Abstract
We isolated two nitrilase genes, ZmNIT1 and ZmNIT2, from maize (Zea mays) that share 75% sequence identity on the amino acid level. Despite the relatively high homology to Arabidopsis NIT4, ZmNIT2 shows no activity toward β-cyano-alanine, the substrate of Arabidopsis NIT4, but instead hydrolyzes indole-3-acetonitrile (IAN) to indole-3-acetic acid (IAA). ZmNIT2 converts IAN to IAA at least seven to 20 times more efficiently than AtNIT1/2/3. Quantitative real-time polymerase chain reaction revealed the gene expression of both nitrilases in maize kernels where high concentrations of IAA are synthesized tryptophan dependently. Nitrilase protein and endogenous nitrilase activity are present in maize kernels together with the substrate IAN. These results suggest a role for ZmNIT2 in auxin biosynthesis.
Indole-3-acetic acid (IAA) is the most abundant natural auxin and was isolated from plants more than 50 years ago (Luckwill, 1952). Yet, the details of IAA biosynthesis are not fully understood, probably because there are parallel pathways that may work together or could be differentially regulated dependent on organs, developmental stages, or environmental conditions (for review, see Normanly and Bartel, 1999; Bartel et al., 2001). These pathways might form a metabolic network and change dynamically to keep auxin homeostasis or to supply IAA for local demands.
Originally, the amino acid Trp was identified as the precursor of IAA, and IAA synthesis was suggested to occur by deamination and decarboxylation of Trp (for review, see Bandurski et al., 1995). The conversion of Trp to IAA was reported in many systems, including cell cultures (Michalczuk et al., 1992) and excised hypocotyls (Ribnicky et al., 1996) of carrot (Daucus carota), roots and leaves of Arabidopsis (Müller et al., 1998; Müller and Weiler, 2000a), and in coleoptile tips (Koshiba et al., 1995) and kernels (Glawischnig et al., 2000) of maize (Zea mays). Characterization of Trp auxotrophs and stable isotope labeling experiments led to the postulation of additional Trp-independent IAA biosynthetic pathways starting from indole or indole-3-glycerolphosphate in maize (Wright et al., 1991; Östin et al., 1999) and Arabidopsis (Normanly et al., 1993). Now there is growing evidence that Trp-dependent and Trp-independent pathways operate in a single plant (Michalczuk et al., 1992; for review, see Bartel et al., 2001).
Furthermore, two separate intracellular pools of Trp were suggested by the fact that exogenously added Trp can be converted to IAA more readily than endogenous Trp (Rapparini et al., 1999). By using [U-13C]Glc and [1,2-13C]acetate as general precursors and retrobiosynthetic NMR analysis (Bacher et al., 1999) as the analytical tool, it was found that only Trp-dependent pathways operate in developing maize kernels (Glawischnig et al., 2000).
Although details of IAA synthesis from Trp are still unclear, two major pathways have been postulated based on the detection of proposed intermediates and enzymes (for review, see Normanly and Bartel, 1999). The indole-3-pyruvate pathway (Tam and Normanly, 1998) might be completed by the aldehyde oxidase-catalyzed reaction in maize (Sekimoto et al., 1997) and Arabidopsis (Seo et al., 1998). Alternatively, indole-3-acetaldoxime, which can be formed by CYP79B homologs (Hull et al., 2000; Mikkelsen et al., 2000), can be converted in vitro to indole-3-acetonitrile (IAN; Helminger et al., 1985).
Nitrilases (EC 3.5.5.1) that hydrolyze nitriles (e.g. IAN) to the corresponding carboxylic acids (as IAA) received particular interest due to their potential role in IAA biosynthesis. Nitrilase activity is observed in the crude extract of some plant families, including Crucifereae, Gramineae, and Musaceae (Thimann and Mahadevan, 1964a). In Arabidopsis, four nitrilase genes were identified and classified into two groups following their gene cluster and substrate specificities (Bartling et al., 1992, 1994; Bartel and Fink, 1994; Hillebrand et al., 1998). AtNIT1, AtNIT2, and AtNIT3 are able to convert IAN to IAA. For AtNIT2 and AtNIT3, a specific role has been suggested for IAA biosynthesis in breaking seed dormancy (Vorwerk et al., 2001) and under sulfurdeficient conditions (Kutz et al., 2002), respectively. IAN is not a substrate for Arabidopsis NIT4, but this enzyme shows high activity toward β-cyano-Ala, the intermediate of cyanide detoxification (Piotrowski et al., 2001). The Arabidopsis mutant nit1 is insensitive to exogenously applied IAN but is morphologically normal (Normanly et al., 1997). Transgenic tobacco (Nicotiana tabacum) expressing Arabidopsis NIT2 gained sensitivity to IAN (Schmidt et al., 1996). However, in Arabidopsis, IAN is implicated in the metabolism of indole glucosinolates in addition to auxin biosynthesis (Bak et al., 2001). Therefore, studying the role of nitrilases in IAA biosynthesis in a plant species lacking indole glucosinolates, e.g. maize, is meaningful.
As maize kernels accumulate high amounts of IAA conjugates (Epstein et al., 1980), they are a suitable model system for biochemical analysis of IAA biosynthesis. In this paper, we report the cloning of two nitrilase genes, ZmNIT1 and ZmNIT2. ZmNIT2 converts IAN to IAA seven to 20 times more efficiently than Arabidopsis NIT1, NIT2, and NIT3 (Vorwerk et al., 2001). The expression of ZmNIT1 and ZmNIT2 in maize kernels, together with the presence of the substrate IAN, implicates a role for nitrilases in auxin biosynthesis.
RESULTS
Two genes encoding nitrilases were isolated from maize, ZmNIT1 (GenBank accession no. AY156979) and ZmNIT2 (GenBank accession no. AY156978). ZmNIT1 was mapped (Burr and Burr, 1991) on chromosome 5L (approximately 163) and ZmNIT2 was located on chromosome 4L (approximately 111). ZmNIT1 and ZmNIT2 encode 351 and 361 amino acid residues, respectively. The two amino acid sequences are 75% identical (Table I). The maize nitrilases show about 60% amino acid identity to Arabidopsis NIT1, NIT2, and NIT3, and about 69% identity with AtNIT4 (Table I). Maize nitrilase 2 revealed especially high degrees of amino acid identity (89.9%) with rice (Oryza sativa) nitrilase (Table I). A cysteine residue located in the active site (Kobayashi et al., 1993) is conserved in both maize nitrilases.
Table I.
Homology of ZmNIT1 and ZmNIT2 to nitrilases from plants
| ZmNIT1 | ZmNIT2 | |
|---|---|---|
| % | ||
| ZmNIT1 (maize) | 75.0 | |
| ZmNIT2 | 75.0 | |
| AtNIT1 (Arabidopsis) | 59.0 | 59.5 |
| AtNIT2 | 61.1 | 62.8 |
| AtNIT3 | 59.0 | 60.1 |
| AtNIT4 | 69.2 | 69.3 |
| NtNIT4 (tobacco) | 71.9 | 78.1 |
| OsNIT4 (rice) | 75.5 | 89.9 |
Analysis of ZmNIT1 and ZmNIT2 Gene Expression by Quantitative Real-Time PCR
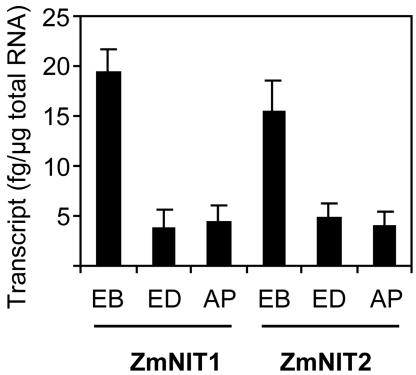
RNA was extracted from isolated embryo, endosperm, and aleurone/pericarp of maize kernels. The expression of ZmNIT1 and ZmNIT2 in maize kernels was quantitatively examined by real-time PCR of cDNA (Fig. 1). The kernel-specific expression pattern was similar for ZmNIT1 and ZmNIT2; the steady-state transcription levels were highest in embryo tissue and increased until 5 weeks after pollination (data not shown).
Figure 1.
Real-time PCR quantification of ZmNIT1 and ZmNIT2 transcript levels in kernels 5 weeks after pollination. Experiments were repeated three times and data are shown as means ± sd. The values indicate the transcript level in femtogram per micrograms of total RNA. EB, Embryo; ED, endosperm; AP, aleurone/pericarp.
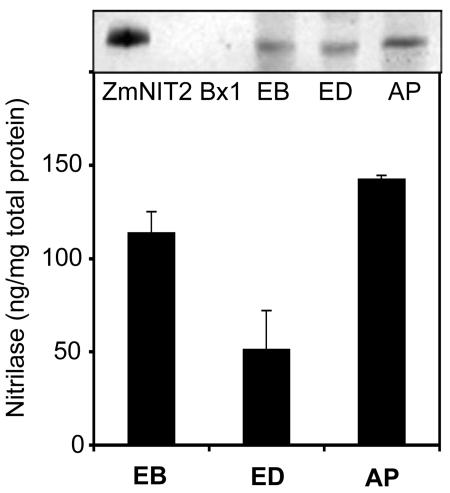
To examine the nitrilase protein expression, we carried out immunoblot analyses with protein extracts of maize kernels (5 weeks after pollination). Anti-ZmNIT1 antibodies recognizing both nitrilases detected strong signals in the aleurone/pericarp layers and in the embryo, and a weaker signal in the endosperm (Fig. 2). Such western signals were not visible in the analysis using preimmune serum. This pattern of tissue-specific expression was further supported by immunoblot tests with kernel tissue prints (data not shown).
Figure 2.
Immunoblot analysis of nitrilase proteins in maize kernels 5 weeks after pollination. Western blots of 80 μg of protein from each fraction of maize kernels were probed with diluted (1:400) anti-ZmNIT1 antibodies recognizing both maize nitrilases. ZmNIT2 (30 ng) and Bx1 (30 ng), a Trp synthase α homolog involved in DIMBOA biosynthesis (Frey et al., 1997) as a negative control, are indicated. The bar graph represents data of four independent western blots.
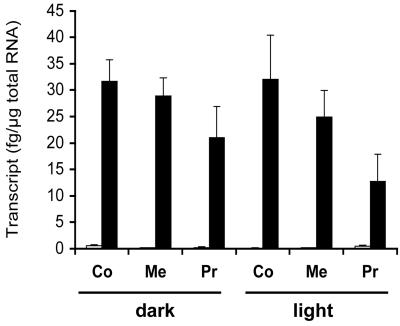
ZmNIT1 and ZmNIT2 expression was also determined in seedlings. In primary root tip 2, 4, and 7 d after germination, ZmNIT2 transcript levels of 4.0 ± 0.4, 15 ± 0.4, and 9.6 ± 0.6 fg μg–1 total RNA, respectively, were detected, whereas ZmNIT1 expression was very low (0.21 ± 0.09, 0.26 ± 0.08, and 0.28 ± 0.03 fg μg–1 total RNA, respectively). As ZmNIT2 mRNA expression was highest 4 d after germination, ZmNIT1 and ZmNIT2 expression was determined at this stage in different tissues of the seedling (Fig. 3). The same expression pattern was observed in light- and dark-grown seedlings.
Figure 3.
Real-time PCR quantification of ZmNIT1 (white bars) and ZmNIT2 (black bars) transcript levels in light- or dark-grown seedlings 4 d after germination. The values indicate the transcript level in femtogram per micrograms of total RNA; n = 3. Co, Coleoptile tip; Me, mesocotyl; Pr, primary root tip.
Characterization of in Vitro Enzyme Activities and Substrate Specificities of Heterologously Expressed Nitrilases
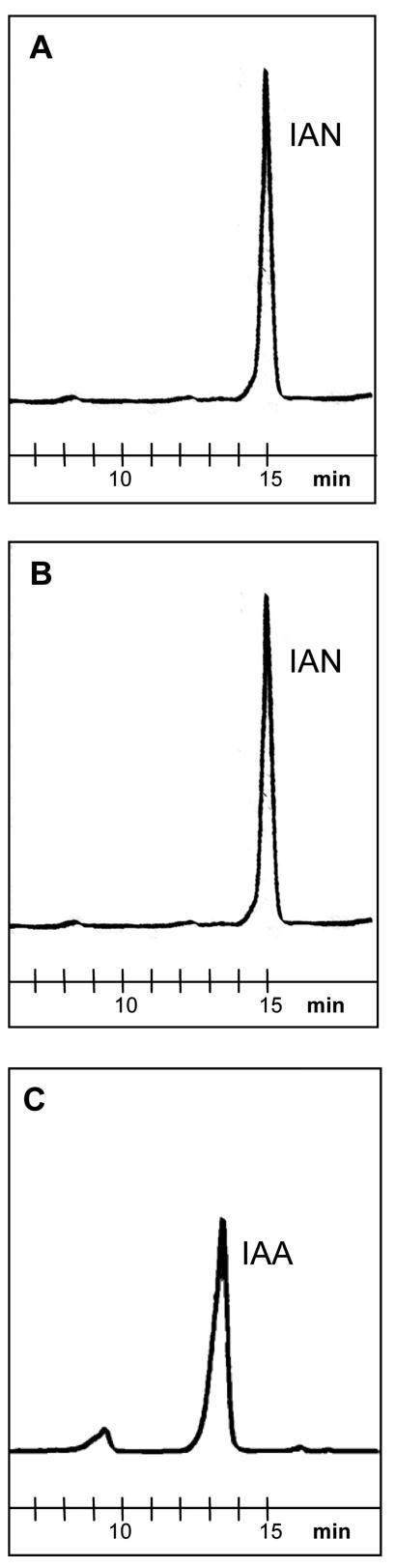
By reverse-phase HPLC, we measured the in vitro enzyme activity of ZmNIT1 and ZmNIT2 toward IAN as substrate using bacterial crude extracts containing heterologously expressed nitrilases. ZmNIT2 efficiently converted IAN to IAA (Fig. 4C). In addition, indole-3-acetamide was formed as a side product. ZmNIT1 revealed only little activity (Fig. 4B) that was not changed in the absence of His-tag (data not shown). Bacterial crude extract containing only empty vectors did not convert IAN to IAA (Fig. 4A).
Figure 4.
HPLC analyses for nitrilase activity toward IAN after 3 h of incubation. Reaction mixtures (100 μL) consisted of 50 mm potassium phosphate buffer (pH 8.0), 1 mm IAN, and bacterial crude extract (50 μg protein) containing no nitrilase (A), heterologously expressed ZmNIT1 (B), or ZmNIT2 (C).
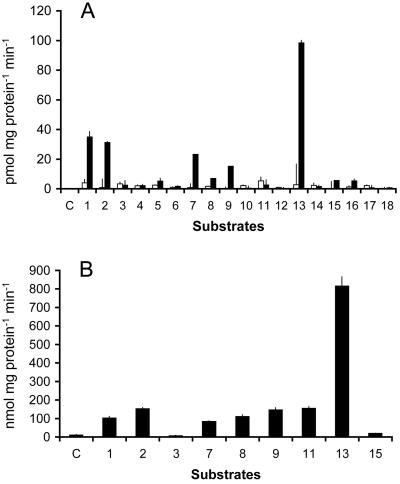
Nitrilase activity was confirmed (Fig. 5A) by the quantification of ammonia released during the reaction (Vorwerk et al., 2001). This simple method for ammonia determination (Bolleter et al., 1961; Vorwerk et al., 2001) made it possible to test nitrilase activities toward 18 nitrile compounds (Fig. 5A) previously tested for Arabidopsis nitrilases (Piotrowski et al., 2001; Vorwerk et al., 2001) or suggested as substrates for plant nitrilases (Thimann and Mahadevan, 1964b). ZmNIT2 showed high activities toward IAN, 3-phenylpropionitrile, allylcyanide, methylthioacetonitrile, and 4-phenylbutyronitrile, which was hydrolyzed most rapidly. In spite of the high homology of ZmNIT2 and Arabidopsis NIT4 (Table I), ZmNIT2 did not show any activity toward β-cyano-Ala, the substrate that implicated a function of AtNIT4 in cyanide detoxification (Piotrowski et al., 2001). ZmNIT1 was hardly active toward the tested nitrile compounds. To examine the substrate specificity of ZmNIT2 in more defined conditions, we carried out in vitro enzyme assays with purified recombinant enzyme (Fig. 5B) and the substrates actively converted by the raw extracts. This assay confirmed that ZmNIT2 is active toward IAN but is inactive toward β-cyano-Ala. For ZmNIT2 with IAN, 3-phenylpropionitrile, and 4-phenylbutyronitrile as substrates, a broad pH optimum with a maximum at pH 7.5 to pH 8.0 was determined. Similar temperature dependency of ZmNIT2 activity with an optimum at approximately 40°C was determined with IAN as well as with 3-phenylpropionitrile. NH4+ formation from 4-phenylbutyronitrile peeked at 25°C. The substrate inhibition by IAN higher than 3 mm reported in Arabidopsis (Vorwerk et al., 2001) was not observed with ZmNIT2 (data not shown).
Figure 5.
Nitrilase activities toward diverse nitrile compounds with bacterial crude extract (50 μg of protein) containing heterologously expressed nitrilases (A; ZmNIT1[white bars]; ZmNIT2 [black bars]) or with 2 μg of purified ZmNIT2 (B). Data originate from at least five measurements and are presented as mean ± se. Background values obtained in the presence of boiled proteins were subtracted from the measurements. C, Control without substrate; S1, IAN; S2, 3-phenylpropionitrile; S3, β-cyano-Ala; S4, 2-cyanopyridine; S5, 3-cyanopyridine; S6, 4-cyanopyridine; S7, allylcyanide; S8, phenylthioacetonitrile; S9, methylthioacetonitrile; S10, 3-hydroxypropionitrile; S11, 3-aminopropionitrile; S12, indole-3-carbonitrile; S13, 4-phenylbutyronitrile; S14, 2-chloroacetamide; S15, cyanoacetamide; S16, cyclopropanecarbonitrile; S17, benznitrile; S18, propionitrile.
To allow direct comparison with nitrilases from Arabidopsis (Table II), the kinetic parameters of ZmNIT2 were determined at 30°C, pH 8.0, and revealed a KM value of 4.1 mm and Vmax of 286 nmol mg–1 protein min–1 for IAN (Table II). For 4-phenylbutyronitrile, ZmNIT2 revealed a higher turnover rate (Vmax = 2,000 nmol mg–1 min–1) but also a higher KM value (13.2 mm).
Table II.
Kinetic parameters of ZmNIT2 with its preferred substrates and of Arabidopsis nitrilases with IAN as substrate (Vorwerk et al., 2001)
KM and Vmax were determined at 30°C, pH 8.0.
| Nitrilase | Substrate | KM | Vmax |
|---|---|---|---|
| mm | nmol mg-1 min-1 | ||
| ZmNIT2 | IAN | 4.1 | 286 |
| 3-Phenylpropionitrile | 4.3 | 345 | |
| 4-Phenylbutyronitrile | 13.2 | 2,000 | |
| AtNIT1 | IAN | 11.1 | 38 |
| AtNIT2 | IAN | 7.4 | 18 |
| AtNIT3 | IAN | 30.1 | 15 |
Test of Endogenous Nitrilase Activity with Protein Extracts from Maize Kernels
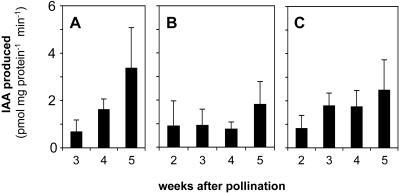
Endogenous nitrilase activity converting IAN to IAA was tested with protein extracts from embryo, endosperm, and aleurone/pericarp of maize kernels (Fig. 6). Nitrilase activity was observed in all the protein samples. Four weeks after pollination, higher activities of nitrilase were determined in embryo (1.62 ± 0.45 pmol mg–1 protein min–1, Fig. 6A) and aleurone/pericarp (1.74 ± 0.70 pmol mg–1 protein min–1, Fig. 6C) than in endosperm (0.77 ± 0.30 pmol mg–1 protein min–1, Fig. 6B). The activity in embryo increased about 5-fold from the 3rd to the 5th week after pollination. IAA was also detected in boiled controls for all the tests, but the level was much lower than that obtained with active proteins (data not shown). All of the data were corrected for the IAA amount observed in the boiled controls that might be generated by nonspecific conversion of IAN to IAA (Ilic et al., 1996) or by release from IAA-protein conjugates (Walz et al., 2002) during boiling.
Figure 6.
Nitrilase activity converting IAN to IAA with maize protein extracts from embryo (A), endosperm (B), and aleurone/pericarp (C). For each test, 40 μg of total protein was included in 250 μL of reaction buffer (pH 8.0) containing 1 mm IAN as substrate. Values show the produced IAA after 3 h determined by fluorescence emission at 360 nm with excitation at 285 nm in HPLC analysis. Nonspecific conversion was corrected, and data are presented as means ± se, n = 4. As proven by t test, the endogenous activity is significantly higher in embryo than in endosperm and significantly increases between weeks 3 and 4.
IAN Is a Natural Compound in Maize Kernels
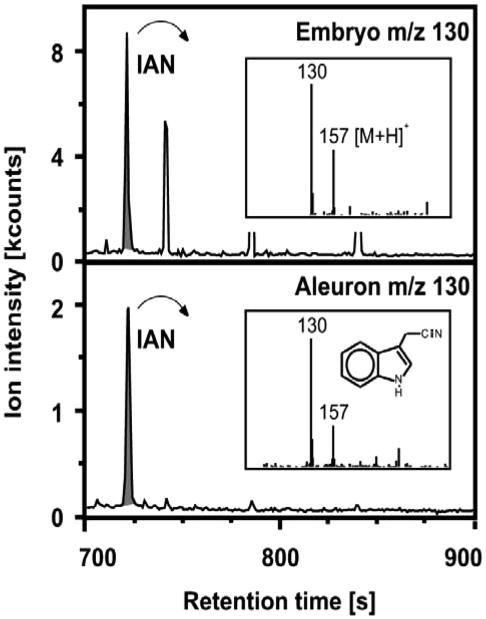
To evaluate the role of maize nitrilases in the conversion of IAN to IAA in vivo, we tested whether the substrate IAN was actually present in maize kernels. Extracts from subfractions of maize kernels were subjected to gas chromatography (GC)/mass spectrometry (MS), and IAN was identified by retention time, molecular mass, and typical fragmentation pattern (Müller and Weiler, 2000a). IAN was unambiguously detected in maize kernels, as exemplified for embryo and aleurone/pericarp fraction 5 weeks after pollination (Fig. 7), and was quantified in the different kernel fractions 4 and 5 weeks after pollination (Table III). IAN has also been detected in seedlings (4 d after germination). Coleoptiles contain approximately 100 pmol g–1 fresh weight (16 ng), and primary roots contain approximately 60 pmol g–1 fresh weight (9 ng) (Table III).
Figure 7.
Detection of endogenous IAN by GC-MS in methanolic extracts of embryo and aleurone/pericarp tissue.
Table III.
GC-MS measurement of IAN concentration in different maize kernel and seedling tissues (w.a.p., Weeks after pollination; d.a.g., Days after germination)
| Tissue | Age | IAN concentration |
|---|---|---|
| pmol g-1 fresh weight | ||
| Endosperm | 4 w.a.p. | 24 ± 3 |
| Aleurone/Pericarp | 4 w.a.p. | 71 ± 10 |
| Embryo | 4 w.a.p. | 99a |
| Endosperm | 5 w.a.p. | 24 ± 7 |
| Aleurone/Pericarp | 5 w.a.p. | 73 ± 27 |
| Embryo | 5 w.a.p. | 347 ± 46 |
| Coleoptile | 4 d.a.g. | 100a |
| Primary root | 4 d.a.g. | 60 ± 21b |
n = 3. a n = 1. b n = 2.
DISCUSSION
Two nitrilase genes, ZmNIT1 and ZmNIT2, were identified in maize. The chromosomal regions where the maize nitrilases are located are syntenic and probably result from ancient tetraploidization (Gale and Devos, 1998). Both genes map relatively close to the centromere. In kernel, the expression pattern of ZmNIT1 and ZmNIT2 is similar (see below). The in vitro enzyme activity tests revealed only little activity of ZmNIT1 (Figs. 4 and 5). However, this does not exclude the possibility that ZmNIT1 is functional in planta but might require a specific reaction environment, e.g. a protein partner to form a complex as suggested for a putative IAA synthase by Müller and Weiler (2000b).
Nitrilase Expression in Maize Kernels
Quantitative real-time PCR (Fig. 1) revealed the expression of ZmNIT1 and ZmNIT2 in maize kernels where Trp-dependent auxin biosynthesis prevails (Glawischnig et al., 2000). The expression of ZmNIT2 was higher in embryo (16 ± 3 fg μg–1 total mRNA) than in endosperm (5 ± 1 fg μg–1) and aleurone/pericarp (4 ± 2 fg μg–1) 4 weeks after pollination. However, the nitrilase protein distribution pattern (Fig. 2) was slightly different from the pattern of transcript accumulation (Fig. 1). In comparison with endosperm (51 ± 26 ng mg–1 total protein), a relatively high nitrilase protein concentration was detected in embryo (114 ± 14 ng mg–1) but also in aleurone/pericarp (143 ± 3 ng mg–1). Differences in translation efficiency or protein stability in the different tissues might account for this observation. In maize endosperm where IAA accumulates (Epstein et al., 1980), nitrilase expression and enzymatic activity were lower than those in embryo and aleurone/pericarp (Figs. 1, 2, and 6). This could indicate that IAA is synthesized in embryo and aleurone/pericarp tissue, where nitrilase activity is high, and transported to the endosperm.
Is ZmNIT2 Involved in IAA Biosynthesis?
ZmNIT2 shows high activity with several substrates in vitro, including IAN (Fig. 5). According to their kinetic parameters, ZmNIT2 hydrolyzes IAN to IAA more efficiently than AtNIT1, AtNIT2, and AtNIT3 (Table II). This property and the fact that ZmNIT2 is expressed in the maize kernel where the substrate IAN is present (Figs. 1, 2, and 7) suggest that ZmNIT2 has the capacity to synthesize IAA in maize kernels. The Arabidopsis nitrilases, especially the comparatively highly expressed AtNIT1, could be involved in indole glucosinolate metabolism in addition to IAA biosynthesis (for review, see Glawischnig et al., 2003). This complication is avoided here because there are no indole glucosinolates present in maize (Fahey et al., 2001).
As expected for a biosynthetic intermediate, the concentration of IAN in maize kernels is relatively low. It ranges from 24 pmol (3.7 ng) g–1 fresh weight in endosperm to 347 pmol (54 ng) g–1 fresh weight in embryo, 5 weeks after pollination (Table III), which is one order of magnitude lower than that reported for Arabidopsis seedlings (Normanly et al., 1997). In consequence, ZmNIT2 has to act efficiently in spite of its high apparent KM value determined in vitro, if IAN, present only in low overall concentrations, is its natural substrate. If maize nitrilase is a component of an IAA-synthase complex as suggested for Arabidopsis (Müller and Weiler, 2000b), then local concentration of IAN in the protein complex might be high enough or the KM value in the complex might be much lower. The hypothesized involvement of other factors in nitrilase action in vivo is supported by low binding constants of nitrilases partially purified from plant tissue such as barley (Hordeum vulgare) leaves (51 μm; Thimann and Mahadevan, 1964a) or Chinese cabbage (Brassica campestris) seedlings (0.52 mm; Rausch and Hilgenberg, 1980) in comparison with heterologously expressed nitrilases. Further evidence comes from the observation that in Arabidopsis seedlings, the addition of just 30 μm IAN to the growth medium induces a strong auxin phenotype, which is significantly reduced if AtNIT1 is mutated (Normanly et al., 1997). This shows that AtNIT1 efficiently converts IAN to IAA in vivo, despite its high apparent KM value of 11.1 mm (Vorwerk et al., 2001).
Jensen and Bandurski (1994) calculated a rate of IAA biosynthesis in maize kernels of 0.19 μg IAA g–1 h–1 for total kernel and 0.0086 μg IAA g–1 h–1 for isolated endosperm (20–30 d after pollination). It can be estimated whether the observed nitrilase activity is sufficient for this biosynthetic rate. Because we cannot distinguish ZmNIT1 and ZmNIT2 immunologically, the total nitrilase enzyme activity per fresh weight was calculated from the specific activity (Fig. 6) per total protein. In conclusion, the maize nitrilases have the potential to synthesize up to 0.23 μg IAA g–1 h–1 in total kernel and 0.13 μg IAA g–1 h–1 in endosperm under the assumption that the substrate IAN is channeled, as suggested above.
ZmNIT2 mRNA was also detected in maize seedlings, where it is expressed in coleoptile, mesocotyl, and primary root (Fig. 3). This expression is not light inducible. Although ZmNIT2 mRNA is also expressed 7 d after germination, the amount of nitrilase protein rapidly decreased with seedling age (data not shown), indicating that nitrilase expression in seedling is partly regulated at the level of protein expression or stability. The presence of IAN in seedlings opens the possibility of a Trp-dependent IAA biosynthetic pathway via IAN in maize seedlings. However, in contrast to kernels, here, the contribution of Trp-dependent IAA biosynthesis is questioned (Östin et al., 1999). In addition, Jensen and Bandurski (1996) conclude from incorporation experiments with D2O that in this tissue, there is no de novo biosynthesis at all or, alternatively, is very low in comparison with IAA transport from the kernel. Therefore, the role of nitrilase in seedlings remains unclear, and alternative functions for ZmNIT2 have to be considered. There are no reports on the presence of other nitriles in maize that could serve as substrates for ZmNIT2, obscuring unknown biosynthetic or degradative pathways ZmNIT2 may be involved in addition to IAA biosynthesis. Therefore, the analysis of knockout mutants rather than a biochemical approach will be helpful to evaluate the function of maize nitrilases in IAA biosynthesis in vivo. Specific mutants may also reveal alternative biological functions. As ZmNit1 expression is low in seedlings, the analysis of a ZmNit2 knockout mutant will give particular insight in the function of ZmNIT2 in seedlings.
Biosynthetic Origin of IAN in Maize
The enzymes potentially involved in IAN biosynthesis in maize are unknown. CYP79B homologs converting Trp to indole-3-acetaldoxime (IAOx; Hull et al., 2000; Mikkelsen et al., 2000), a precursor of IAN in vitro (Helminger et al., 1985), have not been identified in maize. Alternatively, IAOx could be synthesized by a peroxidase (Ludwig-Müller and Hilgenberg, 1988). The analysis of a potential pathway of IAOx formation in maize involving a flavin-dependent monooxygenase (YUCCA; Zhao et al., 2001) remains speculative, as no close homolog of YUCCA has been identified in maize.
Evolution of Plant Nitrilases
When compared with Arabidopsis nitrilases, ZmNIT2 shows highest homology to AtNIT4. AtNIT4 and ZmNIT2 share an identity of 69.3%, whereas ZmNIT2 and the other Arabidopsis nitrilases share an identity of about 60% (Table I). This observation is in contrast to the substrate specificities associated with these enzymes. Arabidopsis NIT4 is not active with IAN as substrate but reveals activity toward β-cyano-Ala and was therefore suggested to be involved in cyanide detoxification (Piotrowski et al., 2001), whereas ZmNIT2 converts IAN to IAA and shows no activity with β-cyano-Ala (Fig. 5). Therefore, nitrilase substrate specificities cannot be predicted on the basis of protein similarity, when enzymes from monocots and dicots are compared. In conclusion, nitrilases of the AtNIT4 subfamily have been recruited for different metabolic functions during evolution.
MATERIALS AND METHODS
Plant Material
Maize (Zea mays) cultivar Landmark (a sweet corn) was grown on a local field in the vicinity of Munich, and the ears were harvested 2 to 5 weeks after the pollination. The material was deep frozen and kept at –70°C.
Gene Cloning and Characterization
Total RNA was isolated from kernels (Wessler, 1994) and seedlings. Expressed sequence tags (ESTs) coding for nitrilases were identified in a Pioneer Hi-Bred EST database and the sequence was used to amplify specific probes by PCR, which were used to screen an endosperm (5–25 d after pollination) cDNA library (maize cv Line C). Two cDNA clones, containing all of the initial EST sequences, were identified and the corresponding genes were designated ZmNIT1 and ZmNIT2. The completeness of the ZmNIT1 cDNA was confirmed by RACE (Roche, Indianapolis). The genes were sequenced and mapped using a recombinant inbred mapping population (Burr and Burr, 1991).
Quantification of Nitrilase Transcript by Real-Time PCR
Real-time PCR primers specific to ZmNIT1 (forward, GACGATGACTATGTGCAGACCTAA; reverse, CAATCTCGTCCAATCCATGTATA) and to ZmNIT2 (forward, AGCTGCCAAGAGTGATATCGATACTAAG; reverse, CACAAGGAACATAACTGCGGCC) were designed based on the 3′ sequences of ZmNIT1 and ZmNIT2. Q-solution (Qiagen, Valencia, CA) was included for the PCR of ZmNIT2 to increase specificity. For real-time PCR, total RNA was isolated from embryo, endosperm, and aleurone/pericarp of maize kernels with a RNA purification kit (NucleoSpin RNA Plant; Macherey & Nagel, Düren, Germany) and 2 μg of the total RNA was reverse transcribed with a cDNA synthesis kit (Taq-Man; Roche). Quantitative real-time PCR was carried out using the LightCycler (Roche), and the increase of PCR product was monitored using Syb®Green. The mRNA level of ZmNIT1 and ZmNIT2 was automatically calculated based on the standard curve obtained with the plasmid clones in the same batch of PCR. pBluescript (Stratagene, La Jolla, CA) transcripts were added to the realtime PCR as an internal standard, and their amplification levels were examined.
Heterologous Expression of Nitrilases and Purification
The coding regions of ZmNIT1 and ZmNIT2 were cloned into a pET3a expression vector containing a 6-His-tag (Novagen, Madison, WI). As the cDNA sequence of ZmNIT2 coding N terminus was incomplete, the missing part was introduced from the genomic clone of ZmNIT2. Sequencing of cDNA confirmed the endogenous expression of the 5′ sequences. ZmNIT1 and ZmNIT2 proteins were heterologously expressed in Escherichia coli BL21(DE3).
Bacterial cells were harvested by centrifugation at 8,000g for 8 min, resuspended in 50 mm potassium buffer (pH 8.0), and treated with lysozyme (1 mg mL–1) for 30 min on ice. The resulting lysate was sonicated three times at 200 W for 10 s and was centrifuged at 10,000g for 30 min at 4°C. The supernatant was used for enzyme assay (crude extract).
ZmNIT2 containing a C-terminal 6-His-tag was purified under native conditions using a Ni-NTA column (Qiagen). ZmNIT1 was purified under denaturing conditions with 8 m urea. After elution from the Ni-NTA column, pure ZmNIT1 was obtained by preparative SDS-PAGE (PrepCell; Bio-Rad, Hercules, CA). Purified enzymes were used for enzyme activity tests and immunization of rabbits (Eurogentec, Brussels, Belgium).
Western Analyses
Deep-frozen plant materials were ground with mortar and pestles, suspended in extraction buffer (20 mm Tris, 140 mm NaCl, 1 mm EDTA, and 1 mm PMSF, pH 7.5), and cleared by centrifugation at 10,000g for 30 min. The crude protein in the supernatant was (NH4)2SO4 (75%, w/w) precipitated, redissolved, separated on a 10% (v/v) SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with nitrilase-specific antibodies. The membrane was further incubated with anti-rabbit immunoglobulin G conjugated with Cy5 (Amersham Pharmacia Biotech, Piscataway, NJ), and the signal was detected with a phosphoimager (Storm 860; Amersham Pharmacia Biotech) using a red fluorescence filter.
Enzyme Activity Tests with Recombinant Maize Nitrilases
The nitrilase activity of recombinant ZmNIT1 and ZmNIT2 converting IAN to IAA in bacterial crude extract was examined by IAA quantification based on a reverse-phase C18 HPLC. The reaction mixture (50 mm potassium phosphate buffer, pH 8.0, 1 mm IAN, and 50 μg of boiled or active proteins, final volume of 100 μL) was incubated for 3 h at 30°C. Reactions were stopped by the addition of 10% (v/v) acetic acid, and the mixture was immediately partitioned three times with 100 μL of ethyl acetate. The recovered organic phase was evaporated in a speed-vac (H.-Saur, Reutlingen, Germany), and the dried substances were dissolved in 100% (v/v) methanol and analyzed on a HPLC with a reverse-phase column (LiChroCART 125-4, RP-18, 5 μm; Merck, West Point, PA). The mobile phase was delivered at a flow rate of 1 mL min–1 with an initial mixture of 25% (v/v) methanol in 10% (v/v) acetic acid for 2 min and was followed by a 12-min linear gradient to 70% (v/v) methanol. The elution profile was monitored with a photodiode array detector (Detector 158; Beckman Instruments, Fullerton, CA). The peaks eluted at 9.2, 13.0, and 15.1 min were identified as indole-3-acetamide, IAA, and IAN, respectively, by comparison with the standard substances with respect to retention time and UV spectrum.
For determination of substrate specificity and kinetic parameters of recombinant ZmNIT1 and ZmNIT2, nitrilase activity was measured by quantification of ammonia produced during the enzyme action using the Berthelot reaction (Bolleter et al., 1961). Reaction mixtures (1 mL) containing 50 μg of crude protein sample or 5 μg of purified nitrilases and 5 mm substrates indicated were incubated for 2 h for the determination of substrate specificity and for 3 h for the kinetic parameters. A 100-μL-aliquot of the reaction was subjected to Berthelot reaction following Vorwerk et al. (2001). Kinetic parameters were determined in the presence of substrates between 0.5 and 5 mm. A640 was compared with a standard curve obtained with ammonium sulfate. The data were corrected for nonspecific conversion observed in boiled control for each substrate.
Detection of Endogenous Nitrilase Activity with Maize Kernel Extracts
To measure endogenous nitrilase activity, we isolated embryo, endosperm, and aleurone/pericarp extracted in 50 mm potassium phosphate buffer (pH 8.0, 1 mm DTT) and cleared by centrifugation at 17,500g for 30 min. The supernatant was precipitated with 75% (w/v) ammonium sulfate and was centrifuged again. The pellet was resuspended with 1 mL of 50 mm potassium phosphate buffer, pH 8.0, and desalted with a disposable column (NAP-10; Amersham Pharmacia Biotech). The desalted fraction was used for enzyme activity test. Reaction mixture (40 μg of protein, 1 mm IAN, and 50 mm potassium phosphate buffer, pH 8.0, total volume of 250 μL) was incubated at 30°C in the dark for 3 h. The pH was then adjusted higher than 9.5 with 1 m Na2CO3, and the solution was extracted with 400 μL of ethyl acetate. The aqueous lower phase was recovered, and the partitioning procedure was repeated after addition of 200 μL of water. The collected aqueous phase was acidified and partitioned as described for the test with recombinant enzymes. The final sample was separated on an HPLC with a reverse-phase column (LiChroCART 250–4, RP-18, 5 μm; Merck). The mobile phase was delivered at a flow rate of 1.2 mL min–1 with an initial mixture of 25% (v/v) methanol in 10% (v/v) acetic acid for 2 min and followed by a 12-min linear gradient to 70% (w/v) methanol. The elution was monitored with a fluorescence detector (RF-10AXL; Shimadzu, Duisburg, Germany) at the emission wavelength of 360 nm with excitation at 285 nm. The amount of produced IAA was determined by integration of the peak area and was corrected for nonspecific conversion.
Identification and Quantification of IAN
One gram of endosperm, embryo, aleurone/pericarp, coleoptile, and primary root tissue, respectively, was ground in liquid nitrogen and extracted with 5 mL of methanol for 1 h at room temperature and for 5 min at 60°C. The solvent was removed under reduced pressure and the residue was extracted in 500 μL of water, 500 μL of MeOH, and 1 mL of CHCl3. The chloroform fraction was dried and redissolved in 200 μL of 30% (v/v) MeOH in water, sonicated, and centrifuged for 20 min at 100,000g. The supernatant was applied to reverse-phase HPLC using a Luna RP-18 column (250 × 0.4 mm; Phenomenex, Torrance, CA). The column was developed at a flow rate of 1 mL min–1 with a gradient of 10 min isocratic flow at 30% (v/v) aqueous methanol 0.01% (v/v) trifluoroacetic acid, followed by a linear gradient up to 100% (v/v) methanol 0.01% (v/v) trifluoroacetic acid during 10 min. The effluent was monitored photometrically at 280 nm. During separation, fractions of 1 mL were collected and the retention volume of IAN (20.5 mL) was determined afterward using 0.5 mm solution. MeOH was removed from the pooled IAN-containing fractions and the remaining water phase was extracted with EtOAc. The solvent was removed under reduced pressure and the residue was redissolved in 10 μL of CHCl3. One microliter was analyzed by GC-MS, as described in Müller and Weiler (2000a).
Acknowledgments
We thank Dr. Benjamin Burr for processing mapping data, Prof. Udo Wienand for providing an endosperm-specific library, Prof. Elmar W. Weiler, Dr. Monika Frey, and Dr. Ulrich Genschel for critical discussions, and Laura Helming for excellent help in isolating the cDNAs.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026609.
This work was supported by the Deutsche Forschungsgemeinschaft Schwerpunktprogramm 1067.
References
- Bacher A, Rieder C, Eichinger D, Arigoni D, Fuchs G, Eisenreich W (1999) Elucidation of novel biosynthesis pathways and metabolite flux patterns by retrobiosynthetic NMR analysis. FEMS Microbiol Rev 22: 567–598 [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandurski RS, Cohen JD, Slovin J, Reinecke DM (1995) Auxin biosynthesis and metabolism. In JP Davies, ed, Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 39–65
- Bartel B, Fink GR (1994) Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA 91: 6649–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugation hydrolysis, and indole-3-butyric acid β-oxidation. J Plant Growth Regul 20: 198–216 [Google Scholar]
- Bartling D, Seedorf M, Mithoefer A, Weiler EW (1992) Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur J Biochem 205: 417–424 [DOI] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler E (1994) Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc Natl Acad Sci USA 91: 6021–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolleter WT, Bushman CJ, Tidwell PW (1961) Spectrophotometric determination of ammonia as indophenol. Anal Biochem 33: 592–594 [Google Scholar]
- Burr B, Burr FA (1991) Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. Trends Genet 7: 55–60 [DOI] [PubMed] [Google Scholar]
- Epstein E, Cohen JD, Bandurski RS (1980) Concentration and metabolic turnover of indoles in germinating kernels of Zea mays L. Plant Physiol 65: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Zalemann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51 [DOI] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winkelmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP et al. (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Plant comparative genetics after 10 years. Science 282: 656–659 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Mikkelsen MD, Halkier BA (2003) Glucosinolate biosynthesis and metabolism. In Y Abrol, A Ahmad eds, Sulphur in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 145–162
- Glawischnig E, Tomas A, Eisenreich W, Spiteller P, Bacher A, Gierl A (2000) Auxin biosynthesis in maize kernels. Plant Physiol 123: 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helminger J, Rausch T, Hilgenberg W (1985) Metabolism of 14C-indole-3-acetaldoxime by hypocotyls of Chinese cabbage. Phytochemistry 24: 2497–2502 [Google Scholar]
- Hillebrand H, Bartling D, Weiler EW (1998) Structural analysis of the NIT2/NIT1/NIT3 gene cluster encoding nitrilases, enzymes catalyzing the terminal activation step in indole-acetic acid biosynthesis in Arabidopsis thaliana. Plant Mol Biol 36: 89–99 [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Normanly J, Cohen JD (1996) Quantification of free plus conjugated indole-acetic acid in Arabidopsis requires correction for the nonenzymatic conversion of indolic nitriles. Plant Physiol 111: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Bandurski RS (1994) Metabolism and synthesis of indole-3-acetic acid (IAA) in Zea mays (levels of IAA during kernel development and the use of in vitro endosperm systems for studying IAA biosynthesis). Plant Physiol 106: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Bandurski RS (1996) Incorporation of deuterium into indole-3-acetic acid and tryptophan in Zea mays seedlings grown on 30% deuterium oxide. J Plant Physiol 147: 697–702 [Google Scholar]
- Kobayashi M, Izui H, Nagasawa T, Yamada H (1993) Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc Natl Acad Sci USA 90: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Kamiya Y, Iino M (1995) Biosynthesis of indole-3-acetic acid from l-tryptophan in coleoptile tips of maize (Zea mays L.). Plant Cell Physiol 36: 1503–1510 [Google Scholar]
- Kutz A, Müller A, Peter H, Kaiser WM, Piotrowski M, Weiler EM (2002) A role for nitrilase 3 in the regulation of root morphology in sulfur starving Arabidopsis thaliana. Plant J 30: 95–106 [DOI] [PubMed] [Google Scholar]
- Luckwill LC (1952) Application of paper chromatography to the separation and identification of auxins and growth inhibitors. Nature 169: 375. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Hilgenberg W (1988) A plasma membrane-bound enzyme oxidases l-tryptophan to indole-3-acetaldoxime. Physiol Plant 74: 240–250 [Google Scholar]
- Michalczuk L, Ribnicky DM, Cooke TJ, Cohen JD (1992) Regulation of indole-3-acetic acid biosynthesis pathways in carrot cell cultures. Plant Physiol 100: 1346–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275: 33712–33717 [DOI] [PubMed] [Google Scholar]
- Müller A, Hillebrand H, Weiler EW (1998) Indole-3-acetic acid is synthesized from l-tryptophan in roots of Arabidopsis thaliana. Planta 206: 362–369 [DOI] [PubMed] [Google Scholar]
- Müller A, Weiler EW (2000a) Indolic constituents and indole-3-acetic acid biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana. Planta 211: 855–863 [DOI] [PubMed] [Google Scholar]
- Müller A, Weiler EW (2000b) IAA synthase, an enzyme complex from Arabidopsis thaliana catalyzing the formation of indole-3-acetic acid from (S)-tryptophan. Biol Chem 381: 679–686 [DOI] [PubMed] [Google Scholar]
- Normanly J, Bartel B (1999) Redundancy as a way of life: IAA metabolism. Curr Opin Plant Biol 2: 207–213 [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen J, Fink GR (1993) Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA 90: 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B (1997) Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östin A, Ilic N, Cohen JD (1999) An in vitro system from maize seedlings for tryptophan-independent indole-3-acetic acid biosynthesis. Plant Physiol 119: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M, Schönfelder S, Weiler EW (2001) The Arabidopsis thaliana NIT4 and its orthologs in tobacco β-cyano-l-alanine hydratase/nitrilase. J Biol Chem 276: 2616–2621 [DOI] [PubMed] [Google Scholar]
- Rapparini F, Cohen JD, Slovin JP (1999) Indole-3-acetic acid biosynthesis in Lemna gibba studied using stable isotope labeled anthranilate and tryptophan. Plant Growth Regul 27: 139–144 [Google Scholar]
- Rausch T, Hilgenberg W (1980) Partial purification of nitrilase from Chinese cabbage. Phytochemistry 19: 747–750 [Google Scholar]
- Ribnicky DM, Ilic N, Cohen JD, Cooke TJ (1996) The effects of exogenous auxins on endogenous indole-3-acetic acid metabolism: the implications for carrot somatic embryogenesis. Plant Physiol 112: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RC, Müller A, Hain R, Bartling D, Weiler EW (1996) Transgenic tobacco plants expressing the Arabidopsis thaliana nitrilase II enzyme. Plant J 9: 683–691 [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Dohmae N, Takio K, Kamiya Y, Koshiba T (1997) Cloning and molecular characterization of plant aldehyde oxidase. J Biol Chem 272: 15280–15285 [DOI] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T (1998) Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol 116: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Normanly J (1998) Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J Chromatogr A 800: 101–108 [DOI] [PubMed] [Google Scholar]
- Thimann KV, Mahadevan S (1964a) Nitrilase: occurrence, preparation and general mode of action. Arch Biochem Biophys 105: 133–141. [DOI] [PubMed] [Google Scholar]
- Thimann KV, Mahadevan S (1964b) Nitrilase: substrate specificity and possible mode of action. Arch Biochem Biophys 107: 62–68. [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Müller A, Weiler EW, Piotrowski M (2001) Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3 gene cluster. Planta 212: 508–516 [DOI] [PubMed] [Google Scholar]
- Walz A, Park S, Slovin JP, Ludwig-Müller J, Momonoki YS, Cohen JD (2002) A gene encoding a protein modified by the phytohormone indole acetic acid. Proc Natl Acad Sci USA 99: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR (1994) Isolation of RNA from Wx and wx endosperms. In M Freeling, V Walbot, eds, The Maize Handbook. Springer-Verlag, New York, pp 545–546
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD (1991) Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254: 998–1000 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]