Abstract
The nuclear lamina is a meshwork of intermediate-type filament proteins (lamins) that lines the inner nuclear membrane. The lamina is proposed to be an important determinant of nuclear structure, but there has been little direct testing of this idea. To investigate lamina functions, we have characterized a novel lamin B1 mutant lacking the middle ∼4/5 of its α-helical rod domain. Though retaining only 10 heptads of the rod, this mutant assembles into intermediate filament-like structures in vitro. When expressed in cultured cells, it concentrates in patches at the nuclear envelope. Concurrently, endogenous lamins shift from a uniform to a patchy distribution and lose their complete colocalization, and nuclei become highly lobulated. In vitro binding studies suggest that the internal rod region is important for heterotypic associations of lamin B1, which in turn are required for proper organization of the lamina. Accompanying the changes in lamina structure induced by expression of the mutant, nuclear pore complexes and integral membrane proteins of the inner membrane cluster, principally at the patches of endogenous lamins. Considered together, these data indicate that lamins play a major role in organizing other proteins in the nuclear envelope and in determining nuclear shape.
Keywords: intermediate filament, nuclear lamina, nuclear pore complex, lamina-associated polypeptide, nuclear shape
Introduction
The nuclear envelope (NE) is a double membrane system continuous with the ER that forms the nuclear boundary. It is punctuated by nuclear pore complexes (NPCs), which regulate molecular transport into and out of the nucleus (reviewed in Gorlich and Kutay 1999). Underlying the inner surface of the NE is the nuclear lamina, a fibrous meshwork consisting of lamins (Aebi et al. 1986) and a number of more minor lamina-associated polypeptides (LAPs; reviewed in Gerace and Foisner 1994; Stuurman et al. 1998).
Lamins are type V intermediate filament (IF) proteins (McKeon et al. 1986). The B1 and B2 lamin subtypes are present in nuclei of nearly all vertebrate cells, whereas the A and C subtypes appear only at or after differentiation (Rober et al. 1989). All lamin subtypes colocalize by light microscopy (reviewed in Gerace and Foisner 1994; Stuurman et al. 1998). Moreover, A and B subtypes can interact in vitro (Georgatos et al. 1988; Ye and Worman 1995), although there is no in vivo indication of the relevance or existence of such heterotypic associations in the assembled lamina. In addition to interacting with LAPs, lamins bind to chromatin (Glass and Gerace 1990), and this may be responsible for anchoring interphase chromosomes to the NE.
The best characterized LAPs are integral membrane proteins of the inner nuclear membrane (INM). These include LAPs 1 and 2 (Senior and Gerace 1988; Foisner and Gerace 1993), LBR (Ye and Worman 1994), and emerin (Fairley et al. 1999). Collectively, these integral proteins appear to help connect the lamina to the NE (reviewed in Collas and Courvalin 2000). The importance of the lamina has recently been underscored by the finding that certain point mutants of emerin in humans cause Emery-Dreifuss muscular dystrophy, and that some lamin point mutations cause lipodystrophy and cardiac dysfunction as well as Emery-Dreifuss muscular dystrophy (reviewed in Wilson 2000; see also Cao and Hegele 2000). How these defects cause such different diseases remains unclear. Unraveling this mystery will require a further understanding of the complex structure of the assembled lamina and of its functions, many of which remain speculative.
A role for the lamins in NE structure is inferred from their abundance and uniform distribution at the nuclear periphery and from their structural properties, such as their ability to assemble into IF-related structures in vitro (Stuurman et al. 1998), the resistance of the polymer to extraction by Triton X-100, and the skeleton-like appearance of the isolated lamina (Dwyer and Blobel 1976; Aebi et al. 1986). The only functional analysis of the role of lamins in nuclear structure comes from depletion studies. Fragile, growth-inhibited nuclei result from in vitro nuclear assembly with lamin-depleted Xenopus egg extracts, though no conspicuous perturbation of nuclear shape is observed (reviewed in Lourim and Krohne 1994; see also Benavente and Krohne 1986). Furthermore, minor aberrations in nuclear shape in some cell types result from reduction of lamin B levels in Drosophila (Lenz-Bohme et al. 1997) and from disruption of the mouse lamin A/C gene (Sullivan et al. 1999). However, it is not clear from these studies whether the role of the lamina in nuclear structure is direct or indirect (e.g., by effects on NPCs or chromosomes).
Similar to other IF proteins, lamins contain a long central α-helical rod domain flanked by nonhelical head and tail domains. Lamins differ from other IF proteins by having an additional six heptads in the rod domain (resulting in a rod of 354 vs. 310 residues), a nuclear localization signal (NLS), and a carboxyl-terminal CaaX box for farnesylation (reviewed in Stuurman et al. 1998). The basic unit of lamin assembly is a two-stranded coiled coil formed by parallel, unstaggered association of the rod domains due to heptad repeats (reviewed in Stuurman et al. 1998). This dimer further assembles into higher order structures, such as 10-nm filaments, by head-to-tail interactions and by staggered antiparallel lateral interactions (Moir et al. 1991; Heitlinger et al. 1992; Stuurman et al. 1996). The lamina polymer continuously increases in mass during interphase in cycling cells as the NE increases in surface area (reviewed in Gerace and Foisner 1994). It is unclear, however, how new lamin subunits are integrated into the lamina during this process. In contrast, nuclear reassembly at the end of mitosis apparently involves de novo assembly of the lamins at the NE via interactions with nuclear membrane proteins and chromosomes (reviewed in Collas and Courvalin 2000).
Although the precise interactions involved in higher order lamin assembly are not clear, it is apparent that the head, tail, and rod domains all make important contributions. Mutational analysis of lamins and other IF proteins has shown that the highly conserved sequences at both ends of the rod domain are particularly important for assembly in vitro (Coulombe et al. 1990; Heald and McKeon 1990; Hatzfeld and Weber 1991; Stuurman et al. 1998). The only analysis of more internal regions of the lamin rod involves overexpression of a mutant lacking a six-heptad region that is absent from other IF proteins, which yields no significant nuclear structural alterations (Mical and Monteiro 1998).
To understand more about the functions of the lamin rod domain, we engineered a deletion of the internal ∼4/5 of the lamin B1 rod. This mutant is predicted to dimerize and engage in head-to-tail interactions, but would be expected to have reduced lateral interactions involved in higher-order assembly. To our surprise this mutant formed IF-like structures in vitro. Furthermore, when overexpressed in cultured cells, it became enriched in patches at the NE. At the same time, endogenous lamins and other NE proteins also became concentrated in patches, which to varying degrees were separated from the mutant. Accompanying the reorganization of the lamina into a patchwork, nuclear shape was grossly altered by lobulation. This is the first lamin mutant described that perturbs the substructure of the assembled lamina and that causes drastic alterations in nuclear shape. Our findings provide new insight on the role of the lamina in NE organization and the importance of the lamin rod domain for lamina stability and for heterotypic associations among lamin subtypes.
Materials and Methods
Plasmid Construction
The human lamin B1 coding sequence was amplified by PCR with primers that added 5′ BamHI/ Nde1 and 3′ Not1 sites. To produce B1Δrod, these primers were used with internal primers containing Hind3 sites that fused nucleotides 207 and 1017 via an added alanine codon. The individual lamin fragments were also subcloned (B1N and B1C). To produce A/B1Δrod, the equivalent 5′ sequence for human lamin A was fused to the 3' sequence from lamin B1. These genes were moved to vectors for mammalian transfection [CMV-driven pHHS10B; with a hemagglutinin (HA) epitope tag] and protein expression (pET28a; Novagen, with a 6× His tag).
Transient Transfection in Cultured Cells
Adherent HeLa, COS-7, or normal rat kidney (NRK) cells were plated on polylysine-coated slides in DMEM supplemented with 10% FBS and l-glutamine at low confluency (∼15%) as cells were usually harvested as long as 60 h after transfection. Roughly 12 h after plating, DNA was transfected using Fugene 6 (Roche) according to the manufacturer's instructions. To assess cell division, cells were plated onto CELLocate coverslips (Eppendorf) and cotransfected with pEGFP-F (CLONTECH Laboratories, Inc.). Fluorescent cells were counted at 20 and 60 h after transfection.
Antibody Production
Polyclonal antibodies were produced in rabbits and guinea pigs from peptides comprising the principal chromatin binding site of human lamin A (residues 396–429; Taniura et al. 1995), and the predicted chromatin binding sites of human lamin B1 (residues 391–428) and human lamin B2 (SPSPSSRVTVSRATSSSSGSLSATGRLGRSKRKRLEVEEPLGSGPS- VLGTGTGGR—this differs from the reported sequence (Genbank M94362), but was repeatedly recovered by PCR from a HeLa cDNA library). Peptides were generated using the pPEP-T vector system (Kammerer et al. 1998). Western blot analysis indicated that each serum was specific for its lamin isotype.
Immunofluorescence Microscopy and Apoptosis Analysis
Cells were fixed for 7 min in 3.7% formaldehyde, permeabilized for 6 min in 0.2% Triton X-100, blocked with 4% BSA in PBS, and reacted for 40 min at room temperature with antibodies to lamins (above) and/or to the NPC (mAb RL1, Snow et al. 1987; affinity purified Tpr polyclonal IgG, gift of P. Frosst, The Scripps Research Institute), LAP2 (mAb RL29; Foisner and Gerace 1993), LAP1C, (mAb RL13; Senior and Gerace 1988), LBR (rabbit serum provided by H. Worman, Columbia University, New York, NY), and the HA tag of the transfected proteins (mAb HA.11; Covance, or purified polyclonal; Upstate Biotechnology). After washing, fluorophore-conjugated secondary antibodies were added for 30 min, followed by 10 min with DAPI to visualize DNA, and mounting in fluoromount G (EM Sciences). Apoptosis was assessed at 60 and 74 h after transfection using the In Situ Cell Death Detection Kit (TUNEL assay; Roche).
Images were obtained using an Axiovert S100TV microscope (Carl Zeiss, Inc.) interfaced to a confocal system (MRC1024; Bio-Rad Laboratories) with 488, 568, and 647 nm krypton/argon laser lines or using an microscope (1Χ70; Olympus) interfaced with a DeltaVision system (Applied Precision, Inc.) and subsequently deconvolved with DeltaVision version 2.0 software. All fluorescence images and micrographs shown in this study were prepared for figures using Photoshop 5.0.
Nuclear Import Assays
To assess nuclear import, Cy5-labeled BSA conjugated to an NLS was microinjected into the cytoplasm of B1Δrod-transfected cells. This was coinjected with Texas red–labeled BSA (Jackson ImmunoResearch Laboratories) that lacked an NLS or FITC-dextran (150 kD; Molecular Probes) to assess nuclear integrity. After 45 min, cells were fixed and processed for immunofluorescence microscopy. The import competence of transfected cells was also examined using a permeabilized cell assay (Adam et al. 1990).
Thin Section EM
HeLa cells cotransfected with B1Δrod and pEGFP-F were harvested by trypsinization at 60 h after transfection, washed, and resuspended in 1× PBS (Ca2+, Mg2+ free), 1 mM EDTA, 25 mM Hepes, pH 7.0, 1% dialyzed FBS. Fluorescent cells were collected by FACS®, fixed with 2% glutaraldehyde and 1% osmium tetroxide, embedded in Epon, and sectioned according to standard protocols. Micrographs were recorded with a Philips EM-208 at 70 kV.
Protein Purification
Lamin B1 and B1Δrod were purified from inclusion bodies. The proteins were induced in Bl21-(DE3) cells at A595 0.7 for 3 h at 37°C that were lysed by sonication in PBS containing 1.5 mM β-mercaptoethanol and protease inhibitors. The pellets from a 7-min centrifugation at 10,000 g were washed with 0.2% Triton X-100, resuspended in 20 mM Tris, pH 8.0, 300 mM NaCl, 8 M urea, 3 mM β-mercaptoethanol, incubated with nickel resin (QIAGEN), and eluted with the same buffer containing 200 mM imidazole, and dialyzed into 20 mM Tris-HCl, pH 8.0, 8 M urea, 2 mM DTT, 1 mM EDTA with protease inhibitors for storage. B1N and B1C were prepared similarly except that cells were lysed in urea as these proteins did not form inclusion bodies.
In Vitro Assembly of Lamins
The solubility properties of the proteins were compared as in Glass and Gerace 1990. In brief, they were diluted in urea to 250, 100, 30, 10, and 3 μg/ml, and then dialyzed into 10 mM Tris, pH 8.8, 250 mM KCl, 1 mM DTT with protease inhibitors followed by 10 mM Tris, pH 8.8, 70 mM KCl, 1 mM DTT, 5 mM MgCl2, followed by 50 mM Hepes, pH 7.3, 70 mM KCl, 1 mM DTT, 5 mM MgCl2. This was subjected to centrifugation at 10,000 g for 7 min and pellets resuspended in the same volume as supernatants before applying equal volumes to SDS-PAGE.
For cross linking, proteins were diluted in urea storage buffer to 0.05 mg/ml, and then dialyzed into 20 mM Na2HPO4, pH 9, 300 mM KCl, 0.5 mM EDTA, 1 mM DTT. Aggregated material was removed by centrifugation at 10,000 g for 20 min and the supernatants were incubated with 0.01% glutaraldehyde for 0, 2, 10, or 20 min. Reactions were stopped by addition of 1 M glycine and precipitated with 15% TCA before analysis by SDS-PAGE.
To investigate filament assembly, purified lamin B1 and B1Δ rod were dialyzed out of urea into 20 mM Tris pH 8.8, 300 mM NaCl, 1 mM EDTA, 1 mM DTT. Aliquots were removed at different times (5 min to 1 h), applied to glow discharged carbon-coated grids, negatively stained with 1-2% uranyl acetate, and viewed on a Philips CM100 at 100 kV.
Lamin Binding Assays
Purified wild-type (WT) lamin B1 and B1Δrod were coupled in urea to Affi-gel 15 matrix (Bio-Rad Laboratories) as in Georgatos et al. 1988 with ∼3 mg bound/ml matrix. Lamins A and C (prepared as in Glass and Gerace 1990) or lamin B1 were diluted to 20 μg/ml and dialyzed out of urea into 25 mM Hepes, pH 7.5, 300 mM NaCl, 5 mM β-mercaptoethanol. This was centrifuged (10,000 g for 10 min) to remove aggregated proteins and incubated with the matrix overnight at 4°C. The columns were washed in the same buffer and eluted with increasing concentrations of urea. Fractions were analyzed by Western blotting with the antibodies described above.
Biophysical Analysis
For circular dichroism analysis, proteins at 0.3 mg/ml were dialyzed from urea storage buffer into 20 mM Na2HPO4, pH 9, 150 mM KCl to maintain most lamins in solution. Insoluble material was removed by centrifugation at 10,000 g for 10 min shortly before determination of the final protein concentration for conversion to mean residue ellipticity and recording of dichroic spectra with an AVIV CD spectrometer using a 1-nm bandwidth, 4-s scan time, and 5-mm cuvette at 25°C. An average of four scans with buffer subtracted is shown.
For Fourier transform infrared spectroscopic (FTIR) analysis, filaments prepared as above were pelleted by centrifugation at 10,000 g for 10 min and washed in water. The pellet was disrupted with a pipette tip, layered onto a 5-mm-thick CaF lens, dried under nitrogen, and viewed in a Nicolet MAGNA-IR 550 Spectrometer Series II with a spectral resolution of 4 cm−1 using OMNIC analysis software. A water blank was subtracted from the spectra (128 interferograms).
Results
In Vitro Assembly of B1ΔRod
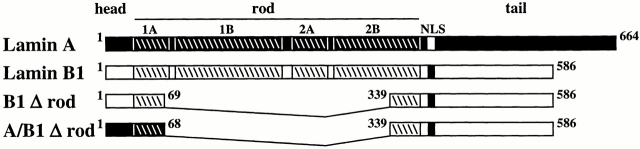
The WT rod domain of lamins contains 48 heptads, which are subdivided into four α-helical regions separated by short spacers (Fig. 1). To analyze the role of the rod domain in lamin assembly and function, we constructed a deletion mutant of lamin B1 containing the entire head and tail domains, but lacking the rod domain except for the first and last five heptads, which were fused in register (Fig. 1, B1 Δ rod). We also generated a chimeric Δrod mutant by fusing the head and first five heptads of the lamin A/C rod with the last five rod heptads and tail of lamin B1 (Fig. 1A/B1 Δ rod). These forms are predicted to have a single α-helix of 10 heptads that should form a coiled coil dimer by computer modeling (Lupas 1996). Consistent with this, recombinant B1Δrod (Fig. 2 A), like WT lamin B1, forms a dimer in solution at pH 9 as determined by cross linking (Fig. 2 B; note this is the same material shown in A) and yields a strong α-helical signal by circular dichroism (Fig. 2 C).
Figure 1.
Design of mutant lamins. Mammalian lamins contain an NH2-terminal head domain (∼33 amino acids) followed by a coiled-coil rod domain (354 amino acids) and a COOH-terminal tail domain (200–300 amino acids). The rod is predicted to contain four separate α-helices: coil 1A, 1B, 2A, and 2B of 6, 20, 6, and 16 heptads, respectively (a slash delineates each heptad). The B1Δrod mutant lacks most of the rod, retaining only five heptads at each end that are fused in register (via an added alanine residue replacing the valine in position g of the fifth NH2-terminal heptad). The A/B1Δrod mutant contains the head and first five heptads of lamin A (black) fused to the last five heptads and tail of lamin B1 (white).
Figure 2.
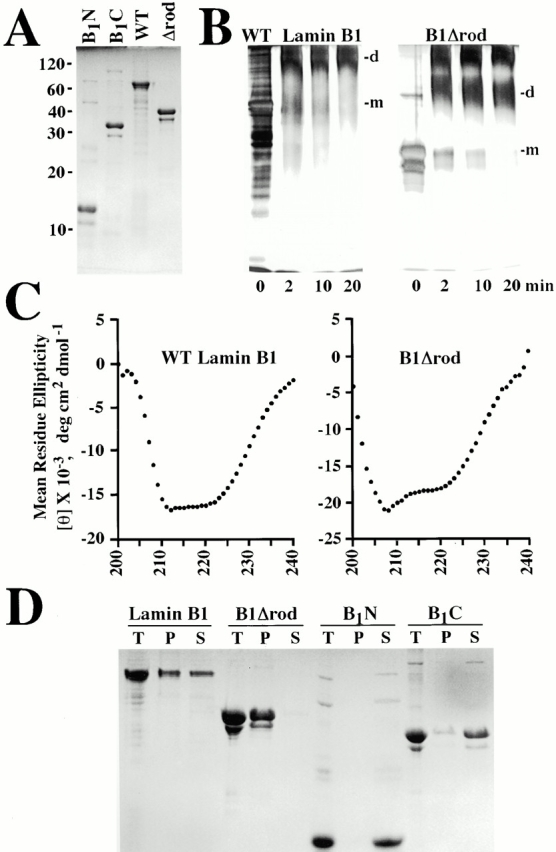
The B1Δrod mutant has characteristics of IF proteins. (A) WT lamin B1, B1Δrod, and the two lamin B1 fragments that were fused in B1Δrod (B1N and B1C) were expressed in Escherichia coli, and were analyzed by SDS PAGE and staining with Coomassie blue. (B) The B1Δrod mutant forms dimers. Soluble WT lamin B1 or B1Δrod (the same material shown in A) were cross linked with glutaraldehyde for 0–20 min and analyzed by SDS-PAGE and staining with silver. No monomeric (m) protein was observed at any time after incubation with cross linker: rather, a diffuse band at the expected Mr for the dimer species (d) was observed that did not increase over time, indicating the stability of this soluble population. An apparent cross-linked tetramer also was seen for B1Δrod, but a tetrameric form of WT lamin B1 was too large to fully migrate into the gel. Note that silver staining does not give a linear representation of the protein species (compare 0-min samples in B to Coomassie blue–stained samples in A). (C) The B1Δrod mutant has considerable α-helical structure. The dichroic spectra of soluble WT lamin B1 and B1Δrod proteins (in a similar buffer as in B) contained strong minima at 208 and 222 nm indicative of α-helical structure. (D) Relative solubility of B1Δrod and its individual segments. Proteins at 250 μg/ml were dialyzed into buffers known to promote lamin assembly. Insoluble material was pelleted (P) and compared with the supernatant (S) and the total starting material (T). This demonstrated that the B1N and B1C fragments are extremely soluble compared with their fusion product, B1Δrod, and WT lamin B1.
In addition to WT lamin B1 and B1Δrod, we prepared separately the two parts of lamin B1 that are fused to form B1Δrod (Fig. 2 A, B1N and B1C) and compared the solubility properties of these fragments and the longer lamin constructs. Proteins were dialyzed from urea into buffers that support lamin assembly in vitro (i.e., an equilibration buffer followed by a polymerization buffer; see Materials and Methods), and insoluble material was pelleted. Some B1Δrod pelleted at the lowest concentration tested (3 μg/ml, data not shown). In contrast, even at 250 μg/ml (where all of B1Δrod and most of WT lamin B1 was pelletable), the two lamin B1 fragments that are fused in B1Δrod were completely soluble (Fig. 2 D). This suggests that the individual NH2- and COOH-terminal segments of B1Δrod do not have a strong capacity for self assembly by themselves, but strongly promote assembly when they are linked together in B1Δrod, which would allow them to drive head-to-tail polymerization of the mutant lamin (see Discussion).
When assembly of the WT and mutant lamins was analyzed by EM using negative staining, ∼10-nm filaments were observed for both proteins, even after dialysis into the equilibration buffer alone (Fig. 3, A–B). These filaments were similar to one another and to filaments formed by other B-type lamins (Heitlinger et al. 1991; Sasse et al. 1997). When WT lamin B1 and B1Δrod were mixed together and dialyzed into the assembly buffers, filaments similar to those formed by each alone were observed (data not shown), indicating that B1Δrod does not disassemble WT lamin B1 filaments nor block their formation. However, we could not distinguish whether these filaments were homo- or heterotypic because we lacked specific reagents to distinguish the two proteins.
Figure 3.
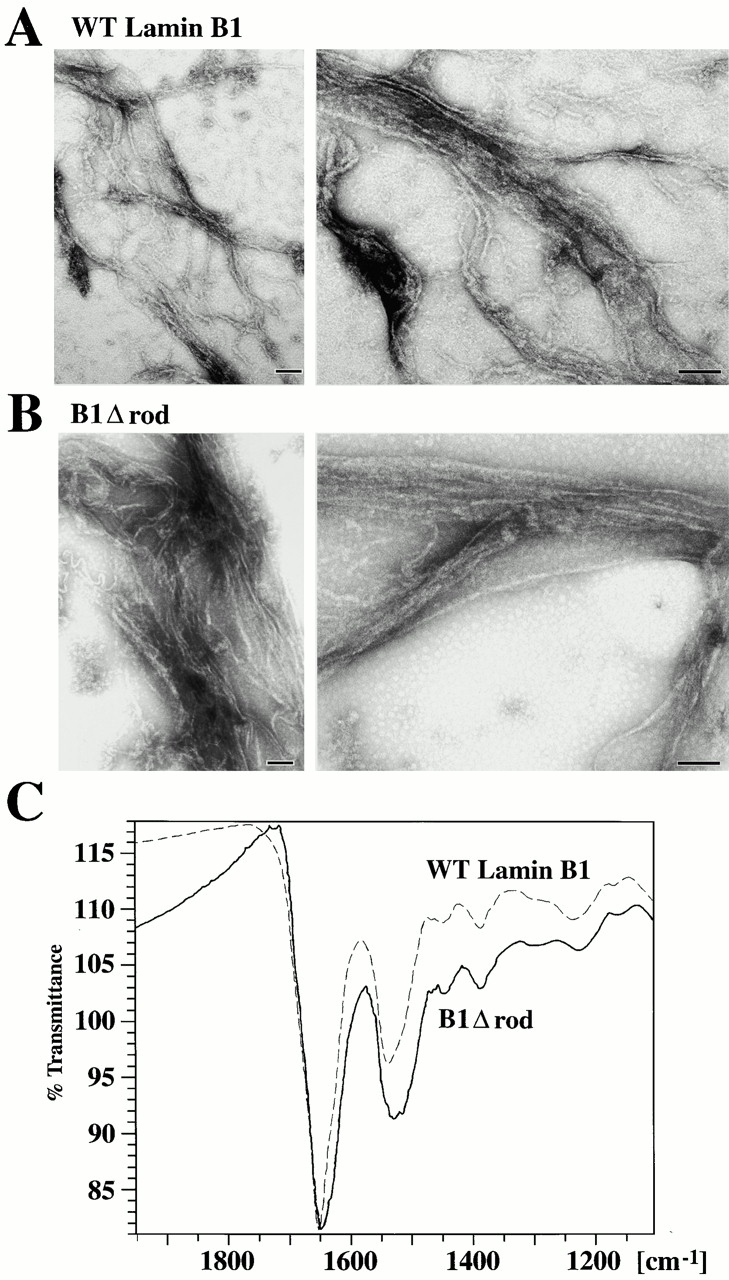
In vitro assembly of B1Δrod. (A–B) WT lamin B1 and B1Δrod that were dialyzed out of urea-formed filaments. The structures formed by each protein were similar in appearance (bars, 100 nm). (C) FTIR indicates α-helical structures occur in the filamentous form of B1Δrod, similar to WT lamin B1. The proteins were dialyzed using conditions that had yielded filaments by EM analysis. Polymerized material was pelleted and analyzed by FTIR. The scans of B1Δrod and WT lamin B1 were very similar although the band around 1650 was wider for B1Δrod than for WT lamin B1. The position of the bands is consistent with α-helical rather than β-sheet characteristics (see text).
Certain short peptides derived from the ends of the rod domain of desmin (a type III IF protein) could assemble into filaments in vitro, but these filaments had a β sheet substructure as revealed by FTIR (Geisler et al. 1993). As an additional criterion to determine whether filaments formed from B1Δrod were IF-like, we analyzed the assembled material by FTIR. The filaments formed by both B1Δrod and WT lamin B1 had a strong, wide band at ∼1,650 cm−1 (Fig. 3 C). This is near the band at 1,658 that is characteristic of α-helical structure, and is distinct from the bands at 1,620 and 1,680 cm−1 that characterize β sheet. The only other IF analyzed by FTIR is desmin, for which the center of the wide peak was shifted to 1,640 cm−1 (Heimburg et al. 1996). The band we see is roughly between these two, indicative of α-helical and inconsistent with β-sheet structures. In summary, based on biochemical, biophysical, and structural criteria, B1Δrod is able to form IF-related filaments in vitro despite the absence of ∼4/5 of the rod domain.
Expression of the Lamin B1Δrod Mutant Leads to Aberrant Nuclear Shape
To examine the effect of B1Δrod polymers on the endogenous lamins, we expressed WT lamin B1 and B1Δrod (both fused to a HA tag) in COS-7, HeLa, and NRK cells by transient transfection. At intermediate to long times after transfection (see below), the nuclei of all cell types transfected with B1Δrod were highly lobulated and irregularly shaped (Fig. 4). Much of the mutant lamin (Fig. 4, red) became localized to the NE, which was identified as the boundary of the DNA staining region (Fig. 4 A, gray) and the border of the nucleus seen by differential interference contrast microscopy (Fig. 4 B). As commonly observed for proteins expressed by transfection, the mutant also appeared in cytoplasmic (Fig. 4 B, top, arrow) as well as intranuclear (not shown) aggregates in some cells. The mutant lamin frequently was concentrated in distinct NE patches of variable sizes, rather than being uniformly distributed throughout the NE (Fig. 4 B, compare the two lobules with arrowheads). In contrast to cells transfected with B1Δrod, most cells transfected with WT lamin B1 had normally shaped, ovoid nuclei (see Fig. 6 A, below). Some had morphological deformations and NE invaginations, but these were relatively minor compared with cells transfected with the mutant.
Figure 4.
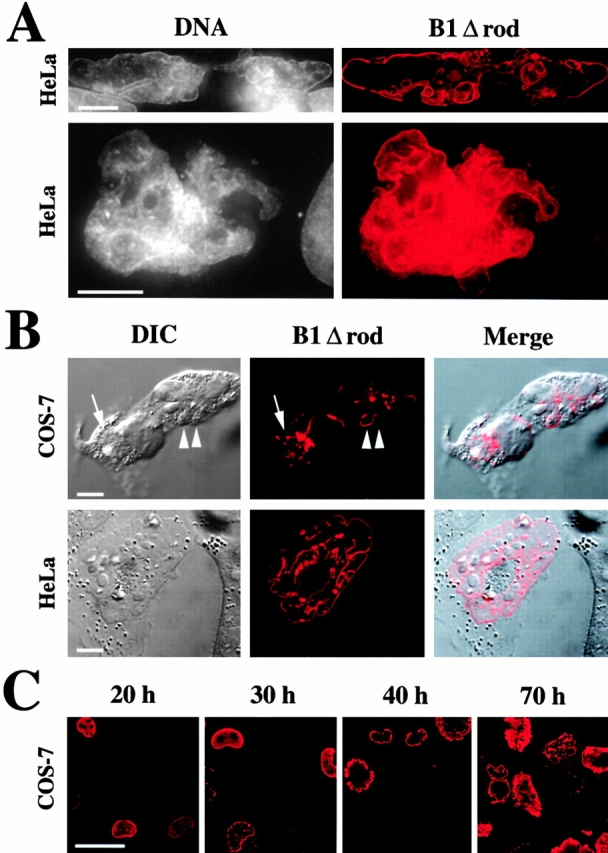
The B1Δrod mutant lamin grossly alters nuclear morphology. (A) Volume projections of deconvolved optical sections reveal that DNA (gray) is bounded by the mutant lamin (red). (B) Differential interference contrast (DIC) images of cells transfected with B1Δrod (left) and confocal sections of the same cells immunostained for the mutant's HA epitope tag (middle, red). The lobules observed by DIC imaging correspond to those observed with mutant lamin staining (Merge). Bars, 10 μm. (C) The lobulation phenotype increases over time. Representative fields of cells immunostained for the HA epitope from a COS7 transfection with B1Δrod are shown at 20, 30, 40, and 70 h after transfection. Initially, the mutant lamin is evenly distributed at the NE, but over time its distribution becomes patchy. The appearance of nuclear lobules correlates with the redistribution of the mutant into patches. This time course was slower for transfections with efficiencies <10%. Bar, 50 μm.
Figure 6.
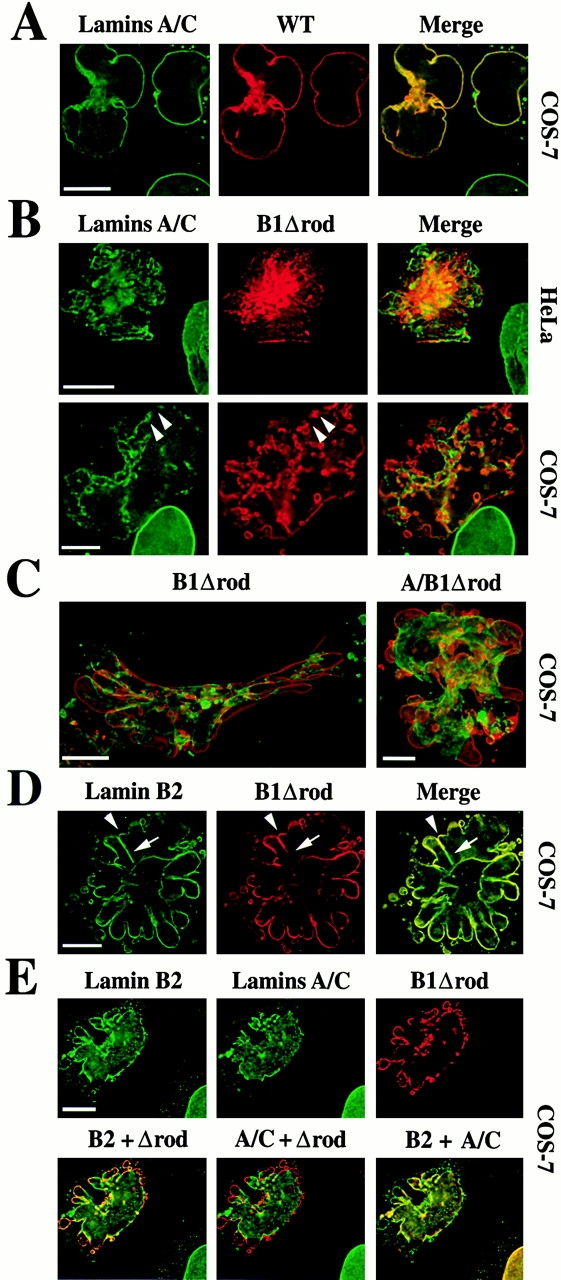
Expression of B1Δrod alters the distribution of endogenous lamins. Transfected cells were analyzed by immunofluorescent staining to detect the exogenously expressed lamin (HA epitope tag, red) and endogenous lamins (green). (A) Localization of lamins A/C in cells transfected with WT lamin B1. Both endogenous and exogenous lamins are uniformly distributed throughout the NE with complete colocalization (yellow). (B) Localization of lamins A/C in cells transfected with B1Δrod. Endogenous A/C lamins lose their normal uniform distribution and cluster at the NE, exhibiting little colocalization with B1Δrod in HeLa (top) and COS-7 (bottom) cells. Deconvolved optical sections are shown in A and B. (C) The A/B1Δrod hybrid mutant yields a similar phenotype to B1Δrod. Volume projections from deconvolved optical sections through COS-7 cells transfected with B1Δrod (left) or A/B1Δrod (right) are shown. Merged images are shown with the Δrod mutants in red and the A/C lamins in green. (D) Localization of endogenous lamin B2 in cells transfected with B1Δrod. More colocalization with the mutant was observed than for A/C lamins. (E) Lack of complete colocalization between A/C and B2 lamins. COS-7 cells triple labeled for lamin B2, lamins A/C, and B1Δrod (top) and the merges of each pair (bottom) revealed that endogenous A/C and B2 lamins become partially segregated in mutant cells. Colocalization is indicated by yellow for lamin B2 (green) and B1Δrod (red), lamins A/C (green) and B1Δrod (red), and lamins B2 (green) and lamins A/C (red). Deconvolved sections are shown in C and D. Bars, 10 μm.
The B1Δrod phenotype developed over time. At 20 h after transfection, the mutant protein was uniformly distributed at the NE and nuclei still had a normal (ovoid) shape (Fig. 4 C). By 30 h, the distribution of the mutant had become patchy in many cells, but the nuclei were still largely ovoid. By 40 h, most nuclei had a patchy distribution of B1Δrod and had assumed an aberrant, highly lobulated shape, which was yet more pronounced at 70 h (Fig. 4 C). Regardless of their aberrant nuclear shape, none of >100 cells analyzed that were expressing B1Δrod exhibited the fragmented and/or condensed DNA characteristic of apoptosis and necrosis (see Materials and Methods).
This time course suggested that the NE deformations occur during interphase nuclear growth rather than upon postmitotic NE reassembly. To test whether cells expressing B1Δrod could still undergo division, HeLa cells were plated onto marked coverslips, WT or mutant lamins were cotransfected with a green fluorescent protein (GFP) marker, and the fluorescent cells were counted at 20 and 60 h after transfection. Cells transfected with WT lamin B1 or GFP alone doubled, while cells transfected with B1Δrod only slightly increased in number (Table ). This indicates that the latter are growth inhibited, implying that the mutant lamin causes disruption of nuclear shape during interphase growth.
Table 1.
Cell Division in WT Lamin B1 and B1Δrod-transfected Cells, Cotransfected with a GFP Marker
| Plasmids transfected | No. cells | No. cells | Fold increase |
|---|---|---|---|
| 20 | 60 | ||
| GFP | 211 | 445 | 2.1 |
| GFP + WT Lamin B1 | 160 | 385 | 2.4 |
| GFP + B1Δrod | 143 | 180 | 1.2 |
The morphology of cells expressing B1Δrod was examined in detail by thin-section EM. Consistent with the light microscopy, the nucleus of some of the transfected cells was highly lobulated (Fig. 5 A), even though chromatin was surrounded by a NE with a double membrane (Fig. 5B, Fig. C, and Fig. E) and NPCs (Fig. 5, arrowheads). Unusual membrane-delimited structures were observed in the cytoplasm, some of which contained several concentric layers of membranes (Fig. 5 A, *, and D). One such structure was observed within the NE (Fig. 5 E, *), possibly reflecting an intermediate in their formation. By contrast, mock or WT lamin B1-transfected cells had an ovoid nucleus, no unusual cytoplasmic membrane structures, and only a thin, uniform zone of condensed chromatin underlying the NE (data not shown). Thus, cells expressing the B1Δrod mutant maintain a double membrane NE with NPCs, even though nuclear shape is drastically altered.
Figure 5.
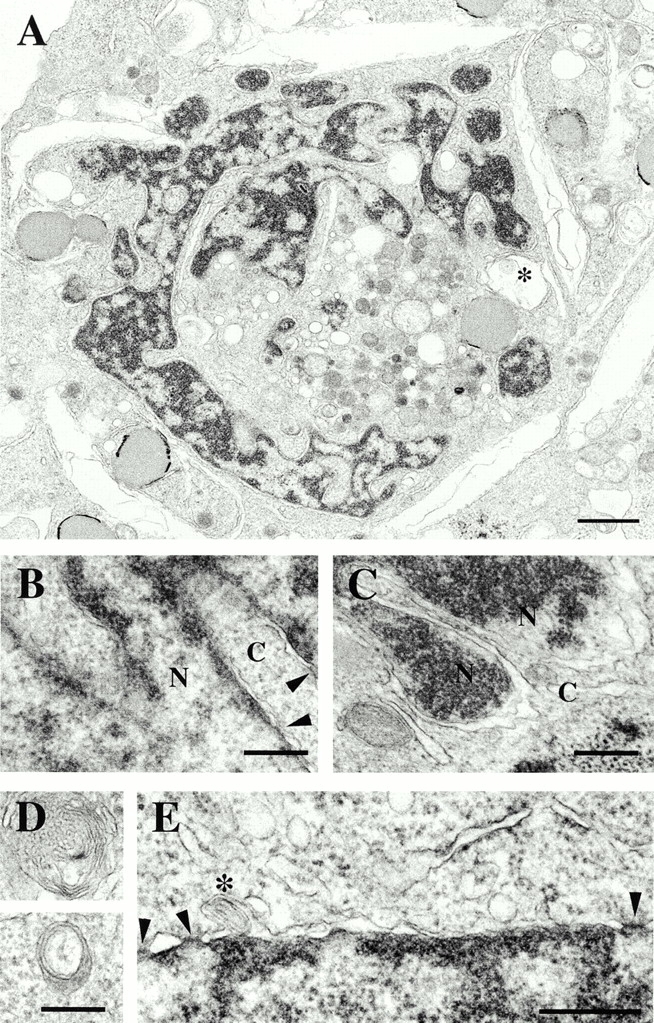
Ultrastructural analysis of a HeLa cell expressing B1Δrod by thin section EM. (A) Low magnification view of a whole nucleus. Bar, 1 μm. *Concentric membrane structure. (B–E) High magnification views. Bars, 300 nm. (B and C) A double membrane encloses the dense chromatin regions within lobules. Cytoplasm (C) and nucleus (N) are indicated. Arrowheads delineate nuclear pores. (D) Views of two concentric membrane structures in the cytoplasm. (E) Arrowheads delineate nuclear pores; *concentric membrane contained within the NE.
Organization of Lamins in Cells Expressing Δrod Mutants
To gain further insight into the mechanism by which expression of the mutant lamin alters nuclear shape, we compared the localization of endogenous and mutant lamins in B1Δrod-transfected cells. In cells transfected with WT lamin B1, all endogenous and exogenous lamins exhibited the continuous distribution throughout the NE that is normally seen in untransfected cells, even if minor aberrations in shape occurred (Fig. 6 A, yellow denotes colocalization). Other NE proteins (lamin B2, LAP2, LBR, and NPC antigens) also exhibited a normal uniform NE distribution in cells transfected with WT lamin B1 (data not shown).
In contrast, lamins A/C were concentrated in NE patches in B1Δrod-transfected cells, and these patches only partially overlapped with B1Δrod patches (Fig. 6 B). The patches varied in size and distribution. In some cells, large NE domains encompassing entire lobules were dominated by one or the other protein (Fig. 6 B, top). In other cells, small patches of one or the other protein alternated within a single lobule (Fig. 6 B, arrowheads). The alternating patches were sometimes seen more clearly in volume projections of nuclei (Fig. 6 C, left).
We also transfected cells with a mutant in which the head domain and first five heptads of lamins A/C were fused to the last five heptads of the lamin B1 rod plus the B1 tail domain (A/B1Δrod; Fig. 1). The A/B1Δrod hybrid (Fig. 6 C, right) was very similar to the B1Δrod mutant in terms of its patchy localization pattern and its effects on nuclear shape. Thus, incorporating the head domain of lamin A in the mutant did not alter the extent of mutant colocalization with endogenous lamins A/C.
Endogenous lamin B2 also was concentrated in patches in cells expressing the B1Δrod mutant. However, the separation of the patches from the mutant was not as pronounced as for lamins A/C. Either lamin B2 or B1Δrod were enriched in some patches (Fig. 6 D, arrows), whereas both proteins appeared to be colocalized in others at the resolution of light microscopy (Fig. 6 D, arrowheads). To directly evaluate the colocalization of lamins A/C, B2, and the mutant, we carried out triple labeling experiments in cells transfected with B1Δrod. Consistent with the finding that each endogenous lamin subtype overlapped to different degrees with B1Δrod patches, lamins A/C and B2 did not completely overlap with each other in these cells (Fig. 6 E). This is in contrast to nontransfected cells, where lamins A/C and B2 exhibit complete colocalization (Fig. 6 E, nucleus at bottom right).
We found that there were some patches of endogenous lamin B1 that lacked a substantial concentration of the mutant (data not shown). However, it was not possible to evaluate the degree of separation of WT lamin B1 and B1Δrod patches, because our lamin B1 subtype-specific antibodies recognize B1Δrod as well. In summary, expression of a mutant lamin lacking most of the rod domain, regardless of whether it contains an A- or B-type NH2 terminus, grossly alters the normal uniform distribution of all lamin subtypes at the NE.
Binding Interactions of B1Δrod
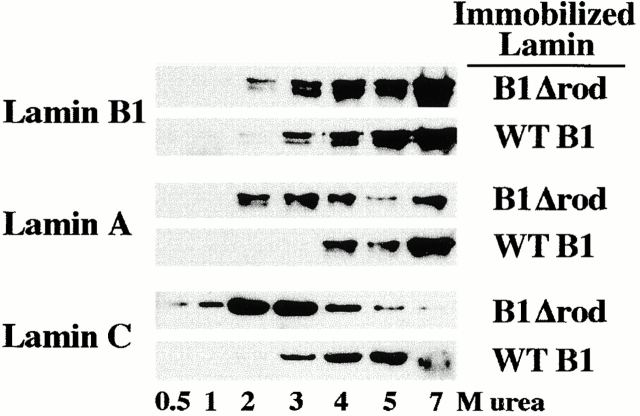
Our observation that B1Δrod colocalized with lamin B2 to a greater extent than with lamins A/C suggests that the portion of the lamin rod deleted in the mutant is more important for association with certain lamin subtype(s) than with others. To test this, we analyzed the relative affinities of lamin A, B1, and C for immobilized WT lamin B1 vs. B1Δrod by incubating a dilute lamin solution with each immobilized lamin matrix, and then eluting the bound protein with increasing concentrations of urea (Fig. 7). Lamin B1 had a minimal difference in affinity for the two matrices, with most protein eluting from both matrices between 4 and 7 M urea. By contrast, lamin A exhibited a significant difference in binding to the two matrices. The majority eluted from the B1Δrod matrix between 2 and 4 M urea, whereas the majority eluted from the WT matrix at 7 M. Lamin C showed even more striking differences. It began to elute from the B1Δrod matrix at 0.5 M urea with the majority eluting between 2 and 3 M, while it began to elute from the WT matrix at 3 M urea with the majority eluting between 5 and 7 M (Fig. 7). Unfortunately lamin B2 could not be tested in this analysis, as no full-length human cDNA has been cloned. The soluble lamins did not bind to a matrix that had been coupled to BSA, nor did BSA bind to the lamin matrices (data not shown). This analysis shows that the affinity of lamins A and C was drastically reduced for B1Δrod compared with WT lamin B1, correlating with the minimal degree of overlap between lamin A/C patches and mutant patches. By contrast, the affinity of WT lamin B1 is similar for itself and the mutant. This suggests that the internal region of the lamin B1 rod is more important for heterotypic interactions with lamins A/C than for homotypic interactions.
Figure 7.
Relative affinities of lamins for B1Δrod and WT lamin B1. WT or mutant lamin B1 were coupled with an Affi-gel matrix to which soluble WT lamins were bound. Shown are Western blots of fractions eluted with increasing concentrations of urea, probed for each specific lamin isotype.
Dominant Role of Lamins in Organizing other NE Proteins
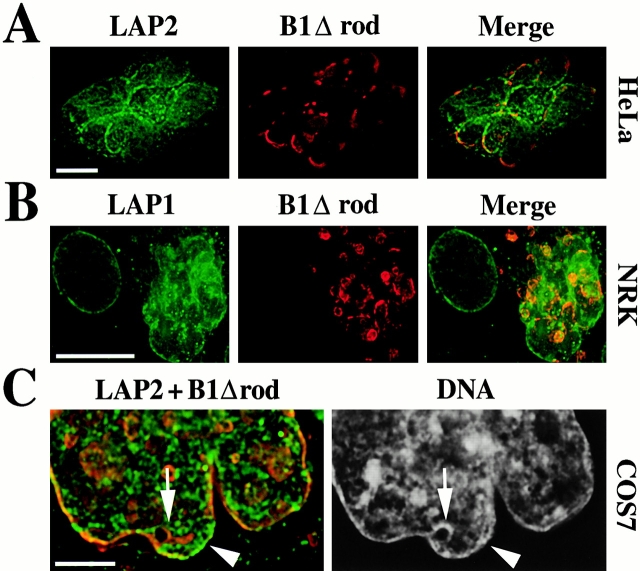
The localization of integral membrane proteins of the INM also was examined in cells expressing the B1Δrod mutant. Immunofluorescent staining revealed that LAP2β, like the endogenous lamins, lost its normal uniform distribution throughout the NE (Foisner and Gerace 1993) and collected in patches. The distribution of LAP2β in the NE varied in different cells. In some cells, it was enriched in discrete patches that had very little overlap with B1Δrod-enriched patches (Fig. 8 A), similar to the pattern seen for lamins A/C. However, in other cells there were regions of the NE with extensive colocalization between LAP2β and the mutant lamin, although these also contained endogenous lamin B2 (data not shown). In some cells, a small amount of the LAP2β appeared to be outside of the NE and this colocalized with ER dyes (data not shown). The patchy localization of two other inner membrane proteins, LAP1C (Fig. 8 B) and LBR (data not shown), in cells transfected with B1Δrod was similar to that of LAP2β. Thus, several INM proteins become enriched in patches at the NE of B1Δrod-transfected cells.
Figure 8.
Changes in localization of integral membrane proteins of the INM induced by B1Δrod. (A) Immunofluorescent staining to detect LAP2 (green) and B1Δrod (red). Similar results were observed when LAP2 was visualized with a polyclonal antiserum, a monoclonal antibody, and using a LAP2-YFP fusion protein (data not shown). A single deconvolved optical section is shown. (B) NRK cells transfected with B1Δrod (red) indicated that LAP1 (green) also is redistributed as a result of expressing the mutant lamin. A deconvolved volume projection is shown. Bars, 10 μm. (C) Distribution of NE proteins relative to chromatin. NE proteins LAP2 in green and B1Δrod in red are on the left and DAPI staining for DNA on the right of the deconvolved section shown. Bar, 5 μm.
The finding that LAP2β was often depleted in regions of the NE containing patches of B1Δrod is consistent with the finding that LAP2β binds to a central part of the lamin B1 rod (Furukawa and Kondo 1998) and that this binding is important for its localization at the NE (Furukawa et al. 1998). Nevertheless, in all cases examined, we observed that chromatin uniformly lined the NE of cells transfected with B1Δrod, and was as strongly associated with regions of the NE deficient in LAP2β (Fig. 8 C, arrow) as with those enriched in LAP2β (arrowhead). Similarly, chromatin was not preferentially associated with patches of LAP1C that arose in B1Δrod-transfected cells (data not shown). Since the major chromatin binding site of lamin B1 is present in the tail domain (Taniura et al. 1995) and thus is retained in B1Δrod, this suggests that chromatin binding to the interphase NE is determined primarily by lamins (mutant plus wild type), which collectively line the NE in the transfected cells, rather than integral proteins of the INM, which assume a highly patchy distribution in the transfected cells.
The distribution of NPC antigens also was examined in cells transfected with B1Δrod using the RL1 monoclonal antibody (Snow et al. 1987). RL1 recognizes multiple O-glycosylated NPC proteins on both the nucleoplasmic and cytoplasmic faces and characteristically shows an evenly dispersed, but punctate, distribution at the NE. In cells expressing B1Δrod, NPC antigens lost this distribution and clustered in patches (Fig. 9, left; compare staining in the untransfected cells). The same pattern was found for Tpr, an NPC component that is localized to the nucleoplasmic face of NPCs (data not shown). Interestingly, as was observed for lamins A/C, the NPC-enriched patches were largely distinct from B1Δrod-enriched patches (Fig. 9 A, Merge) and, in fact, exhibited considerable colocalization with the lamin A/C-enriched patches (B, Merge). Thus, like INM proteins, NPCs tend to cluster with endogenous lamins.
Figure 9.
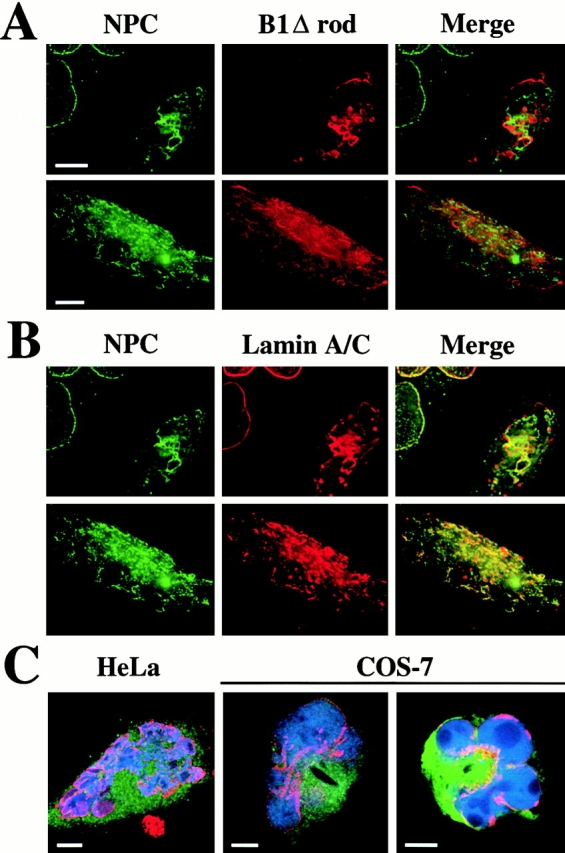
Effects of B1Δrod expression on NPCs. (A) Immunofluorescent staining to detect the NPC (green) and the mutant protein (red). (B) Immunofluorescent staining of the NPC (green) and endogenous lamins A/C (red). (Merge) Colocalization is indicated by yellow. Deconvolved volume projections are shown. (C) B1Δrod cells are capable of nuclear import. HeLa cells (left) were microinjected in the cytoplasm with a fluorescently labeled NLS-containing import substrate (blue) and a fluorescent 150-kD dextran (green). COS-7 cells (right) were injected with the same import substrate (blue) and fluorescent BSA that did not carry an NLS (green). HA staining for the B1Δrod mutant (red) traces the nuclear boundary. Single confocal sections are shown. Bars, 10 μm.
Finally, we functionally analyzed B1Δrod-expressing cells for signal-mediated protein import and the integrity of the NE as a barrier between the nucleus and cytoplasm. Fluorescently labeled BSA conjugated to an NLS was injected into the cytoplasm of HeLa and COS-7 cells expressing the mutant (Fig. 9 C, red). The import substrate (Fig. 9 C, blue) was efficiently concentrated in the nucleus in both cell types, indicating that the cells were competent for signal-mediated import. A coinjected marker protein that lacked an NLS remained outside the nucleus (Fig. 9 C, green), confirming that the NE was intact despite the deformation of the nucleus. Similarly, we found that cells expressing B1Δrod accumulated NLS-BSA in the nucleus in an ATP-dependent manner when analyzed in vitro after digitonin permeabilization (data not shown). This further validates the functional integrity of the NE and NPCs in the transfected cells.
Discussion
We found that the lamin B1Δrod mutant, when expressed in cultured cells, becomes efficiently incorporated in the NE, where it causes the redistribution of other NE proteins including endogenous lamins into patches, and induces dramatic nuclear lobulation. As the NE remains intact and functional for nuclear import in cells expressing B1Δrod and the cells are not undergoing apoptosis, the changes in nuclear structure induced by the mutant are likely to be a direct consequence of changes in NE structure. Our results strongly argue for a role of lamins in controlling the organization of other NE proteins in interphase and in defining nuclear shape. Furthermore, they provide in vivo data indicating the importance of heterotypic interactions in the assembled lamina and suggest that the rod domain contributes to the specificity of heterotypic lamin binding.
The B1Δrod mutant stands out among the many lamin mutants tested in the past as being the only protein that disrupts the uniform distribution of endogenous lamin subtypes at the NE and that drastically alters nuclear shape. The only two other lamin mutants tested that had a significant effect on the endogenous lamina involved deletions of the head and tail domains (Ellis et al. 1997; Spann et al. 1997). These caused the disassembly of endogenous lamins and the formation of intranuclear lamin aggregates, but did not induce a conspicuous perturbation of nuclear shape and the cells were not tested for redistribution of other NE proteins.
The Role of the Lamin Rod Domain in Lamina Structure
The ends of the lamin rod, which are intact in the B1Δrod mutant, have been suggested to be important for dimerization and, together with the head and tail domains, for head-to-tail polymerization of dimers (Stuurman et al. 1998). The importance of head-to-tail interactions in lamin assembly is supported by our finding that the NH2- and COOH-terminal segments derived from B1Δrod do not self assemble into filaments when analyzed individually, but strongly promote self assembly when they are linked together in B1Δrod. The more internal regions of the rod that are absent in B1Δrod are thought to contribute to the lateral packing of dimers in filaments (reviewed in McLean and Lane 1995; Stuurman et al. 1998), yet the mutant still could assemble in vitro into long filaments and filament bundles similar to those formed by WT lamin B1. Moreover, these filaments had biophysical properties characteristic of IFs. This indicates that the central ∼4/5 of the rod is less critical than other regions of the lamin molecule for self assembly into filaments per se.
Nevertheless, the internal rod region of lamin B1 that is deleted in B1Δrod appears to be important for interactions with other lamin molecules, especially with other lamin subtypes. However, it doesn't contribute to the same degree to interactions with different subtypes, as is apparent from the in vivo and in vitro studies reported here. In B1Δrod-transfected cells, different lamin subtypes overlapped to varying degrees with the mutant lamin patches. In vitro, the affinity of different lamin subtypes for the mutant as compared with WT lamin B1 was reduced to varying degrees. The reduction in the association of B1Δrod with the A/C lamins cannot be due simply to mismatched rod lengths (∼10 nm for B1Δrod vs. 48 nm for WT lamins) because WT lamin B1 bound both mutant and WT lamin B1 with similar affinity in vitro. Furthermore, the similarity of the in vivo phenotypes for B1Δrod and the A/B1Δrod hybrid mutant suggests that these internal regions are more important than the head domain for distinguishing heterotypic interactions. This concurs with a very recent study to address lamin heterotypic interactions in vivo. This involved expression of a head deletion of lamin A, which caused disassembly of lamin A, but not lamin B, from the NE (Izumi et al. 2000). Previous work had indicated that A and B lamin heterotypic interactions are stronger than their homotypic interactions in vitro (Georgatos et al. 1988; Ye and Worman 1995). Here we have extended these studies by showing that the lamin rod domain contributes to heterotypic interactions of lamin B1 and, further, have demonstrated for the first time that these interactions are relevant to lamina structure and organization in vivo.
Role of the Lamina in NE Stability and Nuclear Organization
NPCs clustered in cells expressing B1Δrod, yet remained functional for import. The clustering of both NPCs and integral membrane proteins of the INM into patches that largely colocalized with endogenous lamins provides strong evidence that WT lamins play a key role in anchoring these components at the NE during interphase. This complements subcellular fractionation studies indicating a physical connection between lamins and NPCs (Dwyer and Blobel 1976), and in vitro binding studies showing an interaction between lamins and various integral proteins of the INM (Foisner and Gerace 1993; Ye and Worman 1994; Fairley et al. 1999).
Several integral membrane proteins of the INM have been shown to bind directly to chromatin as well as to lamins (reviewed in Collas and Courvalin 2000). Despite the clustering of three INM proteins into patches in the B1Δrod-transfected cells, including two (LAP2β and LBR) that bind to chromatin, chromatin was uniformly associated with the NE and did not segregate preferentially with the INM protein patches. Conversely, the endogenous lamins and B1Δrod, although often segregated in patches, together formed a continuous lamin zone around the NE. These data argue that the principal basis for association of chromatin with the NE is binding to lamins, not to integral proteins of the INM. Consistent with this notion, the major chromatin binding site of lamin B1 (Taniura et al. 1995) was retained in B1Δrod.
Interestingly, although the NE was intact in B1Δrod-expressing cells, we observed a fraction of the LAP2β in cytoplasmic foci by fluorescence microscopy and also saw unusual concentric membrane structures in the cytoplasm by EM. We suggest that these structures might arise by the budding off of NE membranes from regions overlaying B1Δrod patches, where tethering of the membrane to the lamina might be weakened. This is consistent with the notion that the structural scaffolding of the lamina is essential for stabilizing the INM, analogous to functions of spectrin scaffolds at the plasma and Golgi membranes (Lorra and Huttner 1999).
The distortion of nuclear structure caused by assembly of B1Δrod in the NE argues that lamins directly influence nuclear shape. We suggest that the extensive lobulation of nuclei that occurs in these cells is due to the reduction in heterotypic lamin–lamin interactions and the patchwork lamina that is formed as a consequence. Although the B1Δrod mutant could self assemble into filamentous structures, these structures would lack most of the interactions that normally occur along the length of the rod, and are predicted to be less thermodynamically stable than WT lamin filaments. Thus, mutant-enriched patches of the lamina would be expected to have lower mechanical strength than WT patches. Moreover, interfaces between the mutant and WT lamin patches are expected to be weaker on the basis of our in vitro binding results. The weakened areas of the lamina would be expected to be more sensitive to forces imposed over the surface of the NE by the dynamics of attached chromatin and the cytoplasmic cytoskeleton. This could lead to disruption of the normal curvature of the NE, causing the lobulation and gross distortion of nuclear shape that occurred in cells expressing the mutant. Future studies using stably transfected cells may discern whether a specific concentration of the mutant is required for these effects and if levels of endogenous NE proteins are altered by expression of the B1Δrod mutant.
Although no notable changes in nuclear shape were reported in previous studies in which the normal lamin organization at the NE was disrupted, these results are not inconsistent with a role of the lamina in nuclear shape determination. In one type of study, lamin-depleted nuclei were assembled in vitro using Xenopus egg extracts (reviewed in Lourim and Krohne 1994; see also Ellis et al. 1997; Spann et al. 1997). Since these nuclei were replication deficient, transcriptionally inactive, and not associated with a cytoplasmic cytoskeleton, the unevenly distributed forces from attached nuclear and cytoplasmic components that would distort nuclear shape in our model (above) would be absent. In another type of study, a particular lamin subtype was depleted from the nuclei of certain cells by a lamin gene disruption (Lenz-Bohme et al. 1997; Sullivan et al. 1999). Gross distortions in nuclear shape would not be predicted with our model in these cases since the cells still retained an assembled lamina in which the remaining lamins were uniformly distributed at the NE. By altering the substructure of the intact, assembled lamina rather than by depleting lamins from the NE, expression of the B1Δrod mutant provides a novel approach to the question of how nuclear shape is determined. Our results directly support the notion that heterotypic associations among lamin subtypes are important for the molecular organization of the NE and, correspondingly, for higher order nuclear architecture.
Acknowledgments
We thank S. Lyman for nuclear import reagents, and S. Lyman and H. Wodrich for critical reading of the manuscript. We especially thank R. Ghadiri and D. Bonn for assistance with circular dichroism and FTIR analyses, M. Wood for EM assistance, U. Aebi for the pPEPT vector system, and H. Worman for LBR antiserum.
This work was supported by a National Institutes of Health (NIH) postdoctoral fellowship to E.C. Schirmer (F32 GM19085) and an NIH grant to L. Gerace (GM28521).
Footnotes
Abbreviations used in this paper: FTIR, Fourier transform infrared spectroscopy; GFP, green fluorescent protein; HA, hemagglutinin; IF, intermediate filament; INM, inner nuclear membrane; LAP, lamina-associated polypeptide; NE, nuclear envelope; NLS, nuclear localization signal; NPC, nuclear pore complex; NRK, normal rat kidney; WT, wild type.
References
- Adam S.A., Marr R.S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Cao H., Hegele R.A. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- Collas I., Courvalin J.C. Sorting nuclear membrane proteins at mitosis. Trends Cell Biol. 2000;10:5–8. doi: 10.1016/s0962-8924(99)01697-9. [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., Chan Y.M., Albers K., Fuchs E. Deletions in epidermal keratins leading to alterations in filament organization in vivo and in intermediate filament assembly in vitro. J. Cell Biol. 1990;111:3049–3064. doi: 10.1083/jcb.111.6.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N., Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D.J., Jenkins H., Whitfield W.G., Hutchison C.J. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J. Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Fairley E.A., Kendrick-Jones J., Ellis J.A. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J. Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- Foisner R., Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Fritze C.E., Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region, but is distinct from its chromatin interaction domain. J. Biol. Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Kondo T. Identification of the lamina-associated-polypeptide-2-binding domain of B-type lamin. Eur. J. Biochem. 1998;251:729–733. doi: 10.1046/j.1432-1327.1998.2510729.x. [DOI] [PubMed] [Google Scholar]
- Geisler N., Heimburg T., Schunemann J., Weber K. Peptides from the conserved ends of the rod domain of desmin disassemble intermediate filaments and reveal unexpected structural featuresa circular dichroism, fourier transform infrared, and electron microscopic study. J. Struct. Biol. 1993;110:205–214. doi: 10.1006/jsbi.1993.1023. [DOI] [PubMed] [Google Scholar]
- Georgatos S.D., Stournaras C., Blobel G. Heterotypic and homotypic associations between the nuclear laminssite-specificity and control by phosphorylation. Proc. Natl. Acad. Sci. USA. 1988;85:4325–4329. doi: 10.1073/pnas.85.12.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L., Foisner R. Integral membrane proteins and dynamic organization of the nuclear envelope. Trends Cell Biol. 1994;4:127–131. doi: 10.1016/0962-8924(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Glass J.R., Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J. Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M., Weber K. Modulation of keratin intermediate filament assembly by single amino acid exchanges in the consensus sequence at the C-terminal end of the rod domain. J. Cell Sci. 1991;99:351–362. doi: 10.1242/jcs.99.2.351. [DOI] [PubMed] [Google Scholar]
- Heald R., McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Heimburg T., Schunemann J., Weber K., Geisler N. Specific recognition of coiled coils by infrared spectroscopyanalysis of the three structural domains of type III intermediate filament proteins. Biochemistry. 1996;35:1375–1382. doi: 10.1021/bi9515883. [DOI] [PubMed] [Google Scholar]
- Heitlinger E., Peter M., Haner M., Lustig A., Aebi U., Nigg E.A. Expression of chicken lamin B2 in Escherichia colicharacterization of its structure, assembly, and molecular interactions. J. Cell Biol. 1991;113:485–495. doi: 10.1083/jcb.113.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitlinger E., Peter M., Lustig A., Villiger W., Nigg E.A., Aebi U. The role of the head and tail domain in lamin structure and assemblyanalysis of bacterially expressed chicken lamin A and truncated B2 lamins. J. Struct. Biol. 1992;108:74–89. doi: 10.1016/1047-8477(92)90009-y. [DOI] [PubMed] [Google Scholar]
- Izumi M., Vaughan O.A., Hutchison C.J., Gilbert D.M. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell. 2000;11:4323–4337. doi: 10.1091/mbc.11.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer R.A., Schulthess T., Landwehr R., Lustig A., Fischer D., Engel J. Tenascin-C hexabrachion assembly is a sequential two-step process initiated by coiled-coil alpha-helices. J. Biol. Chem. 1998;273:10602–10608. doi: 10.1074/jbc.273.17.10602. [DOI] [PubMed] [Google Scholar]
- Lenz-Bohme B., Wismar J., Fuchs S., Reifegerste R., Buchner E., Betz H., Schmitt B. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorra C., Huttner W.B. The mesh hypothesis of Golgi dynamics. Nat. Cell Biol. 1999;1:E113–E115. doi: 10.1038/12939. [DOI] [PubMed] [Google Scholar]
- Lourim D., Krohne G. Lamin-dependent nuclear envelope reassembly following mitosisan argument. Trends Cell Biol. 1994;4:314–318. doi: 10.1016/0962-8924(94)90228-3. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- McKeon F.D., Kirschner M.W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- McLean W.H., Lane E.B. Intermediate filaments in disease. Curr. Opin. Cell Biol. 1995;7:118–125. doi: 10.1016/0955-0674(95)80053-0. [DOI] [PubMed] [Google Scholar]
- Mical T.I., Monteiro M.J. The role of sequences unique to nuclear intermediate filaments in the targeting and assembly of human lamin Bevidence for lack of interaction of lamin B with its putative receptor. J. Cell Sci. 1998;111:3471–3485. doi: 10.1242/jcs.111.23.3471. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Donaldson A.D., Stewart M. Expression in Escherichia coli of human lamins A and Cinfluence of head and tail domains on assembly properties and paracrystal formation. J. Cell Sci. 1991;99:363–372. doi: 10.1242/jcs.99.2.363. [DOI] [PubMed] [Google Scholar]
- Rober R.A., Weber K., Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animala developmental study. Development (Camb.) 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Sasse B., Lustig A., Aebi U., Stuurman N. In vitro assembly of Drosophila lamin Dm0—lamin polymerization properties are conserved. Eur. J. Biochem. 1997;250:30–38. doi: 10.1111/j.1432-1033.1997.t01-1-00030.x. [DOI] [PubMed] [Google Scholar]
- Senior A., Gerace L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol. 1988;107:2029–2036. doi: 10.1083/jcb.107.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow C.M., Senior A., Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T.P., Moir R.D., Goldman A.E., Stick R., Goldman R.D. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N., Heins S., Aebi U. Nuclear laminstheir structure, assembly, and interactions. J. Struct. Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Stuurman N., Sasse B., Fisher P.A. Intermediate filament protein polymerizationmolecular analysis of Drosophila nuclear lamin head-to-tail binding. J. Struct. Biol. 1996;117:1–15. doi: 10.1006/jsbi.1996.0064. [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H., Glass C., Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- Ye Q., Worman H.J. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- Ye Q., Worman H.J. Protein–protein interactions between human nuclear lamins expressed in yeast. Exp. Cell Res. 1995;219:292–298. doi: 10.1006/excr.1995.1230. [DOI] [PubMed] [Google Scholar]