Abstract
Using a combination of in vivo and in vitro assays, we characterized the sorting pathway and molecular targeting signal for the Arabidopsis 22-kD peroxisome membrane protein (PMP22), an integral component of the membrane of all peroxisomes in the mature plant. We show that nascent PMP22 is sorted directly from the cytosol to peroxisomes and that it is inserted into the peroxisomal boundary membrane with its N- and C-termini facing the cytosol. This direct sorting of PMP22 to peroxisomes contrasts with the indirect sorting reported previously for cottonseed (Gossypium hirsutum) ascorbate peroxidase, an integral PMP that sorts to peroxisomes via a subdomain of the endoplasmic reticulum. Thus, at least two different sorting pathways for PMPs exist in plant cells. At least four distinct regions within the N-terminal one-half of PMP22, including a positively charged domain present in most peroxisomal integral membrane-destined proteins, functions in a cooperative manner in efficient peroxisomal targeting and insertion. In addition, targeting with high fidelity to peroxisomes requires all four membrane-spanning domains in PMP22. Together, these results illustrate that the PMP22 membrane peroxisomal targeting signal is complex and that different elements within the signal may be responsible for mediating unique aspects of PMP22 biogenesis, including maintaining the solubility before membrane insertion, targeting to peroxisomes, and ensuring proper assembly in the peroxisomal boundary membrane.
Peroxisomes are multifunctional organelles that are generally defined as containing at least one hydrogen peroxide-generating oxidase and catalase. Some peroxisomal functions are virtually universal among evolutionarily diverse organisms such as the β-oxidation of fatty acids and defense against oxidative stresses. Other peroxisomal functions are more specialized and depend upon the organism in which the organelle resides, e.g. key steps in the synthesis of ether-linked phospholipids and bile salts in mammalian peroxisomes, and portions of the glycolate and glycerate pathways of photorespiration in plant leaf and leaf-type peroxisomes. Recent studies have shown that peroxisomes also are involved in the biosynthesis of important signaling molecules, including indole acetic acid (Zolman et al., 2000) and jasmonate (Stintz and Browse, 2001), nitric oxide, and various reactive oxygen species (Corpas et al., 2001) that modulate many aspects of the plant's life cycle. In Arabidopsis, the isolation of mutants with defects in a peroxisomal ATP-binding cassette transporter (Zolman et al., 2001; Footitt et al., 2002; Hayashi et al., 2002) or the peroxisome biogenesis protein factor (a peroxin) Pex2p (Hu et al., 2002) have suggested unexpected links between the organelle and fundamental processes, including the breaking of seed dormancy (Footitt et al., 2002) and light-regulated gene expression (Hu et al., 2002). Although the molecular mechanisms of these and other plant peroxisomal functions remain to be clarified, it is apparent that the peroxisome is a source and sensor of molecules that can affect plant growth and development in profound ways.
The biogenesis of peroxisomes is proposed to take place in three distinct steps: the formation of a “nascent” or “preperoxisomal” vesicle that, depending upon the organism, is thought to be derived from the endoplasmic reticulum (ER) or another endomembrane source, or from a pre-existing peroxisome; and the targeting and insertion/assembly of peroxisome membrane proteins (PMP), which include many of the components required for the targeting and import of matrix proteins (Sacksteder and Gould, 2000; Purdue and Lazarow, 2001; Sparkes and Baker, 2002). Overall, the mechanisms governing matrix protein targeting and import are best understood, although specific differences exist between evolutionary distant organisms. Peroxisomal matrix proteins are synthesized on free polyribosomes in the cytosol and contain one of two different peroxisomal targeting signals (PTS), i.e. the type I PTS (PTS1) consisting of a carboxy-terminal tripeptide motif (-SKL) and the type 2 PTS (PTS2) an amino terminal nonpeptide motif (-R-X6-H/Q-A/L/F-; Mullen, 2002). The PTS1 and PTS2 on nascent matrix proteins are recognized in the cytosol by their cognate receptors, Pex5p and Pex7p, respectively, and the resulting receptor-cargo-protein complexes are targeted to the surface of the peroxisomal boundary membrane. Subsequent docking and translocation of the PTS-bearing cargo, as well as possibly the bound receptor, and then the recycling of the receptor back to the cytosol for additional rounds of targeting and import requires the participation of several other soluble and membrane-bound peroxins.
In comparison with our understanding of the targeting and import of matrix proteins, much less is known about the targeting and insertion/assembly of PMPs. Although it is generally accepted that many PMPs are synthesized in the cytosol and inserted posttranslationally into the peroxisome single boundary membrane, a consensus has not been reached on the functional role(s) for some of the protein components (peroxins) involved in PMP targeting/insertion or what constitutes a prototypic membrane peroxisomal targeting signal (mPTS). PMPs do not possess a PTS1 or PTS2 and, depending upon the protein and/or organism, the mPTS can vary from a short stretch of three to six positively charged amino acids residues to large nonoverlapping segments that do not contain obvious consensus motifs (Subramani et al., 2000; Sparkes and Baker, 2002; Trelease, 2002). In addition, mPTSs characterized seem to be inconsistent in terms of whether they are orientated topologically on the matrix or cytosolic side of the peroxisomal boundary membrane and whether one or more transmembrane domains (TMDs) are required for their proper functioning. Also a matter of question is whether the ER serves as the initial sorting site for at least a subset of PMPs. For example, in tobacco (Nicotiana tabacum) Bright Yellow (BY)-2 suspension culture cells, transiently expressed cottonseed (Gossypium hirsutum) ascorbate peroxidase (APX) targets to a subdomain of the ER before its sorting to pre-existing peroxisomes (Mullen et al., 1999). In Yarrowia lipolyitica, Pex2p and Pex16p are sorted to the ER while en route to peroxisomes (Titorenko and Rachubinski, 1998), whereas no evidence for the delivery of these proteins to peroxisomes via ER could be found in mammalian cells (Voorn-Brouwer et al., 2001). Saccharomyces cerevisiae Pex15p has been reported also to sort to peroxisomes via the ER (Elgersma et al., 1997) but this has been disputed (Hettema et al., 2000). The conflicting data published for different PMPs in different organisms have prevented the formation of a consistent working model on the nature of the mPTS, the sorting pathways used by PMPs, or the manner in which peroxins mediate PMP insertion and assembly in the peroxisomal boundary membrane. This is further exemplified in plants, where a paucity of information on PMPs exists because only a few authentic PMPs have been described, and only one of these, APX, has been characterized in terms of its mPTS and sorting pathways (Mullen et al., 1999, 2000; Nito et al., 2001; Lisenbee and Trelease, 2003).
Arabidopsis PMP22 is an integral membrane protein that is prominent in all organs of the mature plant (Tugal et al., 1999). Related proteins include PMP22 in rat, PMP22, Mpv17 and M-LP in mouse, and PMP22 and Mpv17 in human (Tugal et al., 1999; Iida et al., 2003 and refs. therein). Although the precise molecular function of PMP22 and PMP22-like proteins remains to be elucidated, recent studies with mouse Mpv17 (Wagner et al., 2001) and M-LP (Iida et al., 2003) suggest that they are involved in enzymatic antioxidant defense systems. Studies on the in vitro insertion of PMP22 revealed that the rat and Arabidopsis proteins are inserted into isolated peroxisome membranes (Diestelkötter and Just, 1993; Tugal et al., 1999). Studies of the targeting information in mammalian PMP22 and PMP22-like proteins (Pause et al., 2000; Brosius et al., 2002; Iida et al., 2003) have yielded radically different conclusions on the nature of the mPTS(s), most likely because large deletions were used to determine the targeting signals, a strategy that is unreliable if multiple signals act cooperatively and are distributed throughout the protein.
Here, we describe the results of a comprehensive study of molecular signals involved in the targeting and insertion of Arabidopsis PMP22 in vivo and in vitro. We show that, unlike the sorting of cottonseed APX to peroxisomes via the ER, newly synthesized PMP22 is sorted directly from the cytosol to peroxisomes, and the protein is inserted into the peroxisome boundary membrane with N- and C-terminal parts facing the cytosol. We also demonstrate, using a combination of fusion proteins and modified versions of PMP22 (e.g. site-specific substitutions, internal deletions, and truncations), that at least four distinct regions within PMP22 are required for efficient peroxisomal targeting and integration with high fidelity. Efficient targeting of PMP22 to peroxisomes also requires all four of the protein's TMDs. The implications of these results and nature of the mPTS in Arabidopsis PMP22 are discussed.
RESULTS
Intracellular Sorting and Membrane Insertion of Epitope-Tagged Arabidopsis PMP22
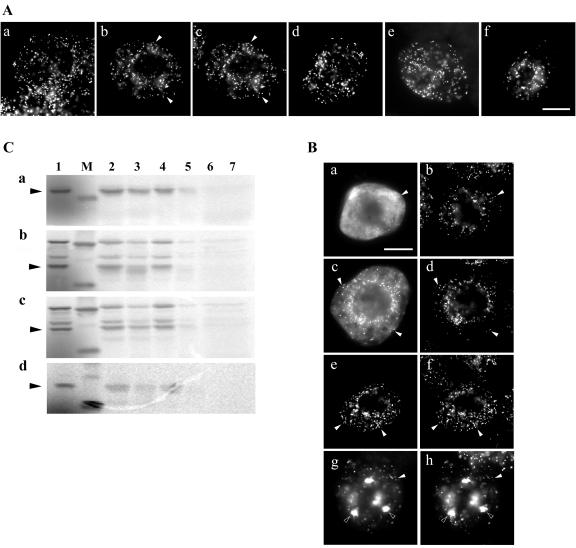
When nontransformed tobacco BY-2 suspension-cultured cells stained with anti-Arabidopsis PMP22 immunoglobulin (Ig) Gs were examined by immunofluorescence microscopy, a punctate fluorescence pattern was observed, characteristic of an antigenic protein, presumably a PMP22, localized to individual peroxisomes (Fig. 1A, a). To distinguish between this endogenous BY-2 PMP22 and ectopically expressed Arabidopsis PMP22, a single copy of the myc epitope tag was fused to the N terminus of Arabidopsis PMP22. Figure 1A (b and c) illustrates that myc-PMP22 was localized exclusively to peroxisomes, as evidenced by its colocalization with the endogenous peroxisomal matrix enzyme catalase. Several other epitope-tagged versions of Arabidopsis PMP22 also localized to BY-2 peroxisomes, including an N-terminal hemagglutinin (HA)-tagged PMP22 (HA-PMP22), C-terminal myc-tagged PMP22 (PMP22-myc), and a double-epitope-tagged version of PMP22 whereby HA and myc epitopes were fused to the N and C terminus of PMP22, respectively (HA-PMP22-myc; Fig. 1A, d–f).
Figure 1.
Peroxisomal targeting and membrane insertion of eptiope-tagged-PMP22s. A, Subcellular localization of endogenous PMP22 and different versions of epitope-tagged Arabidopsis PMP22 in BY-2 cells. Nontransformed (a) or transiently transformed (b-f) BY-2 cells were fixed in formaldehyde, permeabilized with pectolyase and Triton X-100, and incubated in appropriate antibodies. a, Punctate immunofluorescence pattern in nontransformed BY-2 cells incubated with anti-Arabidopsis PMP22 IgGs. b, Transient-expressed myc-PMP22 and endogenous catalase (c) in the same transformed cell; solid arrows indicated obvious colocalizations. Punctate immunofluorescence patterns attributable to expressed HA-PMP22 (d), PMP22-myc (e), and HA-PMP22-myc (f) in transformed cells; colocalizations with endogenous catalase in peroxisomes are not shown. No fluorescence was detected in control experiments including omission of anti-Arabidopsis PMP22, anti-myc, or anti-HA IgGs or mock transformations with pRTL2 vector alone (data not shown). Bar in f = 10 μm. B, Insertion of wild-type and epitope-tagged versions of Arabidopsis PMP22 into isolated peroxisomes in vitro. PMP22 (a), myc-PMP22 (b), HA-PMP22-myc (c), and PMP22-myc (d) were translated in vitro in the presence 35S-Met with the wheat (Triticum aestivum) germ extract system and soluble radiolabeled translation products used in an in vitro import assay with isolated sunflower (Helianthus annuus) peroxisomes. Solid arrows to the left of each panel indicate the location of wild-type or epitope-tagged PMP22 species; the latter was confirmed by immunoprecipitation reactions with anti-myc IgGs (data not shown). Lane 1, Translation products equivalent to 40% of the amount shown in the other lanes. Lane 2, Reisolated radiolabeled protein from an import reaction containing ATP and peroxisomes. Lane 3, The same as lane 2 except that import reactions were treated with the protease thermolysin before reisolation of peroxisomes. Lane 4, Radiolabeled protein reisolated with peroxisomes after an import reaction in the absence of ATP. Lane 5, The same as lane 4 except that import reactions were treated with thermolysin before reisolation of peroxisomes. Lane 6, After an import reaction was carried out in the presence of ATP peroxisomes were reisolated, lysed with the detergent Triton X-100, and treated with thermolysin. Lane 7, The same as lane 5 except that, as a control, peroxisomes were omitted from the mock import reaction. M, Molecular mass markers; upper band (where shown) is 30.1 kD, and the lower band is 20 kD. C, Intracellular sorting of nascent myc-PMP22 from the cytosol to peroxisomes in BY-2 cells. Transiently expressed myc-PMP22 (a, c, e, and g) and endogenous catalase (b, d, f, and h) in transformed cells 5 h (a and b), 12 h (c and d), 20 h (e and f), or 45 h (g and h) after biolistic bombardment. Obvious colocalizations of myc-PMP22 with catalase in individual or globular peroxisomes are indicated with black and white arrows, respectively. Bar in a = 10 μm.
Different epitope-tagged versions of PMP22 were also imported into peroxisomes in vitro. Figure 1B shows the results of representative in vitro import reactions in which radiolabeled wild-type PMP22, myc-PMP22, PMP22-myc, and HA-PMP22-myc were incubated independently with or without isolated sunflower peroxisomes, and in the presence or absence of ATP and an ATP-regeneration system, the protease thermolysin, and/or the detergent Triton X-100. After all import reactions, peroxisomes were reisolated through a 0.7 m Suc cushion and polypeptides were analyzed by SDS-PAGE and phosphorimaging. Consistent with previously published results (Tugal et al., 1999), wild-type PMP22 bound to peroxisomes in a largely ATP-independent manner. That is, the majority of the radiolabeled PMP22 reisolated with peroxisomes in the pellet fraction after incubations in the presence or absence of ATP (Fig. 1C, a; compare lanes 2 and 4). However, a greater degree of protease protection for PMP22 was observed in the presence of ATP (Fig. 1C, a; lane 3) than in the absence of ATP (Fig. 1C, a; lane 5), indicating that the energy may facilitate the protein, acquiring its final protease-resistant state within the peroxisomal boundary membrane (Tugal et al., 1999). Similar data were obtained for the import of myc-PMP22 (Fig. 1C, panel b), HA-PMP22-myc (Fig. 1C, panel c), and PMP22-myc (Fig. 1C, panel d); all three epitope-tagged PMP22 proteins acquired a greater degree of protection to applied thermolysin in the presence of ATP. When import reactions with each of the PMP22 proteins were treated with thermolysin in the presence of Triton X-100 (Fig. 1C, a-d, lane 6) or when peroxisomes were omitted (Fig. 1C, a-d, lane 7), no protected wild-type or epitope-tagged PMP22 proteins were observed in the pellet fractions. This is consistent with protease protection being due to integration of the proteins into the lipid bilayer and not due to protein misfolding and/or aggregation. Separate immunoprecipitation experiments with the soluble fraction from each translation reaction (Fig. 1C, lane 1, b–d) and anti-myc IgGs confirmed the identity of each epitope-tagged PMP22 protein (data not shown). Taken together, the results presented in Figure 1 (A and B) indicate that epitope tags fused to the N and/or C terminus of PMP22 did not disturb peroxisomal targeting or insertion. Because all epitope-tagged proteins behaved in a similar manner to native PMP22, except PMP22-myc, which imported poorly, subsequent experiments to determine the location of the peroxisomal targeting information in PMP22 made use of myc-PMP22.
Figure 1C illustrates the localization myc-PMP22 in representative transformed BY-2 cells at 5, 12, 20, and 45 h postbombardment. At the earlier stages of expression and sorting (i.e. 5 h postbombardment), the majority of nascent myc-PMP22 resided in the nonorganelle cytosol (Fig. 1C, a), with only a small proportion localized to catalase-containing peroxisomes (Fig. 1E, compare a and b). However, after 12 and 20 h of transient expression, myc-PMP22 localized almost exclusively to individual peroxisomes distributed throughout the cell (Fig. 1C, e and f). The lack of fluorescence staining attributable to myc-PMP22 localized in other subcellular compartment(s) beside peroxisomes indicates that nascent PMP22 is sorted directly from its site of synthesis in the cytosol to peroxisomes, and not indirectly to peroxisomes via the ER.
At 45 h postbombardment, myc-PMP22 remained localized to peroxisomes (Fig. 1C, g and h), but the morphology and distribution of myc-PMP22-containing peroxisomes was altered by this time point. Most of the peroxisomes in a myc-PMP22-transformed cell 45 h after bombardment were redistributed into large globular structures up to 6 μm in diameter and that usually numbered 10 to 20 per cell. These globular-like peroxisomes were not apparent in surrounding nontransformed cells (Fig. 1C, h). Large globular peroxisomes also accumulated by 45 h postbombardment in PMP22-myc and HA-PMP22-myc-transformed cells (data not shown).
The N and C Termini of PMP22 Are Exposed to the Cytosol
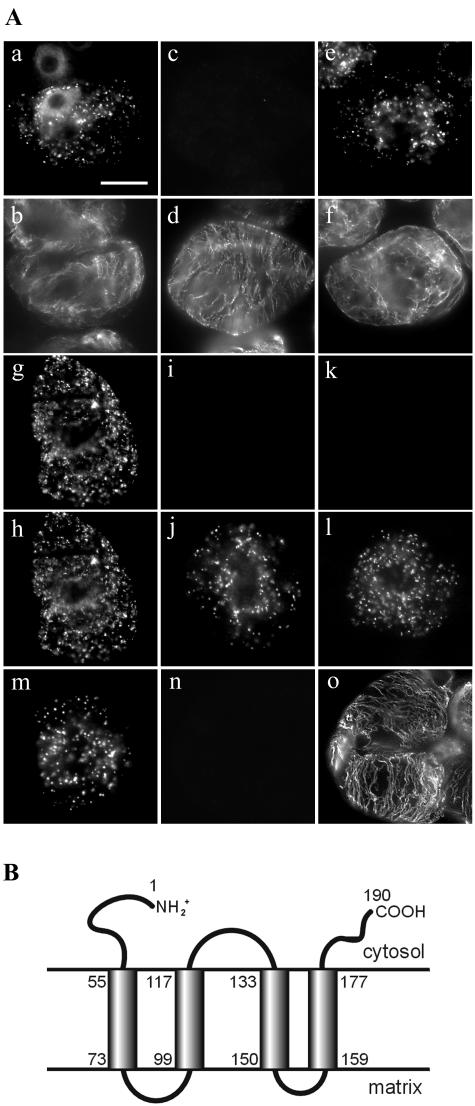
The topology of PMP22 within the peroxisome membrane was determined by the differential permeabilization/immunofluorescence method (Lee et al., 1997; Mullen et al., 2001b). Myc-PMP22-transformed cells at 20 h postbombardment were treated with digitonin, which selectively permeabilizes plasma membranes, leaving intraorganellar antigenic sites inaccessible to antibodies. Figure 2A (a and b) shows that transiently expressed myc-PMP22 and endogenous α-tubulin in cytosolic microtubules were immunostained in the same cell, indicating that the N terminus of myc-PMP22 is accessible to anti-myc antibodies. A simultaneous control experiment verified that only the plasma membrane was permeabilized in the same batch of myc-PMP22-transformed cells, i.e. the peroxisomal matrix enzyme catalase was not immunostained, whereas cytosolic microtubules were readily visualized in the same cell (Fig. 2A, c and d). However, permeabilization of cells with Triton X-100 resulted in peroxisomal catalase and cytosolic microtubules being immunodetected (Fig. 2A, e and f). Identical results were obtained with digitonin-permeabilized PMP22-myc-transformed cells (data not shown), indicating that the C-terminal part of PMP22-myc was also accessible to antibodies.
Figure 2.
Topological orientation of PMP22. A, Immunostaining attributable to transiently expressed myc-PMP22, HA-PMP22-myc, HA-catalase, or to endogenous α-tubulin or catalase in differential permeabilized BY-2 cells. BY-2 cells were formaldehyde fixed, permeabilized with pectolyase and with digitonin (a–d, g–l, n, and o) or Triton X-100 (e, f, and m), and then incubated with appropriate antibodies. Transiently expressed myc-PMP22 (a) and endogenous α-tubulin (b) in the same digitonin-permeabilized transformed cell. Myc-PMP22-bombarded, digitonin-permeabilized cells used for (a and b) immunostained for endogenous peroxisomal matrix catalase (c) and cytosolic α-tubulin (d). Immunostaining of endogenous catalase (e) and α-tubulin (f) in myc-PMP22-bombarded cells permeabilized with Triton X-100. Immunostaining of HA (g) and myc (h) epitopes in a HA-PMP22-myc-transformed cell permeabilized with digitonin. HA-PMP22-myc-bombarded, digitonin-permeabilized cells used for (g and h) immunostained for endogenous catalase (i and k) and the HA (j) or myc epitope (l). Immunostaining of expressed HA-catalase (m) in cells permeabilized with Triton X-100. HA-catalase-bombarded, digitonin-permeabilized cells used for (m) immunostained for the HA epitope (n) and endogenous tubulin (p). Bar in a = 10 μm. B, Predicted topological map of PMP22. Regions of PMP22 proposed to be hydrophobic membrane-spanning domains or hydrophilic domains facing the cytosol or peroxisomal matrix were identified using the TMHMM program (version 2.0) (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Shaded rectangles denote the four membrane-spanning domains (TMD 1–4) and the numbers of their first and last amino acid residues of each TMD are also indicated.
When HA-PMP22-myc-transformed cells were permeabilized with digitonin, identical (superimpossible) staining patterns attributable to myc and HA were observed in the same cell (Fig. 2A, g and h). In contrast, endogenous peroxisomal catalase was not detected in HA-PMP22-myc transformed cells that were permeabilized with digitonin and double-stained with anticatalase and anti-HA or anti-myc antibodies (Fig. 2A, I–l). Control experiments with transiently expressed HA-tagged catalase, a well-established matrix enzyme (Mullen et al., 1997), revealed that the HA epitope was not exposed to the cytosolic face of the peroxisomal boundary membrane (Fig. 2A, m–o).
Collectively, results obtained from differential permeabilization experiments with various epitope-tagged PMP22 constructs indicated that the N- and C-terminal portions of PMP22 are exposed to the cytosol. These data are consistent with the model for the topology of Arabidopsis PMP22 based on primary sequence and hydrophobicity. In this model (Fig. 2B), PMP22 is predicted to consist of four TMDs (TMD1-4; residues 55–73, 99–117, 133–150, and 159–177), two matrix-exposed hydrophilic sequences or “loops” (residues 74–98 and 151–158), and at least three cytosolic-exposed hydrophilic sequences, including one loop region (residues 118–133), and the N- and C-terminal portions (residues 1–54 and 178–190) of the protein.
The Peroxisomal Targeting Information Is Located in the N-Terminal Region of PMP22
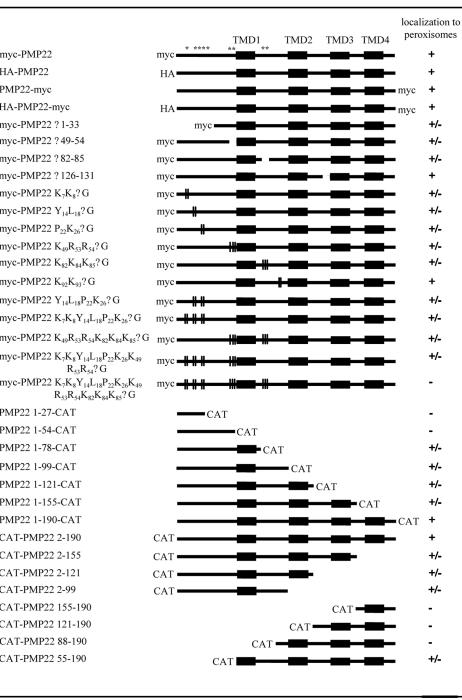
To define the peroxisomal targeting information in PMP22, a series of fusion proteins in which different portions of PMP22 were appended to the N or C terminus of the bacterial passenger protein chloramphenicol acetyltransferase (CAT) were generated. All chimeric proteins as well as mutant versions of epitope-tagged PMP22 described below are listed in Figure 3. The efficiency with which proteins sorted to peroxisomes in BY-2 cells was compared with wild-type myc-PMP22 and was assessed or scored by colocalization with endogenous peroxisomal catalase. That is, PMP22 constructs that colocalized exclusively with catalase in the same cell but no other organelles were designated “+.” Those that targeted partially to peroxisomes and to other organelles and/or cytosol in the same cell were designated “±,” and those that showed no apparent colocalization with catalase but instead localized to other organelles and/or cytosol were designated “–“(Fig. 3).
Figure 3.
Peroxisomal targeting of PMP22 mutant and fusion proteins. Black boxes represent the four predicted TMDs (1–4) in PMP22. Other regions of PMP22 containing putative mPTSs (i.e. amino acids 7 and 8, 14–26, 49–54, and 82–85) are marked with asterisks. Epitope tags and CAT fused to the N or C terminus of PMP22 are indicated. Schematic representations also show deletions or truncations in PMP22 proteins as spaces and site-specific Gly substitutions are marked with vertical bars. The numbers in the name of each fusion construct or myc-PMP22 mutant denotes the specific amino acid residues from PMP22 (1–190 residues) that were fused to the N or C terminus of CAT or delete/replaced with Gly residues. Targeting of PMP22 mutant and fusion proteins to peroxisomes in BY-2 cells was scored as follows: +, exclusively localized to peroxisomes; ±, partially localized to peroxisomes; -, not localized to peroxisomes. Results shown for each construct are a representative of all the transformants (>50) observed from at least two independent biolistic bombardment experiments.
Figure 4A (a–d) shows that CAT alone, as well as fusion proteins PMP22 1-27-CAT and PMP22 1-54-CAT accumulated in the cytosol of individual transformed BY-2 cells. In contrast, PMP22 1-78-CAT, which includes the N-terminal hydrophilic domain and the first putative membrane-spanning domain (TMD1) of PMP22, was sufficient, albeit in an inefficient manner, in redirecting the passenger protein to peroxisomes. Figure 4A (e and f) shows that at least a portion of transiently expressed PMP22 1-78-CAT colocalized with endogenous catalase, and that the peroxisomes in the transformed cell possessed a dramatically altered distribution and morphology similar to myc-PMP22-transformed cells at later stages of expression (Fig. 1C, g). These globular peroxisomal structures in PMP22 1-78-CAT-transfomed cells were strikingly similar to the aggregated peroxisomes in cells expressing a CAT-APX fusion protein. Figure 4A (g and h) shows, for comparison, replicate images of those shown previously (Mullen et al., 1999, 2001b) of the localization of CAT-APX (CAT plus the 36 C-terminal residues of cottonseed peroxisomal APX) to various subcellular structures, including catalase-containing aggregated peroxisomes.
Figure 4.
Subcellular localization of fusion proteins consisting of different portions of PMP22 fused to the N or C terminus of CAT. BY-2 cells transiently expressing CAT alone, a PMP22-CAT fusion protein, or a CAT-PMP22 (or CAT-APX) fusion protein were formaldehyde fixed, permeabilized with pectolyase and Triton X-100, and processed for immunofluorescence microscopy. A, Subcellular localization of N-terminal PMP22-CAT fusion proteins. Immunostaining attributable to transiently expressed CAT (a), PMP22 1-27-CAT (b), and PMP22 1-54-CAT (c). d, Endogenous peroxisomal catalase staining in the same PMP22 1-54-CAT-transformed cell and neighboring nontransformed cells shown in c. Expressed PMP22 1-78-CAT (e) and CAT-APX (g) and corresponding endogenous catalase (f and h) in transformed cells; black arrows indicate obvious colocalizations. PMP22 1-78-CAT (i) and endogenous ER calreticulin (j) in the same transformed cell; white arrows indicate obvious noncolocalizations. PMP22 1-120 CAT (k), PMP22 1-155 CAT (m), and PMP22 1-190CAT (o) and corresponding endogenous catalase (l, n, and p) in transformed cells; black arrows indicate obvious colocalizations. Bar in a = 10 μm. B, Subcellular localization of C-terminal CAT-PMP22 fusion proteins. Immunostaining attributable to transiently expressed CAT-PMP22 155-190 (a), CAT-PMP22 120-190 (c), CAT-PMP22 88-190 (e), and endogenous peroxisomal catalase (b, d, and f) in the transformed cells; black arrows in b, d, and f denote catalase-containing globular peroxisomes in transformed cells. CAT-PMP22 120-190 (g) and endogenous ER calreticulin (h) in the same transformed cells; black arrows indicated obvious colocalizations CAT-PMP22 2-99 (i), CAT-PMP22 2-121 (k), CAT-PMP22 2-155 (m), CAT-PMP22 2-190 (o), and corresponding endogenous catalase (j, l, n, and p) in transformed cells; black arrows indicate obvious colocalizations.
Transiently expressed PMP22 1-78-CAT did not colocalize with endogenous calreticulin, an ER marker (Fig. 4A, i and j). The reticular/circular fluorescence pattern attributable to the fusion protein that was not localized to catalase-containing globular peroxisomes is similar to reticular/circular structures observed in CAT-APX-transformed cells (Fig. 4A, compare e, g, and i). CAT-APX and other APX fusion proteins have been shown previously localized to pER, before their sorting to (globular) peroxisomes (Mullen et al., 1999, 2001b), as well as to plastids and mitochondria due to overexpression and/or mislocalization (Lisenbee et al., 2003).
Similar to the results presented above for PMP22 1-78-CAT, other PMP22-CAT fusion proteins that consisted of larger N-terminal portions of PMP22, e.g. PMP22 1-99-CAT, PMP22 1-120-CAT, and PMP22 1-155-CAT showed only partial colocalization with endogenous catalase in globular peroxisomes (Fig. 4A, k–n). However, full-length PMP22 (residues 1–190) fused to the N terminus of CAT (PMP22 1-190-CAT) was efficiently targeted to peroxisomes because the fusion protein colocalized entirely with endogenous catalase (Fig. 4A, o and p). Individual peroxisomes, and not globular or aggregated peroxisomal structures, were observed in the majority of PMP22 1-190-CAT-transformed cells.
Next, the sufficiency of various C-terminal regions of PMP22 for redirecting CAT from the cytosol to peroxisomes was tested. No apparent colocalizations were observed between endogenous peroxisomal catalase and several transiently expressed CAT-PMP22 fusion proteins (Fig. 4B), including CAT-PMP22 155-190 (Fig. 4B, a and b), CAT-PMP22 121-190 (Fig. 4B, c and d), and CAT-PMP22 88-190 (Fig. 4B, e and f). Each of these fusion proteins instead localized to a reticular network that consisted of ER, as evidenced by colocalizations with endogenous calreticulin (data shown only for CAT-PMP22 121-190; Fig. 4B, g and h). Although CAT-PMP22 155-190, CAT-PMP22 121-190, and CAT-PMP22 88-190 did not appear to contain peroxisomal targeting information, globular peroxisomes were observed in cells transiently expressing each one of these three fusion proteins. We noted also that the size and/or number of the peroxisomal structures that were formed seemed to vary depending on proportion of the C terminus of PMP22 that was appended to CAT; globular peroxisomes were more prevalent in cells expressing CAT-PMP22 88-190 or CAT-PMP22 121-190 than in cells expressing CAT-PMP22 155-190. At least partial localization to catalase-containing globular peroxisomes was observed for several other CAT-PMP22 fusion proteins, including CAT-PMP22 55-190, CAT-PMP22 2-99, CAT-PMP22 2-121, and CAT-PMP22 2-155 (Figs. 3 and 4B, i–n). Each of these fusion proteins, like the N-terminal PMP22-CAT fusions shown in Figure 4A also localized to other subcellular compartment(s) with a reticular/circular appearance that did not colocalize with calreticulin in the ER (data not shown). Figure 4B (o and p) shows that CAT-PMP22 2-190, consisting of full-length PMP22 fused to the C terminus of CAT localized exclusively to catalase-containing globular peroxisomes. Overall, the results presented in Figure 4 indicate that although the N-terminal one-half of PMP22 contains the peroxisomal targeting information, efficient sorting of the passenger protein CAT from the cytosol to peroxisomes required that it was appended to nearly the entire PMP22 sequence, including all four TMDs.
PMP22 Contains Several Distinct Regions That Are All Necessary for Efficient Peroxisomal Targeting
To define more precisely the region(s) within PMP22 responsible for targeting to peroxisomes, amino acid sequences were sought within the protein that resemble the so-called prototypic mPTS found in most other PMPs. These consist of a cluster of three to five positively charged amino acid residues adjacent to at least one TMD (for review, see Subramani et al., 2000; Purdue and Lazarow, 2001; Trelease et al., 2002).
Figure 5 shows an alignment of deduced amino acid sequences for PMP22s from Arabidopsis, mouse, rat, and human. Overall, the Arabidopsis sequence was similar (45%–57%) to mammalian PMP22s, with the most notable difference being that the relative positions of the first three (of four) predicted TMDs varied between Arabidopsis PMP22 and the mammalian proteins. At least three amino acid sequences within Arabidopsis PMP22 closely resembled the prototypic mPTS (bold and underlined in Fig. 5). These included the basic cluster -KIQLRR-(residues 49–54) that is immediately adjacent to amino-terminal end of TMD1 and conserved among other PMP22s, and the basic clusters -KGKK-(residues 82–85) and -RERIKK-(residues 126–131) that appear to be unique to the Arabidopsis protein and are adjacent to TMD1 and TMD3, respectively.
Figure 5.
Sequence comparison of PMP22s from rat, mouse, human, and Arabidopsis. Deduced amino acid sequences were obtained from GenBank (accession nos.: rat Q07066; mouse P42925; human AY044439; and Arabidopsis AJ006053) and were aligned using ClustalW and visual inspection. Identical amino acid residues in each of the aligned PMP22s are indicated by asterisks, and similar residues are indicated by dots. TMDs were identified using the TMHMM program (version 2.0). The four predicted TMDs in each of the proteins are shaded and regions tested in Arabidopsis PMP22 in this study to function as mPTSs are in bold and underlined.
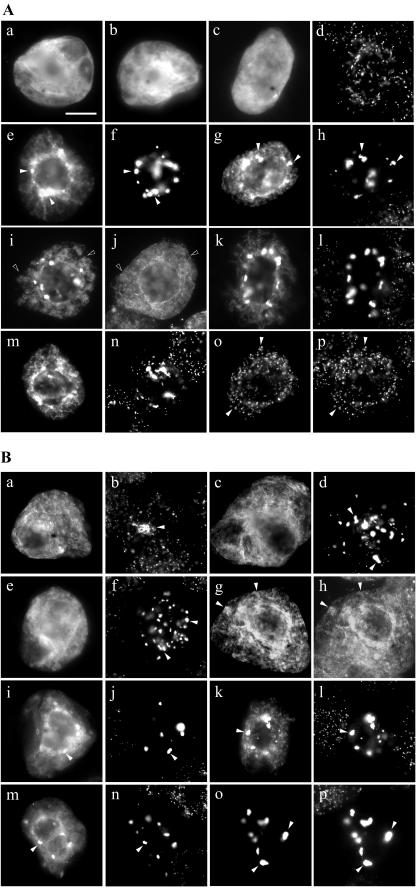
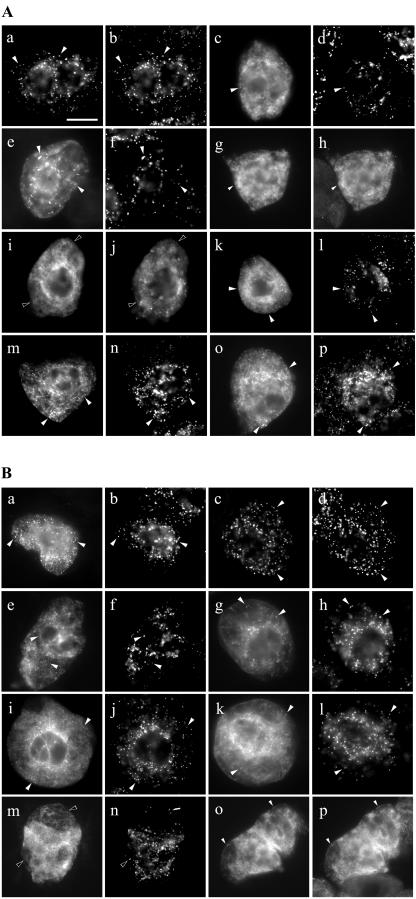
To determine whether any of these three clusters of basic amino acid residues in PMP22 confer necessary peroxisomal targeting information, they were independently deleted or altered in myc-PMP22, and the localization of resulting mutant proteins was examined in transformed BY-2 cells. As shown in Figure 6A (a and b), myc-PMP22Δ126-131, which lacks the amino acids -RERIKK-immediately upstream of TMD3, colocalized entirely with endogenous peroxisomal catalase. In contrast, myc-PMP22Δ49-54 (Fig. 6A, c and d) and myc-PMP22Δ82-85 (Fig. 6A, e and f) only partially colocalized with endogenous catalase and a proportion of each expressed protein mislocalized to the cytosol and ER. Partial localization of myc-PMP22Δ49-54 and myc-PMP22Δ82-85, as well several other myc-PMP22 mutant proteins described below, to the ER was evident by their colocalization with endogenous calreticulin; data are shown only for myc-PMP22Δ49-54 (Fig. 6A, g and h). Interestingly, no myc-PMP22 mutant protein tested was mislocalized readily to pER; no apparent colocalization was observed, for example, when myc-PMP22Δ49-54 and CAT-APX, as marker for pER, were coexpressed in the same BY-2 cell (Fig. 6A, i and j).
Figure 6.
Subcellular localization of modified versions of myc-PMP22. Transiently transformed BY-2 cells were formaldehyde fixed, permeabilized with pectolyase and Triton X-100, and processed for immunofluorescence microscopy. A, Subcellular localization of myc-PMP22 mutants with alterations in amino acids sequences that resemble positively charged mPTSs in other PMPs. Transiently expressed myc-PMPΔ126-131 (a), myc-PMP22Δ49-54 (c), myc-PMP22Δ82-85 (e), and corresponding endogenous catalase (b, d, and f) in transformed cells; black arrows indicate obvious colocalizations. myc-PMP22Δ49-54 (g) and endogenous ER calreticulin (h) in the same transformed cell; black arrows indicate obvious colocalizations. Coexpressed myc-PMP22Δ49-54 (i) and CAT-APX (j) in the same transformed cell; white arrows indicate obvious noncolocalizations. Myc-PMP22K49R53R54ΔG (k), myc-PMP22K82K84K85ΔG (m), PMP22K49R53R54K82-K84K85ΔG (o), and corresponding endogenous catalase (l, n, and p) in transformed cells; black arrows indicate obvious colocalizations. Bar in a = 10 μm. B, Subcellular localization of myc-PMP22 mutants with Gly substitutions of amino acids sequences that have been proposed to function as mPTSs in mammalian PMP22s. Transiently expressed myc-PMP22K7K8ΔG (a), myc-PMP22K92K93ΔG (c), myc-PMP22Y14L18P22K26ΔG (e), myc-PMP22K7K8Y14L18-P22K26ΔG (g), myc-PMP22Δ1-33 (i), myc-PMP22K7-K8Y14L18P22K26K49K53K54ΔG (k), and corresponding endogenous catalase (b, d, f, h, j, and l) in transformed cells; black arrows indicate obvious colocalizations. Expressed myc-PMP22K7K8Y14L18P22-K26K49K53K54K82K84K85ΔG (m and o) and endogenous catalase (n) or endogenous calreticulin (p) in transformed cells; white arrows in m and n indicate obvious noncolocalization and black arrows in o and p indicate obvious colocalizations.
We next tested whether peroxisomal targeting of PMP22 was affected when all the basic residues at position 49 through 54 (underlined, KIQLRR) or position 82 through 85 (KGKK) or at both positions were replaced with noncharged glycines. Figure 6A (k–p) shows that the resulting proteins (myc-PMP22K49R53R54ΔG, myc-PMP22K82K84K85ΔG, and myc-PMP22 K49R53R54K82K84K85ΔG) were only partially localized to peroxisomes (Fig. 6A, k–p), and all three mutants mislocalized to a similar extent to the ER. Taken together, the results presented in Figure 6A indicate that although at least two regions in PMP22 that resemble a prototypic mPTS, namely regions 49 through 54 and 82 through 85, are necessary for efficient sorting to peroxisomes, additional targeting information exists within the protein.
Because the C-terminal one-half of PMP22 was insufficient in sorting CAT to peroxisomes (Fig. 5), it was reassured that any peroxisomal targeting information in PMP22, in addition to regions 49 through 54 and 82 through 85, was contained in the N-terminal region of the protein. From the results of previous studies of mammalian PMP22s (Pause et al., 2000; Brosius et al., 2002), at least three separate regions within the N-terminal one-half of the proteins were speculated, but not experimentally proven, to function as mPTSs, and each of these regions are divergent from the prototypic “basic cluster” mPTS described above. Inspection of the Arabidopsis PMP22 sequence revealed that all three of these putative mPTSs were conserved in the plant protein, -KK- at positions 7 and 8, -KK- at positions 92 and 93, and the motif -Y-x3-l-x3-P-x3-K- at positions 14 through 26 (bold and underlined in Fig. 5).
Figure 6B (a and b) shows that when the two Lys residues at positions 7 and 8 in PMP22 were each replaced with a Gly residue, the resulting mutant protein (myc-PMP22K7K8ΔG) was not completely localized to peroxisomes. Instead, a portion of myc-PMP22K7K8ΔG was mislocalized to the ER, as evidenced by colocalization with endogenous calreticulin (data not shown). In contrast, expressed myc-PMP22K92K93ΔG colocalized entirely with endogenous catalase (Fig. 6B, c and d). These data suggest that the di-Lys residues at positions 7 and 8, but not those at positions 92 and 93, are necessary for PMP22 to be targeted efficiently to peroxisomes. The targeting function of the proposed Y-x3-l-x3-P-x3-K motif identified in rat PMP22 was tested by replacing the conserved Y, L, P, and K residues at positions 14 through 26 in Arabidopsis PMP22 with Gly. Figure 6B (Fig. 6B, e and f) shows that the mutant myc-PMP22Y14L18P22K26ΔG only partially localized to peroxisomes. Targeting to peroxisomes was also diminished when residues at position 14 through 26 were deleted (myc-PMP22Δ14-26) or when the Y and L (myc-PMP22Y14L18ΔG) or the P and K (myc-PMP22P22K26ΔG) alone were replaced with Gly residues (data not shown and Fig. 3), suggesting that all four conserved residues in this motif were equally important for efficient peroxisomal localization of PMP22.
The effect of disrupting several regions at once in PMP22 on the peroxisomal targeting of the protein was investigated. Figure 6B (g and h) shows that when Lys 7 and 8 as well as the conserved residues in the -Y-x3-l-x3-P-x3-K-motif at positions 14 through 26 were replaced with Gly, the resulting mutant protein (myc-PMP22K7K8Y14L18P22K26ΔG) was still partially localized to peroxisomes. Similarly, deletion of the first 33 residues of PMP22 (myc-PMP22Δ1-33) did not abolish peroxisomal targeting completely (Fig. 6B, i and j). However, these results for the partial targeting of myc-PMP22K7K8Y14L18P22K26ΔG and myc-PMP22Δ1–33 to peroxisomes were not entirely unexpected because at least two other putative targeting regions within PMP22 (i.e. -KIQLRR- and -KGKK- at positions 49–54 and 82–85, respectively) remained intact in each of these mutants. Partial localization to peroxisomes was still observed when all three of the putative targeting regions in N-terminal hydrophilic domain of PMP22 were mutated (myc-PMP22K7K8Y14L18P22K26K49K53K54ΔG; Fig. 6B, k and l). Only when all four of the regions considered to be important for targeting PMP22 were altered by substitutions of specific residues with Gly was the resulting mutant protein (myc-PMP22K7K8Y14L18P22K26K49K53K54K82K84K85ΔG) not localized to peroxisomes, but instead localized to ER (Fig. 6A, m–p).
Import of myc-PMP22 Mutants in Vitro
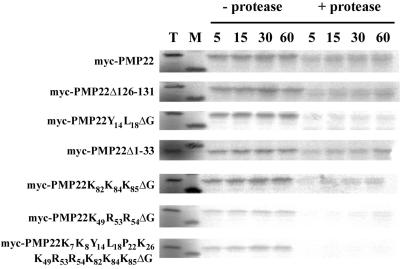
The ability of selected myc-PMP22 mutants to insert into isolated peroxisomes in vitro was also tested (Fig. 7). In vitro-translated proteins were incubated in duplicate with isolated sunflower peroxisomes in the presence of ATP and an ATP-regeneration system for 5, 15, 30, or 60 min. Reactions were stopped by transferring the samples to ice and one aliquot was then treated with the protease thermolysin. A direct comparison of samples treated or not treated with protease allowed discrimination between myc-PMP22 mutants that have achieved the protease-resistance characteristic similar to that of wild-type myc-PMP22 (and native PMP22; Tugal et al., 1999) and mutant myc-PMP22s that had simply reisolated with peroxisomes. Moreover, an analysis of selected PMP22 mutants ability to acquire protease resistance in time-course experiments allowed us to determine whether they inserted in vitro into peroxisomes with slower kinetics than wild-type myc-PMP22.
Figure 7.
Kinetics of import of various myc-PMP22 mutants in vitro. Selected mutants were tested for their ability to bind to and insert into isolated sunflower peroxisomes. Each mutant myc-PMP22 protein was prepared by in vitro transcription and translation in the presence of 35[S] Met and was incubated with isolated peroxisomes in the presence of ATP at 26°C for the time indicated. For each experiment, two mutant proteins were compared with the parental myc-PMP22 construct to control for any variations in import efficiency between different peroxisome preparations. At the end of the incubation, reactions were returned to ice and treated (+ protease) or not treated (–protease) with thermolysin as described in “Materials and Methods.” After inactivation of the protease, peroxisomes were reisolated through a 0.7 m Suc cushion and were processed for SDS-PAGE and phosphorimaging. “T” translation products equivalent to 40% of the radiolabeled protein added to each of the other incubations. M, Molecular mass markers; the 20-kD marker is shown.
The maximum amount of protease-protected myc-PMP22 was typically observed after 15 to 30 min of incubation with isolated peroxisomes (Fig. 7). After 60 min, the amount of imported myc-PMP22 protein remained constant or even declined slightly. Figure 7 also shows that the mutant protein myc-PMP22Δ126-131 imported into isolated peroxisomes at a maximum level within approximately 30 min in a manner similar to myc-PMP22. These in vitro data for myc-PMP22 and myc-PMP22Δ126-131 are consistent with the in vivo data where both proteins localized exclusively to BY-2 peroxisomes (see Figs. 1, A and E and 7, a and b). Also consistent with the in vivo data presented above, site-specific mutations within one or more of the putative targeting sequences in PMP22 negatively affected, but to different extents, the kinetics of in vitro import relative to wild-type myc-PMP22. For instance, the mutant myc-PMP22Y14L18ΔG showed low levels of imported, protease-protected protein that did not increase over the 60-min time course. On the other hand, import of the mutants myc-PMP22Δ1-33 and mycPMP22 K82K84K85ΔG was only slightly impaired relative to myc-PMP22. Perhaps the most pronounced effects for the kinetics of import in vitro was observed for myc-PMP22K49R53R54ΔG and the multiple mutant myc-PMP22K7K8Y14L18L18P22K26K49K53K54K82K84K85 ΔG. Figure 7 shows that only a small amount of protease-protected myc-PMP22K49R53R54ΔG was detected at the later time points of incubation (i.e. 60 min) and no myc-PMP22K7K8Y14L18L18P22K26K49K53 K54K82K84K85ΔG protein was imported at any of the time points examined, consistent with in vivo data (Fig. 6B, m–p) that disruption of all four of these regions in PMP22 completely abolished peroxisomal targeting.
DISCUSSION
In this paper, a comprehensive analysis of the targeting information in the Arabidopsis 22-kD integral PMP (PMP22) was carried out. Using BY-2 cells as a well-characterized in vivo system for defining the molecular targeting signals in ectopically expressed proteins, and isolated sunflower peroxisomes to examine protein insertion into the peroxisomal boundary membrane in vitro, we demonstrated that at least four distinct regions within PMP22 are important for efficient targeting of the protein from its site of synthesis in the cytosol directly to peroxisomes and for efficient insertion of the protein into the peroxisomal boundary membrane. These observations that multiple regions mediate the trafficking of PMP22 were not entirely surprising because several other PMPs from different organisms have been reported recently to possess more than one mPTS. However, what was unexpected from the results of this study was the cooperative targeting action of the different regions identified in PMP22, as well as the overall lack of sequence similarities among some of these regions and among mPTSs previously identified in other PMPs. As discussed below, the nature of the mPTS overall seems to be more complex than has been suggested and the specific characteristics of this targeting signal, such as the number and location, varies depending upon, for example, the overall structural characteristics of the PMP.
Wild-Type PMP22 Sorts Directly to Peroxisomes, But Some PMP22 Mutants Mislocalize to Other Compartments and Can Affect Peroxisome Morphology
Newly synthesized PMP22 sorts directly from the cytosol to peroxisomes (Fig. 1C), unlike nascent APX, which sorts initially to a subdomain of the ER, termed peroxisomal ER, and then to peroxisomes (Mullen et al., 1999). Based on these observations, it appears that at least two separate intracellular sorting pathways for PMPs exist in plant cells. It is important to note that although some PMP22 CAT fusion proteins such as PMP1-78-CAT localized to subcellular structures that resembled those containing CAT-APX (a marker for pER), we do not consider them to be physiological sorting intermediates, but rather mislocalized forms arising from the loss of peroxisomal targeting information and/or exposure of a cryptic (pER) targeting signal due to experimental manipulation. Similarly, several myc-PMP22 mutants mislocalized to the more general ER rather than pER; experiments to explain these differences in the mislocalization of the various PMP22 mutants to separate regions of the ER are currently under way.
Expression of PMP22 at later time points after biolistic bombardment (e.g. 45 h; Fig. 1C, g and h) and of several PMP22-CAT (or CAT-PMP22) fusion proteins (Fig. 4) resulted in a striking alteration of peroxisome morphology. Typically, a large number of globular peroxisomes were observed that were usually concentrated around the nucleus. The formation of such globular peroxisomal structures can result from the oligomerization or “zippering” of cytosolically exposed CAT moieties after insertion of APX fusion proteins into the peroxisomal boundary membrane (Mullen et al., 2001b), and this is the likely explanation for the formation of the globular peroxisomes seen with the PMP22-1-78 CAT and some other CAT constructs. However, not all fusion proteins in which CAT was located on the cytosolic side of the membrane resulted in this effect. Expression of PMP22-1-190-CAT did not result in formation of globular peroxisomes (Fig. 4A, o and p) even though the CAT moiety was shown by digitonin permeabilization to be cytosolic (data not shown). Even more bizarre was that some CAT-PMP22 fusion proteins that are not themselves targeted to peroxisomes caused this alteration in peroxisome morphology (e.g. CAT-PMP22-155-190, CAT-PMP22-121-190, and CAT-PMP22-88-190; Fig. 4B, a–f), which excludes the zippering mechanism (described in Mullen et al., 2001b) in these cases. We speculate that these targeting-defective PMP22 constructs acted in a dominant-negative manner, sequestering the machinery required for the proper localization of PMPs, including protein components required for normal peroxisome division and segregation.
Features of the Arabidopsis PMP22 mPTS
Two of the four regions we identified as part of the mPTS(s) within Arabidopsis PMP22 corresponded to a pair of Lys residues at positions 7 and 8 (-KK-) and a sequence of 13 amino acids at positions 14 through 26 (-YLSQLQQHPLRTK-) that were previously proposed to be core components of one or two distinct mPTSs in rat and human PMP22s. For instance, Pause et al. (2000) proposed that the core component of a single mPTS in rat PMP22 consisted of the motif -Y-x3-l-x3-P-x3-K-located in the N-terminal hydrophilic region of the protein. In contrast, Brosius and coworkers (2002) speculated that two N- and C-terminal mPTSs, each consisting of a pair of Lys and/or Arg residues, as well as adjacent sequences that included at least two TMDs, functioned in rat and human PMP22. However, as with the -Y-x3-l-x3-P-x3-K-motif identified by Pause et al. (2000), neither pair of basic residues in rat and human PMP22 was shown experimentally to be necessary for peroxisomal targeting in these studies. Therefore, it is possible that different key residues in the N- and C-terminal region of rat and human PMP22 function as the core components of the mPTS(s). Using gain-of-function and loss-of-function experiments, we showed that the di-Lys pair at position 7 and 8 as well as the -Y-x3-l-x3-P-x3-K-motif in the N-terminal cytosolic tail are important for peroxisomal targeting of Arabidopsis PMP22. In contrast, the di-Lys pair at positions 92 and 93 are not necessary. In fact, we found no evidence that any portion of the C terminus of Arabidopsis PMP22 (residues 88–190) was sufficient for peroxisomal sorting. These data suggest that the uncharacterized mPTS reported to exist in the C-terminal one-half of rat and human PMP22 (Brosius et al., 2002) does not exist in Arabidopsis PMP22, although all three proteins probably use an N-terminal basic cluster and -Y-x3-l-x3-P-x3-K-motif as important components of their mPTS.
Two other regions located at positions 49 through 54 (-KIQLRR-) and 82 through 85 (-KGKK-) in the N-terminal one-half of Arabidopsis PMP22 were identified as possible elements of one or more mPTSs due to their resemblance to the basic clusters of four to six amino acid residues located in the mPTSs of other PMPs. These include Candida boidinii PMP47 (-KIKKR-; Dyer et al., 1996), Pex3p (-RHKKK-; Soukupova et al., 1999; Baerends et al., 2000), S. cerevisiae Pex15p (-RKKK-; Elgersma et al., 1997), and cottonseed APX (-RKRMK; Mullen and Trelease, 2000). Consistent with this premise, peroxisomal targeting and membrane insertion were impaired when positively charged residues in either of these basic cluster were replaced with Gly. In addition, certain portions of PMP22 containing one or both of these basic clusters were sufficient, albeit in an inefficient manner, for sorting CAT to peroxisomes. However, another basic cluster located at positions 126 through 131 (-RERIKK-), between TMD2 and TMD3, was not sufficient (CAT-PMP22 88-190; Fig. 4A) or necessary (myc-PMP22Δ126-131; Figs. 6A and 7) for peroxisomal targeting. This indicates that in Arabidopsis PMP22, not all basic clusters are required for efficient peroxisomal targeting. The results with myc-PMP22Δ126-131 and certain other modified versions of PMP22 (e.g. myc-PMP22 K92K93ΔG) also indicate that mutations of the protein did not always affect the fidelity of peroxisomal targeting. This latter point is an important one because a substantive caveat of this study (and all other analyses of PMP targeting) is that aberrant protein folding, rather than disruptions in key components of one or more peroxisomal targeting signals, can result in mislocalization and/or poor membrane insertion. Although the possibility that such mutations caused deleterious affects in the protein's overall structure cannot be excluded, the strength of using a combination of loss-of-function, gain-of-function, and in vitro membrane insertion experiments allowed the definition of the regions in PMP22 that contain bona fide targeting/insertion information.
Although the basic clusters located at positions 49 through 54 and 82 through 85 appear to participate in some way in the peroxisomal targeting of Arabidopsis PMP22, neither sequence was involved in the peroxisomal targeting of mammalian PMP22s. For instance, mammalian PMP22s do not possess sequences similar to the basic cluster located at position 82 through 85 in Arabidopsis PMP22 (refer to Fig. 5). Also, in mammalian PMP22s, the basic cluster equivalent to that cluster at positions 49 through 54 in Arabidopsis PMP22 was not necessary or sufficient for peroxisomal targeting (Pause et al., 2000; Brosius et al., 2002). On the other hand, mouse M-LP, a PMP closely related to mammalian PMP22s, contains a mPTS that includes a basic cluster equivalent to the basic cluster at positions 49 through 54 in Arabidopsis PMP22 (Iida et al., 2003). It is also worth pointing out that other notable differences exist between the basic clusters that form part of the mPTS(s) in Arabidopsis PMP22 and the basic clusters that are a key component of the mPTS identified in most other PMPs. For instance, in PMP47, Pex3p, Pex15p, and APX, the basic cluster is located on the matrix side of the peroxisomal boundary membrane (McCammon et al., 1994; Elgersma et al., 1997; Soukupova et al., 1999; Baerends et al., 2000; Mullen and Trelease, 2000), whereas in PMP22, differential permeabilization studies (Fig. 2A) and computer-based predictions (Fig. 2B) indicated that the basic cluster at position 49 through 54 was located on cytosolic side of the membrane and the basic cluster at position 82 through 85 was matrix orientated. Due to the difference in the position of TMD1 between the plant and mammalian PMP22-like proteins (Fig. 5), the sequence in M-LP equivalent to the basic cluster at position 49 through 54 in Arabidopsis PMP22 is predicted to be matrix oriented. Another difference between the basic clusters in mPTS(s) of Arabidopsis PMP22 and other PMPs is the amount of sequence adjacent to the cluster that together is minimally sufficient for proper (efficient) localization to peroxisomes. We showed that only when full-length PMP22 (190 amino acids), including both basic clusters, the proposed targeting information located at residues 7 and 8 and 14 through 26, and all four predicted TMDs, was fused to CAT (to the N or C terminus of CAT) was the resulting fusion protein(s) sorted efficiently to peroxisomes; all other fusion proteins containing shorter fragments of PMP22 and including those containing one or both of the basic clusters were sorted to peroxisomes in an inefficient manner, i.e. partially mislocalized to other nonperoxisomal structures (Fig. 4). In contrast to the results for PMP22, the mPTSs reported as sufficient for peroxisomal targeting of other PMPs are relatively short, ranging, for example, from 29 amino acids long for APX (approximately 10% of the protein; Mullen and Trelease, 2000) to 83 amino acids long for C. boidinii PMP47 (20% of the protein; Dyer et al., 1996). Several other PMPs such as human PMP34, PMP70, and Pex13p (Sacksteder et al., 2000; Jones et al., 2001) have been shown to contain at least two distinct regions that are minimally sufficient for peroxisomal targeting. However, none of these regions corresponds in position or sequence to the single mPTSs of APX, Pex3p, and PMP47.
Taken together, how can all of the conflicting data described above for the nature of the mPTS in Arabidopsis PMP22, mammalian PMP22s, and other PMPs be easily reconciled? Part of the problem may be defining what is meant by a PMP targeting signal, and that most targeting assays, ours included, measure the end point of a complex multistep process that begins during synthesis on free polyribosomes in the cytosol and ends with assembly in the peroxisomal boundary membrane. For single membrane-spanning PMPs such as Pex3p, APX, and Pex15p, there is general agreement that the targeting information is contained within one distinct region that includes a matrix-orientated basic cluster and adjacent TMD. However, as suggested in models proposed by Wang et al. (2001) and Jones et al. (2001), multispanning PMPs such as PMP22, PMP47, and PMP34 have more complex requirements. These include preventing aggregation of the PMP by having to shield multiple hydrophobic TMDs from the cytosol and ensuring proper assembly of the PMP in the peroxisomal boundary membrane by promoting the correct insertion of multiple TMDs into the lipid bilayer. Each of these functions is likely performed by one or more chaperone/receptor proteins that would interact with multiple sites on the PMP. Interfering with any of these auxiliary sites would adversely affect targeting to peroxisomes whether or not they play a role in determining the specificity of the final sorting destination. It would also seem likely that chaperones/receptors would need to interact with the nascent multispanning PMP during translation to prevent aggregation and misfolding, even though the protein itself is inserted into peroxisomes posttranslationally. Our observations that sequences near the amino terminus of PMP22 including the di-Lys pair (residues 7 and 8) and the -Y-x3-l-x3-P-x3-K-motif (residues 14–26) are important for efficient targeting to peroxisomes are consistent with this idea.
Wang et al. (2001) proposed that a matrix-orientated basic cluster was the key component of the mPTS in all PMPs, although other sequences with less basic characteristics overall can suffice for the basic cluster and thus act as redundant targeting signals. Indirect evidence in support of this premise came from their data showing that a basic cluster in a minimally sufficient PMP47-green fluorescent protein fusion need not contain more than two basic charges, or may function with even fewer in the proper context. Thus, it is possible that for those other PMPs reported to contain multiple mPTSs, but that lack conspicuous basic clusters, there must exist at least some positively charged residues in each region of the protein that is minimally sufficient for peroxisomal targeting. In the case of Arabidopsis PMP22, the requirement for the matrix-orientated basic sequence -KGKK- at positions 82 through 85 would conform to this model.
Model for the PMP22 mPTS
Based on the hypothetical model presented above, we propose that Arabidopsis PMP22 contains a single mPTS consisting of several critical elements including a matrix-orientated basic cluster at position 82 through 85 that serves as a key targeting component, an N-terminal cytosolic-orientated hydrophilic domain that is required to mediate the proper context for the basic cluster to function, and all four TMDs for correct insertion and assembly in the peroxisomal boundary membrane. Only when all of these elements in PMP22 are intact is targeting with high fidelity to peroxisomes achieved.
This working model for the PMP22 mPTS provides a reasonable explanation for the partial colocalization of PMP22 1-78-CAT to peroxisomes, a fusion protein lacking the basic cluster at position 82 through 85. We suspect that this portion of PMP22 is sufficient for sorting CAT to peroxisomes (although in an inefficient manner) because it retains the sequence -KIQLRR- at positions 49 through 54 that suffices as a matrix-localized targeting element in this fusion protein. Digitonin permeabilization experiments with PMP22 1-78-CAT-transformed cells revealed the CAT moiety was exposed to the cytosol (data not shown), suggesting that the N-terminal domain of this fusion protein (residues 1–54, including -KIQLRR-) was topologically orientated in the peroxisomal matrix. Therefore, this fusion has created an artificial single spanning membrane PMP, inverting the topological orientation that the N-terminal segment possesses in native PMP22. This result implies an important role for the basic cluster at position 82 through 85, possibly in conjunction with TMD1 and 2, in determining the topology of the native PMP22. In vitro import experiments (Fig. 7) indicate also that the basic cluster -KIQLRR-can function as a targeting element because the mutant in which -KGKK- at positions 82 through 85 was altered to -GGGG-was still imported, although less effectively than the parental myc-PMP22. Although we cannot rule out the possibility that the second matrix-orientated sequence in PMP22 (-NYKVPLHRF-, residues 151–158) may function also as a redundant mPTS, this possibility seems unlikely because minimal fusion proteins, including the loop sequence at positions 151 through 158, were not sufficient in any way for sorting to peroxisomes. However, the minimally sufficient fragment reported to exist in the C-terminal one-half of mammalian PMP22s (Brosius et al., 2002) suggests the sequence -KMR-in the second matrix loop of these proteins can suffice as a basic cluster with redundant targeting function, and it is noteworthy to point out also that we observed that myc-PMP22 K49R53R54K82K84K85ΔG at least partially localized to peroxisomes (Fig. 6A, o and p). Hence, it may be possible that other yet undetected targeting signals exist in PMP22.
Probably the most difficult component of the PMP22 mPTS to assess a specific function for is the protein's TMDs. It is well known that TMDs are crucial for the proper localization of PMPs. However, because TMDs do not appear to contain specific targeting information (Mullen and Trelease, 2000), the TMDs in PMP22 (and those in other PMPs) are most likely required to form an overall three-dimensional conformation that enables the nascent protein in the cytosol to be efficiently recognized by a receptor(s) and then integrated/assembled into the peroxisomal boundary membrane. Thus, although we are now closer to understanding the nature of the mPTS, further investigation is needed to characterize the mechanisms involved in early steps in PMP folding in the cytosol and subsequent targeting to peroxisomes, including the protein components (e.g. molecular chaperones and receptors) that mediate these events.
MATERIALS AND METHODS
Recombinant DNA Procedures and Reagents
Restriction enzymes and other DNA-modifying enzymes were purchased from Promega (Madison, WI) or New England Biolabs (Beverly, MA). Custom synthetic oligonucleotides were purchased from Invitrogen Canada (Burlington, Ontario, Canada). Site-directed mutagenesis of plasmid DNA was performed using PCR and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). PCRs were performed with a GeneAmp PCR system 2400 programmable thermal controller from Perkin Elmer (Wellesley, MA). Isolation of DNA fragments and plasmids was carried out using Qiagen kit reagents (Mississauga, Ontario, Canada). Automated fluorescent dye-terminator cycle sequencing to confirm all PMP22-derived constructs was performed at the University of Guelph Molecular Supercenter (Guelph, Ontario, Canada) using an ABI Prism 377 Automated Sequencer (Applied Biosystems, Foster City, CA). Standard recombinant DNA procedures were carried out as described by Sambrook et al. (1989).
Construction of Plasmids
All PMP22 constructs used in this study are shown schematically in Figure 3. A complete description of the construction of these constructs along with a list of the sequences of oligonucleotide primers used is available as supplemental information that can be viewed at http://www.plantphysiol.org.
Tobacco (Nicotiana tabacum) BY-2 Cell Cultures, Microprojectile Bombardment, and Immunofluorescence Microscopy
Tobacco suspension cultures were maintained and prepared for biolistic bombardment as described previously (Banjoko and Trelease, 1995). For experiments designed to determine the topological orientation of PMP22 within the peroxisomal boundary membrane, BY-2 cells were prepared for bombardment as described by Mullen et al. (2001b). Transient transformations were carried out using 10 μg of plasmid DNA with a Biolistic Particle Delivery System (Bio-Rad Laboratories, Mississauga, Ontario, Canada; Lee et al., 1997). For cotransient expression experiments, cells were bombarded with 5 μg of each plasmid DNA. After biolistic bombardment, cells were left for 4 to 45 h to allow transient expression of the introduced gene(s). BY-2 cells were then fixed in formaldehyde, incubated with pectolyase Y-23 (Kyowa Chemical Products, Osaka), and permeabilized in Triton X-100 or, for topology experiments, in digitonin (Lee et al., 1997).
Fixed, permeabilized cells were then processed for immunofluorescence microscopy as described previously (Trelease et al., 1996). Antibodies and sources were as follows (IgGs were affinity purified using a protein A-Sepharose column): mouse anti-myc IgGs (clone 9E10) and rabbit anti-myc IgGs (Berkeley Antibody Company, Richmond, CA); mouse anti-HA IgGs (clone 12CA5; Boehringer Mannheim, Mississauga, Ontario, Canada); mouse anti-α-tubulin IgGs (clone DM 1A) and rabbit anti-CAT IgGs (Sigma-Aldrich, St. Louis); mouse anti-CAT hybridoma medium; rabbit anti-cottonseed (Gossypium hirsutum) catalase IgGs (Kunce et al., 1988); rabbit anti-Arabidopsis PMP22 IgGs (Tugal et al., 1999); rabbit anti-castor bean (Ricinus communis) calreticulin (Coughlan et al., 1997); goat anti-mouse and goat anti-rabbit Alexa Fluor 488 IgGs (Cedar Lane Laboratories, Hornby, Ontario, Canada); and goat anti-mouse cyanine 3 (Cy3) IgGs and goat anti-rabbit rhodamine red-X IgGs (Jackson ImmunoResearch Laboratories, West Grove, PA). Controls included omitting primary antibodies and mock transformations with vector (pRTL2) alone.
Labeled cells were mounted on glass slides and were viewed using an epifluorescence microscope (Axioskop 2 MOT; Carl Zeiss, Thornwood, NY). Epifluorescence images were captured using a CCD camera (Retiga 1300; Qimaging, British Columbia, Burnaby, Canada). Images were deconvolved (a computational technique to reduce fluorescence from sources not in the plane of focus) and then adjusted for brightness and contrast using northern Eclipse 6.0 software (Empix Imaging, Mississauga, Ontario, Canada). Images were composed into figures using Adobe Photoshop 6.0 (Adobe Systems, Klamath Falls, OR).
In Vitro Membrane Association Experiments
Plasmids encoding various epitope-tagged and mutant versions of PMP22 were transcribed and translated in vitro using a TNT-coupled wheat (Triticum aestivum) germ extract system (Promega). Translation-grade l-35[S]-Met (specific activity > 1,000 Ci mmol–1) was from ICN Pharmaceuticals (Basingstoke, Hants, UK).
In vitro membrane association assays were performed using radiolabeled proteins and peroxisomes isolated from 3-d postimbibition sunflower (Helianthus annuus) cotyledons as described previously (Horng et al., 1995; Tugal et al., 1999). For each import assay, 150 to 200 μg of peroxisomal protein and 15 μL of translation product were used. After the import reaction, samples were reisolated by centrifugation through a 0.7 m Suc cushion and the supernatant plus Suc cushion was carefully removed. Pellets were solubilized in 1× SDS-PAGE sample buffer (Laemmli, 1970) at 37°C for 60 min. Twenty-five percent of each pellet fraction was separated on 14% (w/v) SDS-polyacrylamide gels alongside 4C-labeled marker proteins (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), and the radioactivity was detected by using a phosphorimager (Fuji1000; Fuji-film Electronic Imaging, Hempstead, Herts, UK).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining permission will be the responsibility of the requestor.
Acknowledgments
We thank Imogen Sparkes for some preliminary results and helpful discussions with in vitro insertion experiments, Barbara Johnson and Yeen Ting Huang for technical assistance, and Derek Bewley for critical reading of the manuscript. We thank also Richard Trelease and Sean Coughlan for providing antibodies. Erin Anderson and Andrew McCartney maintained tobacco BY-2 suspension cell cultures. We are grateful also to John Dyer for insightful discussion during the course of this work.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027870.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant no. 217291), by the Ontario Premier's Research in Excellence Award (to R.T.M.), and by the Biotechnology and Biology Research Council (grant no. 24/C12039 to A.B.).
The online version of this article contains Web-only information.
References
- Baerends RJ, Faber KN, Kram AM, Kiel JAKW, van der Klei IJ, Veenhuis M (2000) A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem 275: 9985–9995 [DOI] [PubMed] [Google Scholar]
- Banjoko A, Trelease RN (1995) Development and application of an in vivo plant peroxisome import system. Plant Physiol 107: 1201–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius U, Dehmel T, Gärtner J (2002) Two different targeting signals direct human peroxisomal membrane protein 22 to peroxisomes. J Biol Chem 277: 774–784 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Rio LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Coughlan SJ, Hastings C, Winfrey R (1997) Congin and characterization of the calreticulin gene from Ricinus communis L. Plant Mol Biol 34: 897–911 [DOI] [PubMed] [Google Scholar]
- Diestelkötter P, Just WW (1993) In vitro insertion of the 22-kD peroxisomal membrane protein into isolated rat liver peroxisomes. J Cell Biol 123: 1717–1725, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, McNew JA, Goodman JM (1996) The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol 133: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF (1997) Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J 16: 7326–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M (2002) Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiol 43: 1–11 [DOI] [PubMed] [Google Scholar]
- Hettema EH, Girzalsky W, Van den Berg M, Erdmann R, Distel B (2000) Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J 19: 223–233, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng JT, Behari R, Burke CA, Baker A (1995) Investigation of the energy requirement and targeting signal for the import of glycolate oxidase into glyoxysomes. Eur J Biochem 230: 157–163 [DOI] [PubMed] [Google Scholar]
- Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Iida R, Yasuda T, Tsubota E, Takatsuka H, Masuyama M, Matsuki T, Kishi K (2003) M-LP, Mpv17-like protein, has a peroxisomal membrane targeting signal comprising a transmembrane domain and a positively-charged loop and up-regulates expression of the manganese superoxide dismutase gene. J Biol Chem 278: 6301–6306 [DOI] [PubMed] [Google Scholar]
- Jones JM, Morrell JC, Gould SJ (2001) Multiple distinct targeting signals in integral peroxisomal membrane proteins. J Cell Biol 153: 1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunce CM, Trelease RN, Turley RB (1988) Purification and biosynthesis of cottonseed (Gossypium hirsutum L.) catalase. Biochem J 251: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee MS, Mullen RT, Trelease RN (1997) Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9: 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee CS, Karnik SK, Trelease RN (2003) Overexpression of a tail-anchored GFP fusion protein leads to “zippered” aggregation of plastids, mitochondria, and peroxisomes in plant cells. Traffic 4: 491–501 [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Trelease RN (2003) Peroxisomal ascorbate peroxidase resides within a domain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol 132: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon MT, McNew JA, Willy PJ, Goodman JM (1994) An internal region of the peroxisomal membrane protein PMP47 is essential for sorting to peroxisomes. J Cell Biol 124: 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT (2002) Targeting and import of matrix proteins into peroxisomes. In A Baker, I Graham, eds, Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 339–385
- Mullen RT, Flynn CR, Trelease RN (2001a) How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci 6: 256–261 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lee MS, Trelease RN (1997) Identification of the peroxisomal targeting signal for peroxisomal catalase. Plant J 12: 313–322 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Flynn CR, Trelease RN (2001b) Stable and transient expression of chimeric peroxisomal membrane proteins induces an independent “zippering” of peroxisomes and an endoplasmic reticulum subdomain. Planta 213: 849–863 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN (2000) The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J Biol Chem 275: 16337–16344 [DOI] [PubMed] [Google Scholar]
- Nito K, Yamaguchi K, Kondo M, Hayashi M, Nishimura M (2001) Pumpkin peroxisomal ascorbate peroxidase is localized on peroxisomal membranes and unknown membranous structures. Plant Cell Physiol 42: 20–27 [DOI] [PubMed] [Google Scholar]
- Pause B, Saffrich R, Hunziker A, Ansorge W, Just WW (2000) Targeting of the 22 kDa integral peroxisomal membrane protein. FEBS Lett 471: 23–28 [DOI] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (2001) Peroxisome biogenesis. Annu Rev Cell Dev Biol 17: 701–752 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ (2000) The genetics of peroxisome biogenesis. Annu Rev Genet 34: 623–652 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol 148: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Soukupova M, Sprenger C, Gorgas K, Kunau W-H, Dodt G (1999) Identification and characterization of the human peroxin PEX3. Eur J Cell Biol 78: 357–374 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Baker A (2002) Peroxisome biogenesis and protein import in plants animals and yeasts: enigma and variations. Mol Membrane Biol 19: 171–185 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S, Koller A, Snyder WB (2000) Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem 69: 399–418 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (1998) Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol 18: 2789–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelease RN (2002) Peroxisomal biogenesis and acquisition of membrane proteins. In A Baker, I Graham, eds. Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 305–337
- Trelease RN, Lee MS, Banjoko A, Bunkelmann J (1996) C-terminal polypeptides are necessary and sufficient for in vivo targeting of transiently expressed proteins to peroxisomes in suspension-cultured plant cells. Protoplasma 195: 156–167 [Google Scholar]
- Tugal HB, Pool M, Baker A (1999) Arabidopsis 22-kilodalton peroxisomal membrane protein: nucleotide sequence and biochemical characterization. Plant Physiol 120: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn-Brouwer T, Kragt A, Tabak HF, Distel B (2001) Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J Cell Sci 114: 2199–2204 [DOI] [PubMed] [Google Scholar]
- Wagner G, Stettmaier K, Bors W, Sies H, Wagner EM, Reuter A, Weiher H (2001) Enhanced γ-glutamyl transpeptidase expression and superoxide production in Mpv17–/– glomerulosclerosis mice. Biol Chem 382: 1019–1025 [DOI] [PubMed] [Google Scholar]
- Wang X, Unruh MJ, Goodman JM (2001) Discrete targeting signals direct Pmp47 to oleate-induced peroxisomes in Saccharomyces cerevisiae. J Biol Chem 276: 10897–10905 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal β-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]