed development. However, the starch level declines to about 1% and is replaced by accumulating oil reserves on maturation (Adams et al., 1980). As in soybean, M. truncatula seed development is characterized by a transient accumulation of starch and by the deposition of protein and lipid reserves to a final concentration of about 40% and 10%, respectively (J.P. Boutin, personal communication). The biochemical pathways that produce the storage compounds are well known, but the processes that coordinate their accumulation at specific stages during seed development are not well understood. Our study revealed that the 16-DAP stage was characterized by the onset of storage protein accumulation (Fig. 3) and associated with an accumulation of starch synthase, Suc synthase, and triosephosphate isomerase (spots 246, 272, and 297 and group 01 in Table II; Fig. 5), which could be involved in the supply of carbon substrates for the synthesis of storage compounds in these limiting conditions for the fixation of CO2 (Heim et al., 1993). Interestingly, an oxygen-evolving enhancer protein and two chlorophyll a/b-binding proteins (spots 234, 249, and 363 and group 02 in Table II; see also Fig. 5) accumulated at the same stage. In accordance with this finding, embryonic photosynthesis was proposed to play a role in providing ATP and NADPH for storage compound synthesis during seed filling (Browse and Slack, 1985; Batz et al., 1995; Eastmond and Rawsthorne, 2000; Ruuska et al., 2002). Together, these data suggest that photosynthesis activity in these seeds may respond to the heavy demand by protein/oil synthesis for cofactors, such as ATP and NADPH. Although CO2 fixation has been reported to be low in developing seeds, we observed that the level of a subunit of Rubisco (spot 126 and group 02 in Table II) increased at 20 DAP. A similar observation was reported in developing Arabidopsis seeds, where a role for this enzyme in recycling the CO2 released during the biosynthesis of storage compounds during seed filling was proposed.
Figure 5.
Kinetics of accumulation of proteins involved in carbon and plastidial metabolisms in M. truncatula developing seeds. Some enzymes and substrates leading to the synthesis of starch and/or oil are shown. Not shown are other contributions to oil metabolism, such as the oxidative pentose phosphate pathway. FAS, Fatty acid synthesis; PEP, phosphoenolpyruvate; TAG, triacylglyceride.
Proteins Associated with Cell Expansion Accumulate during Reserve Deposition
During seed filling, the cells continue to grow by enlargement as they accumulate the storage components. Two proteins identified in this study could be involved in this cell enlargement process during protein deposition. The first corresponded to actin (ACT7 and spot 187 in Table IV), which is a fundamental component of the cytoskeleton. Its level increased specifically during protein deposition and decreased after this stage. In Arabidopsis, the gene encoding the same isoform ACT7 is preferentially expressed in vegetative tissues that contain rapidly dividing and expanding cells and appears to be the only actin gene expressed in seed tissues (McDowell et al., 1996a). Actin is involved in a number of cellular processes such as cytoplasmic streaming, cell shape determination, organelle movement, and extension growth (McDowell et al., 1996b). Because of these functions, the M. truncatula seed protein identified as ACT7 could play an important role in cell expansion during the phase of protein deposition.
The second protein (spot 206 and group 07 in Table II) showed more than 88% identity with the reversibly glycosylated polypeptide (RGP1) identified in suspension-cultured cells, roots, and leaves in Arabidopsis (Delgado et al., 1998) and in pea seedlings (Dhugga et al., 1997). In these tissues, this protein is thought to have a role in cell wall polysaccharide synthesis, possibly that of xyloglucan. A similar protein was also found in wheat endosperm (Langeveld et al., 2002), but its function during seed development has not been elucidated yet. The protein RGP1 identified in our study accumulated significantly at the stage 18 DAP, which is characterized by a large increase in the abundance of the storage proteins (Fig. 3). Therefore, it would be of particular interest to examine its possible role in cell wall expansion at this stage.
Enzymes of Met Metabolism May Be Implicated in the Transition from a Highly Active State to a Quiescent State during Seed Development
Interestingly, many proteins whose abundance varied during seed filling corresponded to enzymes involved in Met biosynthesis. Among the essential amino acids synthesized by plants, Met is a fundamental metabolite because it functions not only as a building block for protein but also as the precursor of Ado-Met, the primary methyl-group donor and the precursor of polyamines and the plant ripening hormone ethylene (Ravanel et al., 1998; Kim and Leustek, 2000; Gakière et al., 2002).
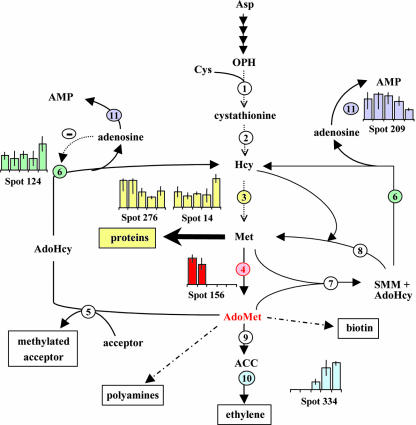
In plants, Met can be synthesized through two pathways (Fig. 6). In the de novo biosynthetic pathway, O-phosphohomo-Ser is transformed to cystathionine in a reaction catalyzed by cystathionine γ-synthase, then to Hcy in a reaction catalyzed by cystathionine β-lyase, and finally to Met in the presence of the cobalamin-independent Met synthase. In the Met-recycling pathway (Hanson and Roje, 2001), SMM, a compound unique to plants, is synthesized by a methyl transfer from Ado-Met to Met, in a reaction catalyzed by Ado-Met:Met S-methyltransferase. SMM can then be reconverted to Met by transferring a methyl group to Hcy in a reaction catalyzed by SMM: Hcy S-methyltransferase. These reactions, together with the reactions catalyzed by Ado-Met synthetase and AdoHcy hydrolase, constitute the SMM cycle, which may be the main mechanism in plants for shortterm control of Ado-Met level. This cycle consumes half of the Ado-Met produced (Ranocha et al., 2001).
Figure 6.
Differential accumulation of enzymes involved in Met de novo biosynthesis during M. truncatula seed filling. Reaction intermediates: ACC, AMP, S-adenosylhomo-Cys (AdoHcy), S-adenosyl-Met (Ado-Met), homo-Cys (Hcy), O-phosphohomo-Ser (OPH), and S-methyl-Met (SMM). Enzymes: 1, cystathionine γ-synthase; 2, cystathionine β-lyase; 3, cobalamine-independent Met synthase; 4, Ado-Met synthetase; 5, Ado-Met-dependent transmethylases; 6, AdoHcy hydrolase; 7, Ado-Met:Met S-methyltransferase (MMT); 8, SMM:Hcy S-methyltransferase (HMT); 9, ACC synthase; 10, ACC oxidase; and 11, adenosine kinase.
Consistent with the high demand for protein synthesis between 12 and 20 DAP, two spots detected in seed extracts throughout this period corresponded to Met synthase (spots 14 and 276 and group 01 in Tables II and V), which catalyzes the last step of the de novo biosynthetic pathway of Met. In addition, one spot (156 in Table III) corresponded to Ado-Met synthetase, which catalyzes the synthesis of Ado-Met from Met and ATP. Interestingly, the level of Ado-Met synthetase fell sharply at the 16-DAP stage (Fig. 6) and remained low up to desiccation (data not shown). This result was supported by the observation that ESTs corresponding to Ado-Met synthetase were only found in cDNA libraries corresponding to early stages of seed development (Table VI). Ado-Met synthetase also was absent from dry mature Arabidopsis seeds (Gallardo et al., 2001) but accumulated in the transition from a quiescent to a highly active state during germination (Gallardo et al., 2002a). In contrast to germination, during seed development, there is a switch from a period of highly active metabolism associated with cell expansion, differentiation, and accumulation of storage products to a period during which the overall biosynthetic activity decreases as the embryo prepares for quiescence. Assuming a functional homology to Ado-Met synthetase found in germinating Arabidopsis seeds, the accumulation pattern of the M. truncatula Ado-Met synthetase may also reflect the metabolic shift in developing seeds.
After the decrease of Ado-Met synthetase, there was an increase in the abundance of two enzymes involved in Ado-Met consumption. The first protein corresponded to AdoHcy hydrolase (spot 124 and group 01 in Table II; Fig. 6). The hypothesis that AdoHcy hydrolase is active during seed development agrees with previous results showing that Met recycling via the Ado-Met/AdoHcy and SMM cycles is not sufficient in mature seeds to maintain an appropriate pool of Met for rapid germination and seedling establishment (Gallardo et al., 2002a). Knowing that AdoHcy hydrolase is being subjected to feedback inhibition by adenosine, it was interesting to note that developing seeds also contained adenosine kinase (spot 209, Table III; Fig. 6) catalyzing the phosphorylation of adenosine to adenine monophosphate (Moffatt et al., 2002). The second protein corresponded to ACC oxidase (spot 334 and group 03 in Table II; Fig. 6). This enzyme is involved in the synthesis of the plant ripening hormone ethylene, which has been shown to control cotyledon expansion during embryo development in rapeseed (Hays et al., 2000), and is implicated generally in ripening processes.
In plants, Ado-Met has an important influence on cell growth and development. Beside its role in ethylene, biotin, and polyamine biosynthesis, Ado-Met is the primary methyl group donor for the methylation of amino acids, lipids, RNA, and DNA, and functions as an effector in the regulation of Thr, Lys, and Met synthesis (Ravanel et al., 1998). The disappearance of Ado-Met synthetase and the accumulation of Ado-Met-consuming enzymes are likely to decrease Ado-Met levels. Given the important regulating influence of Ado-Met, this may promote the repression of the metabolic activities leading to a quiescent state. This suggests that the same mechanism may be implicated in the repression of metabolic activities during seed development and their resumption during germination (Gallardo et al., 2002a).
CONCLUSION
We utilized a proteomic approach to identify 84 M. truncatula seed proteins with characteristic developmental patterns of accumulation during protein deposition. Some of these had previously been shown to play a role during seed filling in other legume species (e.g. legumins, vicilins, and convicilins), confirming the validity of M. truncatula as a model system for analysis of legume seed filling. The present study also revealed the kinetics of storage protein accumulation in M. truncatula and new proteins to be associated with the reserve deposition process, with presumed roles in cell division (annexin), cell expansion (ACT7 and RGP1), or metabolic activities (for example, Ado-Met synthetase and AdoHcy hydrolase). Furthermore, the data revealed nutritionally undesirable components whose elimination should improve the quality of legume seeds, such as lipoxygenases and components of the precursor-accumulating vesicles (PV100), which give rise to a trypsin inhibitor on maturation. These data will facilitate further studies, which investigate the effects of genetic and environmental factors on seed quality. The role of these proteins can be further assessed by a combination of forward and reverse genetics, such as the TILLING (targeting induced local lesions in genomes) methodology (McCallum et al., 2000). This will provide additional information useful for understanding the complexities of seed metabolism and its control during seed development.
MATERIALS AND METHODS
Plant Material
A batch of 20 plants of Medicago truncatula cv Jemalong, line J5 was used for all experiments. Plants were grown in a growth chamber at 22°C/19°C day/night temperatures, under a 16-h photoperiod at 220 μE m2 s–1 light intensity with 60% to 70% relative humidity. To harvest pods at defined stages during development, individual flowers were tagged on the day of flower opening. For each stage, at least 40 flowers were labeled on the first, second, and third nodes of the main ramifications. Pods were harvested between 8 and 44 DAP. Developing seeds were removed from pods at 4°C to prevent dehydration. To determine seed fresh weight, dry weight, and water content during development, three pools of 15 randomly selected seeds were weighed (Sartorius ISO 9001 scale, Quality Control Services, Portland, OR) just after harvest and after drying at 70°C for 24 h. To assess their tolerance to dehydration, three pools of 15 fresh seeds were subjected to germination assays for seed performance analysis, and three other replicates of 15 seeds were dried for 48 h at room temperature (22 ± 3°C). These drying conditions resulted in a similar rate of water loss to that which occurred during drying at 70°C for 24 h. For protein analyses, pools of 35 seeds were frozen in liquid nitrogen and stored at –80°C.
Germination Assays
Germination assays were carried out in a growth chamber under conditions described above. Two seed samples were subjected to germination assays: fresh developing seeds and developing seeds dried at room temperature. For each assay, three replicates of 15 developing seeds were incubated on three sheets of absorbent paper and a black membrane filter with a white grid (45 mm diameter, Schleicher & Schull, Dassel, Germany) wetted with 1.5 mL of distilled water, in covered plastic boxes (50 mm diameter). Germination was scored when the primary root protruded through the surrounding structures.
Preparation of Total Protein Extracts
Total protein extracts were prepared from immature seeds at different stages of seed development. For each stage, a batch of 35 seeds was ground in liquid nitrogen using mortar and pestle. Total proteins were extracted at 4°C in 20 μL mg–1 seed dry matter (see Fig. 1) of a thiourea/urea lysis buffer used previously for Arabidopsis seeds (Gallardo et al., 2002b). After 10 min at 4°C, 14 mm dithiothreitol (Amersham Biosciences, Orsay, France) was added, and the protein extracts were stirred for 20 min at 4°C and then centrifuged (35,000g for 10 min) at 4°C. The supernatant was submitted to a second clarifying centrifugation step as above. The final supernatant corresponded to the total protein extract. Protein concentration was measured according to Bradford (1976).
2-D Electrophoresis
Proteins were first separated by isoelectrofocusing (IEF). For the preparation of 2-D gels for Coomassie Blue staining, IEF was carried out with 30 μL of the various protein extracts; for silver-stained 2-D gels, IEF was performed with 15 μL of protein extracts. Proteins were separated using gel strips forming an immobilized nonlinear 3 to 10 pH gradient (Immobiline DryStrip, 24 cm; Amersham Biosciences). Strips were rehydrated in the IPGphor system (Amersham Biosciences) for 7 h at 20°C with the thiourea/urea lysis buffer containing 2% (v/v) Triton X-100, 20 mm dithiothreitol, and the protein extracts. IEF was performed at 20°C in the IPGphor system for 7 h at 50 V, 1 h at 300 V, 2 h at 3,500 V, and 7 h at 8,000 V. Before the second dimension, each gel strip was incubated at room temperature for 2 × 15 min in 2 × 15 mL equilibration solution as described by Gallardo et al. (2002a). Proteins were then separated in vertical polyacrylamide gels according to Gallardo et al. (2002a). For each stage analyzed, at least four replicated 2-D gels were done.
Protein Staining and Analysis of 2-D Gels
Gels were stained with either Coomassie Brilliant Blue G-250 (Bio-Rad, Hercules, CA) according to Mathesius et al. (2001) or with silver nitrate according to Blum et al. (1987) using the Hoefer Automated Gel Stainer apparatus from Amersham Biosciences. Image acquisition was done using a Sharp JX-330 scanner (Amersham Biosciences) with a resolution of 300 microns pixel–1 and an optical density range from 0.05 to 3.05. Image analysis was carried out on Coomassie Blue gels with the ImageMaster 2-D Elite version 3.1 software (Amersham Biosciences), which allows spot detection and quantification, background subtraction (non-spot mode), and spot matching across the different gels. Protein spots were selected for quantitative analysis if they were consistently visible in the four replicates for at least one stage. Because seed development is associated with many morphological (e.g. embryo size), physiological (e.g. increase in seed dry weight), and molecular (e.g. a high proportion of proteins accumulated) changes, the choice of a reference or normalization procedure to correct experimental variations in 2-D gels was complex. For this purpose, the volume of each spot (i.e. spot abundance) was normalized in the different gels by using several methods. A first procedure, provided by the ImageMaster 2-D Elite software, divided each spot volume value by the sum of all spot volume values to obtain relative spot abundances. Because the most abundant proteins were not present in 2-D gels at 12 DAP and account for more than 30% of the total spot volume at 20 DAP, this method did not reduce any experimental variations in 2-D gels between the different stages. Therefore, we tested two other normalization procedures based either on a single spot (procedure provided by the ImageMaster 2-D Elite software) or on 20 spots showing constant levels during the different stages subjected to proteomics. These spots were selected by comparing qualitatively silver- and Coomassie Blue-stained 2-D gels across developmental stages. The quantitative data showed a greater reduction of experimental variations between replicated 2-D gels and between the different developmental stages by using the scaling procedure based on the 20 reference spots. Therefore, the volume of each spot was normalized to the total volume of these 20 reference spots whose abundance was qualitatively constant from 12 to 20 DAP (Fig. 4B, diamonds). To compare differences in protein abundance among the different samples, a one-way ANOVA and a Student-Newman-Keuls test were performed for each spot, using the SAS software package (SAS Institute, 1999).
Protein Identification by MS
Spots of interest were excised from Coomassie Blue 2-D gels and digested by sequence grade trypsin (Promega, Madison, WI). After digestion, the supernatant-containing peptides were concentrated by batch adsorption on beads (POROS 50 R2; Roche Molecular Biochemicals, Basel) and analyzed on a MALDI-TOF mass spectrometer (Reflex II; Bruker, Bremen, Germany) after on-target desorption with matrix solution (Gevaert et al., 1998). Before each analysis, the instrument was externally calibrated using two synthetic peptides spotted as near as possible to the biological sample. All searches were done using MASCOT (http://www.matrixscience.com) against the M. truncatula EST (approximately 181,000 entries in September 2002) from the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Entrez/) and TIGR (http://www.tigr.org/tdb/tgi/mtgi/). To qualify as a positive identification, the following criteria were used: protein scores should have P ≤ 0.05, coverage of the protein by the matching peptides must reach a minimum of 10%, and at least four independent peptides should match within a stringent 10-ppm maximum deviation of mass accuracy. In some cases, protein identities were further confirmed from PSD spectra generated from selected peptides. A number of protein spots with uncertain identities were selected and analyzed by electrospray ionizationtandem MS (ESI-MS/MS) at the Unité Mixte de Recherches de Génétique Végétale, INRA/Université de Paris-Sud/Institut National Agronomique Pares-Grignon (Gif-sur-Yvette, France). Search for protein sequence homology was carried out by submitting the EST (or TC) translation product to a BLASTP search (http://www.ncbi.nlm.nih.gov/blast/). Theoretical masses and isoelectric points of the homologous proteins were predicted by entering the sequence at http//:www.expasy.ch/tools/peptide-mass.html/.
EST Counting in Data Sets of Two cDNA Libraries from Developing Seeds
The relative expression level of the mRNAs encoding the identified proteins was studied by using the Medicago EST Navigation System (http://medicago.toulouse.inra.fr/) release of January 2003. In this program, the ESTs are clustered based on sequence similarity and assembled into contigs reflecting the transcripts. The cluster accession numbers corresponding to the ESTs identified by mass spectrometry were extracted from the sequence retrieval system (SRS search) and used to determine the EST frequencies by electronic northern in two different data sets: a cDNA library of immature seeds collected from pods ranging in age from 11 to 19 DAP (MtGESD, 4,525 ESTs) and a cDNA library of immature seeds collected from pods ranging in age from 25 to 35 DAP (MtGLSD, 4,866 ESTs). The data were then compared with the proteomic results.
Acknowledgments
We thank Gerard Duc (Unité de Gènetique et Ecophysiologie des Legumineuses, INRA, Dijon, France) for initiating this project, for his constant support, and for critical reading of the manuscript. We are grateful to Françoise Moussy (Unité et Ecophysiologie des Legumineuses, INRA, DIJON, FRANCE) for her valuable help in the collection of the plant material. We thank H. Demol and M. Puype (Flanders Interuniversity Institute for Biotechnology, Department of Biochemistry, Gent University, Belgium) for excellent work regarding MALDI-TOF mass spectrometry analysis. We sincerely thank Luc Negroni (INRA, Gif-sur-Yvette, France) for ESI-MS/MS analyses. We also thank Dominique Job (Bayer CropScience Joint Laboratory Unité Mixte de Recherche Centre National de la Recherche Scientifique, Lyon, France) for helpful discussions, in particular regarding Met metabolism.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025254.
This work was supported by the INRA-Action Transversale Structurante program on Medicago truncatula and by INRA (postdoctoral fellowship to K.G.).
References
- Adams CA, Rinne RW, Fjerstad MC (1980) Starch deposition and carbohydrate activities in developing and germinating soybean seeds. Ann Bot 45: 577–582 [Google Scholar]
- Batz O, Scheibe R, Neuhaus HE (1995) Purification of chloroplasts from fruits of green pepper (Capsicum annuum L.) and characterization of starch synthesis: evidence for a functional chloroplastic hexosephosphate translocator. Planta 196: 50–57 [Google Scholar]
- Baud S, Boutin J, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW et al. (2001) The Medicago Genome Initiative: a model legume database. Nucleic Acids Res 29: 114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirske W, Van Staveren M, Stiekema W et al. (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391: 485–488 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination. Plenum Press, New York
- Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93–99 [Google Scholar]
- Bove J, Jullien M, Grappin P (2002) Functional genomics in the study of seed germination. Genome Biol 3: reviews 1002.1–1002.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Browse J, Slack C (1985) Fatty acid synthesis in plastids from maturing sunflower and lineseed cotyledons. Planta 166: 74–80 [DOI] [PubMed] [Google Scholar]
- Casey R (1999) Distribution of some properties of seed globulins. In PR Shewry, R Casey, eds, Seed Proteins. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 159–169
- Casey R, Christou P, Domoney C, Hedley C, Hitchin E, Parker M, Stoger E, Wang T, Zasiura C (2001) Expression of legumin and vicilin genes in pea mutants and the production of legumin in transgenic plants. Nahrung 45: 385–387 [DOI] [PubMed] [Google Scholar]
- Croy RR, Gatehouse JA, Tyler M, Boulter D (1980) The purification and characterization of a third storage protein (convicilin) from the seeds of pea (Pisum sativum L.). Biochem J 191: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro RD, van Lammeren AAM, Groot SPC, Bino RJ, Hilhorst HWM (2000) Cell division and subsequent radicle protrusion in tomato seeds are inhibited by osmotic stress but DNA synthesis and formation of microtubular cytoskeleton are not. Plant Physiol 122: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado IJ, Wang Z, de Rocher A, Keegstra K, Raikhel NV (1998) Cloning and characterization of AtRGP1: a reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol 116: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM (1997) A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA 94: 7679–7684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol 122: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129: 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakière B, Denis L, Droux M, Job D (2002) Over-expression of cystathionine γ-synthase in Arabidopsis thaliana leads to increased levels of methionine and S-methylmethionine. Plant Physiol Biochem 40: 119–126 [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2002a) Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant 116: 238–247 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002b) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Demol H, Sklyarova T, Vandekerckhove J, Houthaeve T (1998) A peptide concentration and purification method for protein characterization in the subpicomole range using matrix assisted laser desorption/ionization-postsource decay (MALDI-PSD) sequencing. Electrophoresis 19: 909–917 [DOI] [PubMed] [Google Scholar]
- Gruis DF, Selinger DA, Curran JM, Jung R (2002) Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 14: 2863–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hatano K, Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I (1997) A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol 38: 344–351 [DOI] [PubMed] [Google Scholar]
- Hays DB, Reid DM, Yeung EC, Pharis RP (2000) Role of ethylene in cotyledon development of microspore-derived embryos of Brassica napus. J Exp Bot 51: 1851–1859 [DOI] [PubMed] [Google Scholar]
- Heim U, Weber H, Baumlein H, Wobus U (1993) A sucrose-synthase gene of Vicia faba L.: expression pattern in developing seeds in relation to starch synthesis and metabolic regulation. Planta 191: 394–401 [DOI] [PubMed] [Google Scholar]
- Higgins TJ, Chandler PM, Randall PJ, Spencer D, Beach LR, Blagrove RJ, Kortt AA, Inglis AS (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J Biol Chem 261: 11124–11130 [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O et al. (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung R, Scott MP, Nam YW, Beaman TW, Bassuner R, Saalbach I, Muntz K, Nielsen NC (1998) The role of proteolysis in the processing and assembly of 11S seed globulins. Plant Cell 10: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainuma K, Ookura T, Kawamura Y (1995) Purification and characterization of protein disulfide isomerase from soybean. J Biochem 117: 208–215 [DOI] [PubMed] [Google Scholar]
- Kermode AR, Bewley JD, Dasgupta J, Misra S (1986) The transition from seed development to germination: a key role for desiccation. HortScience 21: 1113–1118 [Google Scholar]
- Kim J, Leustek T (2000) Repression of cystathionine γ-synthase in Arabidopsis thaliana produces partial methionine auxotrophy and developmental abnormalities. Plant Sci 151: 9–18 [Google Scholar]
- Komeda Y, Tanaka M, Nishimune T (1988) A th-1 mutant of Arabidopsis thaliana is defective for a thiamin-phosphate-synthesizing enzyme: thiamine phosphate pyrophosphorylase. Plant Physiol 88: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld SM, Vennik M, Kottenhagen M, Van Wijk R, Buijk A, Kijne JW, de Pater S (2002) Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiol 129: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CP, Larkins BA (1996) Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol Biol 30: 873–882 [DOI] [PubMed] [Google Scholar]
- Li SL, Redei GP (1969) Thiamine mutants of the crucifer, Arabidopsis. Biochem Genet 3: 163–170 [DOI] [PubMed] [Google Scholar]
- Mathesius U, Keijzers G, Natera SH, Weinman JJ, Djordjevic MA, Rolfe BG (2001) Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1: 1424–1440 [DOI] [PubMed] [Google Scholar]
- Mazur B, Krebbers E, Tingey S (1999) Gene discovery and product development for grain quality traits. Science 285: 372–375 [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18: 455–457 [DOI] [PubMed] [Google Scholar]
- McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB (1996a) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 111: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB (1996b) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142: 587–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C (2002) Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol 128: 812–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust J, Houlne G, Schantz ML, Shen WH, Schantz R (1999) Regulation of biosynthesis and cellular localization of Sp32 annexins in tobacco BY2 cells. Plant Mol Biol 39: 361–372 [DOI] [PubMed] [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD (2001) The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J 25: 575–584 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95: 7805–7812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Robinson DS, Wu Z, Domoney C, Casey R (1995) Lipoxygenases and the quality of foods. Food Chem 54: 33–43 [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (1999) SAS/STAT User's Guide. SAS Institute Inc., Cary, NC
- Tabe LM, Wardley-Richardson T, Ceriotti A, Aryan A, McNabb W, Moore A, Higgins TJ (1995) A biotechnological approach to improving the nutritive value of alfalfa. J Anim Sci 73: 2752–2759 [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Asirvatham VS, Wang L, Sumner LW (2003) Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol 131: 1104–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel M, Gers-Barlag H, Schimpl A, Rudiger H (1993) Time course of lectin and storage protein biosynthesis in developing pea (Pisum sativum) seeds. Biol Chem Hoppe Seyler 374: 887–894 [DOI] [PubMed] [Google Scholar]
- Yamada K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (1999) Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J Biol Chem 274: 2563–2570 [DOI] [PubMed] [Google Scholar]
- Zhang JX, Braakman I, Matlack KE, Helenius A (1997) Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol Biol Cell 8: 1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]