Abstract
The large cytoplasmic DNA viruses such as poxviruses, iridoviruses, and African swine fever virus (ASFV) assemble in discrete perinuclear foci called viral factories. Factories exclude host proteins, suggesting that they are novel subcellular structures induced by viruses. Novel perinuclear structures, called aggresomes are also formed by cells in response to misfolded protein (Johnston, J.A., C.L. Ward, and R.R. Kopito. 1998. J. Cell Biol. 143:1883–1898; García-Mata, R., Z. Bebök, E.J. Sorscher, and E.S. Sztul. 1999. J. Cell Biol. 146:1239–1254). In this study, we have investigated whether aggresomes and viral factories are related structures. Aggresomes were compared with viral factories produced by ASFV. Aggresomes and viral factories were located close to the microtubule organizing center and required an intact microtubular network for assembly. Both structures caused rearrangement of intermediate filaments and the collapse of vimentin into characteristic cages, and both recruited mitochondria and cellular chaperones. Given that ASFV factories resemble aggresomes, it is possible that a cellular response originally designed to reduce the toxicity of misfolded proteins is exploited by cytoplasmic DNA viruses to concentrate structural proteins at virus assembly sites.
Keywords: virus assembly, aggresomes, African swine fever virus, microtubules, vimentin
Introduction
The assembly of viruses often takes place in specific subcompartments within the cell, a process that concentrates viral structural components and increases the efficiency of assembly. The cytoplasm is favored as a site of assembly by the large DNA viruses such as poxviruses, iridoviruses, and the closely related African swine fever virus (ASFV). Assembly occurs in viral factories that contain high levels of viral structural proteins, viral DNA, and amorphous membranous material used to produce viral envelopes. Interestingly, the factories tend to exclude host proteins, suggesting that these viruses induce the formation of a new subcellular structure to act as a scaffold for virus replication and assembly.
ASFV shares the genomic organization of poxviruses with the striking icosahedral symmetry of the iridoviridae (Goorha and Granoff 1979; Yáñez et al. 1995), suggesting an evolutionary connection to both virus families. The 170-kb genome of ASFV encodes some 150 open reading frames, and as many as 50 viral proteins are assembled into viral particles (Esteves et al. 1986). Assembly of the virus takes place in perinuclear viral factories (Moura Nunes et al. 1975) that contain fully assembled virions seen as 200-nm-diameter hexagons in cross section, and a series of one- to six-sided assembly intermediates (Rouiller et al. 1998). Approximately 35% of the mass of the virion is provided by p73, the major capsid protein, while the ordered processing of a 220-kD polyprotein to structural proteins, p150, p37, p34, and p14, provides another 25% of the virion mass (Andrés et al. 1997). Recruitment of these proteins into viral factories is highly efficient, and they are almost exclusively localized to virus factories when observed by immunofluorescence microscopy (Cobbold et al. 1996; Andrés et al. 1997). At present, it is not known how p73, or other proteins required for virion assembly, are recruited into virus factories. The generation of a specific assembly site within cells could occur actively by targeting viral proteins into the assembly site. Alternatively, they may form passively as a consequence of the localized accumulation of large quantities of protein during virus infection.
It has been shown recently that cells respond to the production of high levels of misfolded proteins by transporting them to perinuclear sites called aggresomes. Aggresomes were first described as sites able to sequester misfolded cystic fibrosis transmembrane conductance receptors or unassembled presenilin (Johnston et al. 1998; Wigley et al. 1999), but it is now thought possible that aggresome formation may be a general cellular response to misfolded or unassembled proteins (García-Mata et al. 1999). Aggresomes are located close to centrosomes and are enclosed in a characteristic vimentin cage. They also recruit cellular chaperones and proteasomes and may regulate protein folding and degradation (Kopito 2000). The ability of aggresomes to concentrate proteins and cellular chaperones make them highly suitable for facilitating virus assembly. In this study we investigated the possibility that viral factories and aggresomes are related structures, and find a striking similarity between them. It is possible that a cellular response originally designed to reduce the potential toxicity of misfolded proteins is exploited by ASFV as a means of concentrating structural proteins at sites of virus assembly.
Materials and Methods
Virus Propagation Metabolic Labeling and Immunoprecipitation
Vero (ECACC 84113001) cells were infected with ASFV as described previously (Cobbold et al. 1996; Cobbold and Wileman 1998). Infected cells were labeled with [35S]methionine and cysteine and viral proteins immunoprecipitated and resolved by SDS-PAGE as described previously (Cobbold et al. 1996).
Antibodies and Reagents
The monoclonal antibody 4H3 binds the major ASFV capsid protein p73 (Cobbold et al. 1996). The antibody specific for vp30 was from Dan Rock (United States Department of Agriculture, Plum Island Animal Disease Center, Greenport, NY). Rabbit antibody specific for p34 was generated from recombinant his-tagged protein using the pTrcHis expression vector (Invitrogen). 9E10 was used to detect myc-tagged p50/dynamitin. Antibody against TCPI α chain was from StressGen Biotechnologies. Nocodazole and all other antibodies were from Sigma-Aldrich.
Cell Culture and Transfections
Vero cells were grown on 12-mm coverslips to 70% confluency and transfected transiently with plasmids using Transfast (Promega). The GFP-250 plasmid was a gift from Dr. Elizabeth Sztul (University of Alabama, Birmingham, AL). The p50/dynamitin plasmid (PCMVH50myc was a gift from Dr. Richard Vallee (University of Massachusetts Medical School, Boston, MA) and is described in Echeverri et al. 1996.
Fluorescence Microscopy
Cells were fixed in −20°C methanol or 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked with 0.2% gelatin and 30% goat serum. The cells were incubated with primary antibodies in the same buffer and visualized with the secondary antibodies conjugated to Alexa dyes (Molecular Probes). Cells were mounted in Fluoromount-G (Southern Biotechnology Associates, Inc.) and viewed at 60×/1.4 NA with a Nikon E800 microscope, and 0.2-μm digital sections were captured with a DCC camera (C-4746A; Hamamatsu) and digitally deconvolved using Openlab software from Improvision. The Vernier calibration tool was used to calculate aggresome and factory cross sectional areas.
Online Supplemental Material
(Fig. S1) Virus assembly can initiate at multiple sites within the virus factory. (Table SI) Distribution of virus assembly sites within virus factories. Available online at http://www.jcb.org/cgi/content/full/153/3/449/DC1.
Results and Discussion
Aggresomes Can Be Produced in Cells Susceptible to Infection with ASFV
Vero cells were transfected with an expression vector encoding a protein chimera composed of green fluorescent protein fused to 250 amino acids of the cytosolic protein p115 (GFP-250). Fig. 1 A shows that the majority of the GFP-250 protein observed 48 h later produced an intense perinuclear fluorescence signal, with the remainder being distributed throughout the cytoplasm as small granules. The merge of GFP-250 and DNA staining shows that the perinuclear fluorescence impinged on the nucleus as previously described for aggresomes generated using this probe (García-Mata et al. 1999).
Figure 1.
Aggresomes and ASFV assembly sites cause redistribution of the intermediate filament protein vimentin. Vero cells were transfected with the GFP-250 plasmid and grown at 37°C for 48 h (A) or infected for 12 h with ASFV (B). Fixed and permeabilized cells were incubated with an antibody specific for vimentin, and antibodies specific for p34 (B). Protein distribution was visualized by using appropriate secondary antibodies conjugated to Alexa Fluor 488 (green) or 594 (red), and the DNA was labeled with DAPI (blue). In the lower images the GFP-250/p34, vimentin and cellular DNA images were digitally merged.
Aggresomes and Viral Factories Are Surrounded by Vimentin Cages
Aggresomes induce a striking redistribution of the intermediate filament protein vimentin into a halo-like cage around aggregates of protein. The distribution of the vimentin around aggresomes containing the GFP-250 protein was therefore studied. The central image in Fig. 1 A shows that expression of GFP-250 caused the collapse of vimentin into a ring-like structure in the perinuclear area of the cell occupied by the aggresome. Assembly of ASFV takes place in similar perinuclear areas, and the effects of ASFV infection on the distribution of vimentin was determined. Viral factories were identified using antibodies specific for the viral structural protein, p34, and two different examples are shown in Fig. 1 B. In the absence of virus, vimentin surrounded the nucleus and extended to the cell periphery. Strikingly, with virus, vimentin surrounded the nucleus and the viral factory. The collapse of vimentin around both aggresomes and virus assembly sites suggested that they might be related structures. Even though there was considerable variation in the size of vimentin cages formed around aggresomes and virus factories, the average cross-sectional areas were quite similar and calculated to be 46 ± 16 and 39 ± 12 μm2, respectively. It is possible that there is an upper limit to the size of these structures. Interestingly, the vimentin cage surrounding the viral factory was thicker than the cage formed around the aggresome. The reasons for this are unknown, but it is possible that aggregated misfolded protein is better able to exclude vimentin from the interior of the aggresome than viral DNA and assembling virions. Most (66%) virus factories contained a single area of virus replication; in some viral factories, however, staining for the major capsid protein was found in two or more clusters, giving the impression of multiple assembly sites. Close inspection of these structures showed, however, that there was always a single patch of viral DNA enclosed in vimentin, and therefore a single aggresome-like structure (see online supplement). Later, during infection, virions appear in large numbers in the cytoplasm and the well-defined vimentin cage, and the virus factory begins to disperse. The use of aggresome-like structures by ASFV appears, therefore, to be important early during infection possibly for localizing viral DNA replication to a single site within the cell and for initiating virus assembly.
The mechanism of vimentin reorganization during aggresome or factory formation is not known. Vimentin cages form during mitosis, and this may be regulated by phosphorylation of vimentin by cell cycle control kinases such as cdc2 and rho (Chou et al. 1990; Inagaki et al. 1987; Goto et al. 1998). Significantly, vimentin cages have long been known to form around the assembly sites of the cytoplasmic iridovirus, frog virus 3 (FV3) (Murti and Goorha 1983), and a fourfold increase in phosphorylation of vimentin occurs in cells infected with FV3 (Chen et al. 1986). This is particularly relevant since ASFV was originally classified as an iridovirus and is structurally related to FV3. Phosphorylation and dephosphorylation of vimentin therefore offer a possible means of regulating cage formation during ASFV infection.
Aggresomes and Viral Factories Locate Near the Microtubule Organizing Center and Recruit Mitochondria and Cellular Chaperones
The perinuclear location of aggresomes has implicated a role for microtubules and the microtubule organizing center (MTOC) in construction of the organelle (Johnston et al. 1998; García-Mata et al. 1999). The location of the MTOC in cells expressing the GFP-250 protein was determined using an antibody specific for γ-tubulin (Fig. 2 A). The centrioles were difficult to image because partial disruption of the MTOC by the aggresome produced diffuse γ-tubulin staining, as reported by Johnston et al. 1998. Two centrioles were, however, distinguished as orange spots in the inset. Viral factories were located using an antibody specific for p73, the major capsid protein (Fig. 2 B). Viral factories did not disperse γ-tubulin staining, and centrioles at the MTOC were clearly visible (red). The merged image is representative of many cells viewed, and in each case the assembly site of ASFV was located close to the MTOC. In contrast to aggresomes, however, virus factories did not localize directly at the MTOC. The reasons for this difference are unknown.
Figure 2.

Aggresomes and viral factories locate close to the MTOC and recruit mitochondria. Vero cells were transfected with the GFP-250 plasmid (A and C) and grown at 37°C for 48 h, or infected for 12 h with ASFV (B and D–F). The green GFP signal indicates the aggresome in A and C, while virions in viral factories are identified in B, D, and E using antibodies specific for p73 and a secondary antibody coupled to Alexa Fluor 488 (green). Cellular and viral DNA was labeled with DAPI (blue). (A and B) Merged images where antibodies specific for γ-tubulin identify centrioles (red). (C–E) Merged images where Mito-Tracker red CMXRos identifies mitochondria (red). (F) Phase contrast image of cells in E.
The distribution of mitochondria, labeled using Mito-Tracker red, in cells expressing the GFP-250 chimera or cells infected with ASFV, are compared in Fig. 2C and Fig. D. Mitochondria were distributed throughout the cytoplasm of the cells that were negative for GFP. In the cell expressing GFP-250, mitochondria clustered to the area of the aggresome. Mitochondria also clustered around the viral factory in the infected cells in Fig. 2 D and, when the stain produced by Mito-Tracker was compared with phase contrast images of infected cells (Fig. 2E and Fig. F), the retraction of mitochondria from the cell periphery towards the factory was clear. It has been proposed that the aggresomes recruit mitochondria as an energy source for protein folding and protein degradation. Aggresomes contain chaperones and the 20S proteasome, all of which need ATP (García-Mata et al. 1999; Wigley et al. 1999). The recruitment of Hsp70 into aggresomes and viral factories was therefore examined. Fig. 3A–D, shows that both structures stained for Hsp70; the effect was, however, less noticeable for virus factories. The distribution of the ring chaperonin TCP1 in infected cells was determined (Fig. 3E and Fig. F) and, in contrast to aggresomes, which are reported to accumulate TCP1 (García-Mata et al. 1999), the ring chaperonin was absent from virus assembly sites. The difference in relative levels of chaperones recruited into these structures may result from their different functions. Aggresomes sequester misfolded proteins that would continuously expose hydrophobic domains susceptible to chaperone binding. An accumulation of chaperones in viral factories, on the other hand, would not be expected because factories recruit proteins that are either folded, or would eventually be folded, during the assembly of virions.
Figure 3.

Recruitment of chaperones into aggresomes and viral factories. Cells containing aggresomes were generated using GFP-250 (A), and distribution of Hsp70 in the same cell is shown in B. Vero cells infected with ASFV for 12 h were fixed and permeabilized and labeled with antibody specific for viral protein p34 (C and E) and Hsp70; antibodies specific for TCP1 identify the ring chaperonin (E and F).
The Integrity of Viral Factories Is Dependent on the Microtubular Network
Aggresomes are dispersed when cells are incubated with nocodazole (Johnston et al. 1998), or after the overexpression of p50/dynamitin to inhibit the microtubule-dependent dynein/dynactin motors (García-Mata et al. 1999). The effect of nocodazole on preformed virus factories was tested by adding the drug 12 h after infection, and observing cells 2 h later. Fig. 4 A shows control cells infected with ASFV. The factory was compact, indicated by bright staining for p34 located over viral DNA (top and bottom). α-Tubulin formed a filamentous network extending throughout the cell (center). There was a partial exclusion of tubulin from the area occupied by the factory, as has been described for the aggresomes (García-Mata et al. 1999). ASFV did not otherwise cause obvious disruption of microtubules. The effect of nocodazole on the viral factories is shown in Fig. 4B and Fig. C. Cells were either stained with antibodies specific for p34 and α-tubulin (B), or two different markers for viral factories, p34 and p73 (C). Nocodazole disrupted the filamentous microtubule network and caused the staining for the two viral structural proteins and extranuclear DNA to extend in a polarized fashion from one side of the nucleus to the cell periphery. These results suggested that, in common with aggresomes, the compact perinuclear location of the viral factory was dependent on an intact microtubule network and, by implication, that microtubules are used to transport proteins to viral factories.
Figure 4.
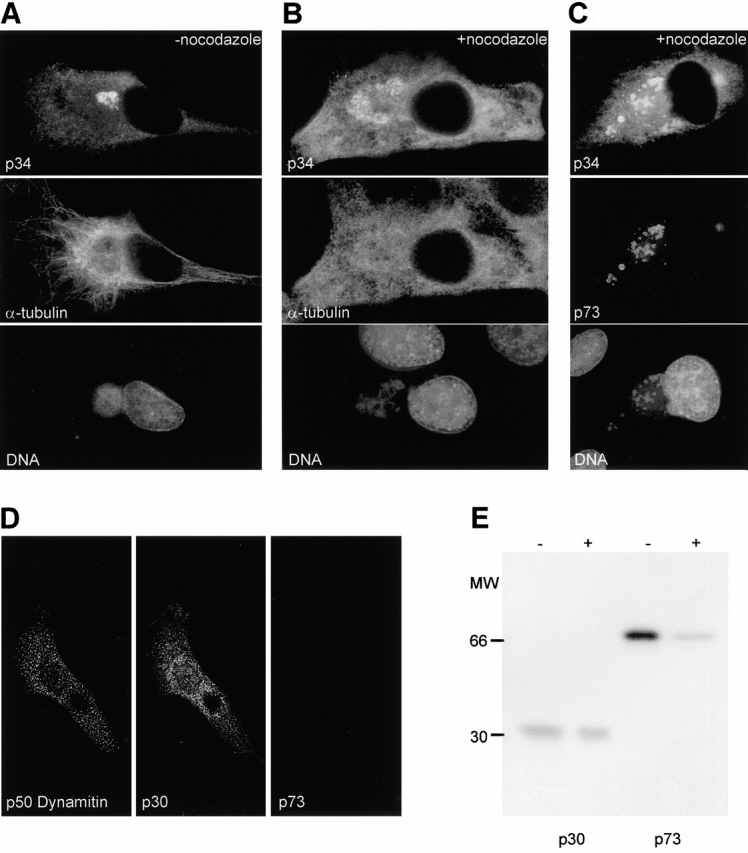
Viral factories require an intact microtubule network. (A–C) Microtubules are required for the integrity of preassembled factories. Vero cells were infected with ASFV for 14 h, and then incubated at 37°C in the absence (A) or presence (B and C) of 10 μg/ml nocodazole for another 2 h, and then fixed and permeabilized. The viral factories were labeled with antibody raised against p34 (top) or antibody specific for p73 (C, middle). Microtubules were labeled with antibody specific for α-tubulin (A and B, middle). The proteins were visualized with the Alexa Fluor 488 or 594 conjugated to appropriate secondary antibodies. (D and E) Late gene expression requires microtubules. (D) Vero cells were transfected with a plasmid expressing p50/dynamitin; 2 h later, cells were infected with ASFV and incubated at 37°C for another 12 h before processing for immunofluorescence microscopy. Cells were triple stained using antibodies specific for the myc-tag in dynamitin, a biotinylated antibody specific for p30, and rabbit polyclonal antibody recognizing p73. The proteins were visualized using Marina blue, Alexa Fluor 488, or 594 conjugated to appropriate secondary antibodies. (E) Vero cells were infected with ASFV for 2 h, and then incubated at 37°C in the absence (−) or presence (+) of 10 μg/ml nocodazole for another 10 h. Cells were pulse labeled for 30 min, lysed, and immunoprecipitated using antibodies specific for p30 or p73. Proteins were resolved by SDS/PAGE and visualized by autoradiography.
The Microtubule Network Is Required for the Initial Assembly of Viral Factories and Transition to Late Gene Expression
The role played by microtubules during the initial delivery of viral components to the aggresome, and subsequent assembly of factories was tested by incubating cells with nocodazole soon after infection with ASFV, or by transfecting cells with p50/dynamitin. Surprisingly, p73 was absent from cells expressing p50/dynamitin (Fig. 4 D). The abundant early viral phosphoprotein p30 was nevertheless expressed at high levels. The results suggested that dynein/dynactin motors were required for DNA replication and transition to late gene expression. To test this, cells were infected with virus for 2 h and incubated with nocodazole for 10 h. They were then analyzed for p30 and p73 expression by immunoprecipitation. Nocodazole had little effect on levels of p30, but expression of p73 was substantially reduced (Fig. 4 E). The results demonstrated that an intact microtubular network was required for the expression of late viral proteins, suggesting that delivery of early viral components to the MTOC, and by implication aggresomes, was required for DNA replication and transition to late gene expression.
Given the striking similarity between aggresomes and ASFV factories, it is interesting to ask how the virus stimulates aggresome formation and then gains access to the organelle. In a model proposed by Johnston et al. 1998, hydrophobic interactions between misfolded proteins lead to aggregation, and once aggregates reach a diameter of 60–80-nm diameter they are delivered to the MTOC by retrograde transport along microtubules. Exit of aggregates by diffusion into the cytoplasm is prevented by reorganization of intermediate filaments into a cage surrounding the aggresome. The ability of the aggresome pathway to recognize large protein aggregates provides one mechanism for recruitment of ASFV into aggresomes. The outer envelopes of ASFV are removed in endosomes and lysosomes as the virus enters the cell (Geraldes and Valdeira 1985), and a compact nucleoprotein core of ∼100-nm diameter enters the cytosol (Valdeira et al. 1998). We propose that viral cores may be recognized by the aggresome pathway and transported by minus end–directed motors to the MTOC along microtubules (Fig. 5). The subsequent reorganization of vimentin into cages around these particles would then prevent release of virion cores into the cytosol. Since virion cores contain the enzymes necessary for genome replication, this mechanism would establish an intracellular site within the aggresome ideally suited for the localized production of viral DNA. Support for this comes from the observation that late gene expression was inhibited if early viral components were denied access to the aggresome pathway by p50/dynamitin or nocodazole (Fig. 4D and Fig. E). At later stages of replication, viral components need to be delivered to aggresomes to initiate assembly. The experiments with nocodazole suggests that these are delivered by microtubules. Interestingly, three viral core proteins of vaccinia virus, another virus that replicates in cytoplasmic factories, bind microtubules (Ploubidou et al. 2000), and microtubules are thought to facilitate virus egress (Sanderson et al. 2000) and also transport viral cores into vaccinia virus factories earlier during assembly. After assembly in factories, ASF virus particles are released into the cytoplasm. Given that the exclusion limit of the aggresome vimentin cage is 60–100 nm, and virus particles are 200 nm in diameter, ASF virus must have some means of breaking down the vimentin cage. It is possible that this is achieved through phosphorylation and dephosphorylation of vimentin.
Figure 5.
Model for the recruitment of ASFV into aggresomes. ASF virions enter the cell by receptor-mediated endocytosis (1 and 2). The virus is uncoated and the virion cores enter the cytoplasm (3). The cores mimic large protein aggregates, initiate aggresome formation, and are transported to the newly established aggresomes on microtubules (4). DNA replication begins leading to late protein synthesis (5). Later, during infection, viral proteins synthesized on cytoplasmic ribosomes are carried by hypothetical viral carrier proteins along microtubules (6) to the aggresomes to facilitate further replication and assembly.
Is aggresome formation a general response to localized virus assembly in cells? This has not been addressed in detail, elements of the aggresome pathway have, however, been reported to be activated during the assembly of several viruses. Vimentin collapses around Theiler's virus (Nédellec et al. 1998), reovirus inclusions (Sharpe et al. 1982), vaccinia virus (Leão-Ferreira et al. 1994), and iridovirus factories (Murti and Goorha 1983). Mitochondrial clustering has been demonstrated around herpes simplex virus (HSV) tegument proteins (Murata et al. 2000), and reovirus inclusions (Sharpe et al. 1982). Finally, many viruses have been shown to move along microtubules, HSV, adenoviruses, and reoviruses, for example, use microtubules for minus end–directed transport into the nucleus (Georgi et al. 1990; Sodeik et al. 1997; Suomalainen et al. 1999). Given these observations, it is possible that exploitation of the aggresome pathway may be a general feature of virus assembly.
Supplemental Material
Acknowledgments
This work was supported by The Biotechnology and Biological Sciences Research Council and The Ministry of Agriculture, Fisheries and Food.
Footnotes
The online version of this article contains supplemental material.
Dr. Heath's present address is Department of Cell Biology, Washington University Medical School, St. Louis, MO 63110.
Abbreviations used in this paper: ASFV, African swine fever virus; MTOC, microtubule organizing center.
References
- Andrés G., Simón-Mateo C., Viñuela E. Assembly of African swine fever virusrole of polyprotein pp220. J. Virol. 1997;71:2331–2341. doi: 10.1128/jvi.71.3.2331-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Goorha R., Murti K.G. Interaction of frog virus-3 with the cytomatrix. IV. Phosphorylation of vimentin precedes the reorganisation of intermediate filaments around the virus assembly sites. J. Gen. Virol. 1986;67:915–922. doi: 10.1099/0022-1317-67-5-915. [DOI] [PubMed] [Google Scholar]
- Chou Y.H., Bischoff J.R., Beach D., Goldman R.D. Intermediate filament reorganisation during mitosis is mediated by p34cdc2 phosphorylation of vimentin. Cell. 1990;62:1063–1071. doi: 10.1016/0092-8674(90)90384-q. [DOI] [PubMed] [Google Scholar]
- Cobbold C., Whittle J.T., Wileman T. Role of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J. Virol. 1996;70:377–384. doi: 10.1128/jvi.70.12.8382-8390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold C., Wileman T. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 1998;72:5215–5223. doi: 10.1128/jvi.72.6.5215-5223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal B.M., Vaughan K.T., Vallee R.B. Molecular characterization of the 50kDa subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves M., Marques M.I., Costa J.V. Two-dimensional analysis of African swine fever virus protein and proteins induced in infected cells. Virology. 1986;152:192–206. doi: 10.1016/0042-6822(86)90384-3. [DOI] [PubMed] [Google Scholar]
- García-Mata R., Bebök Z., Sorscher E.J., Sztul E.S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi A., Mottolahartshorn C., Warner A., Fields B., Chen L.B. Detection of individual fluorescently labeled reovirions in living cells. Proc. Natl. Acad. Sci. USA. 1990;87:6579–6583. doi: 10.1073/pnas.87.17.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A., Valdeira M.L. Effect of chloroquine on African swine fever virus infection. J. Gen. Virol. 1985;66:1145–1148. doi: 10.1099/0022-1317-66-5-1145. [DOI] [PubMed] [Google Scholar]
- Goorha R., Granoff A. Icosahedral cytoplasmic deoxyviruses. In: Fraenkel-Contrat H., Wagner R.R., editors. Newly Characterised Vertebrate Viruses. Comprehensive Virology 14. Plenum Publishing Corp; New York, NY: 1979. pp. 347–399. [Google Scholar]
- Goto H., Kosako H., Tanabe K., Yanagida M., Sakurai M., Amano M., Kaibuchi K., Inagaki M. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J. Biol. Chem. 1998;273:11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- Inagaki M., Nishi Y., Nishizawa K., Matsuyama M., Sato C. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro . Nature. 1987;328:649–652. doi: 10.1038/328649a0. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. Aggresomesa cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Leão Ferreira R.L., Moussatche N., Neto V.M. Rearrangement of intermediate filament network of BHK-21 cells infected with vaccinia virus. Arch. Virol. 1994;138:273–285. doi: 10.1007/BF01379131. [DOI] [PubMed] [Google Scholar]
- Moura Nunes J.F., Vigario D., Terrinha M. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 1975;49:59–66. doi: 10.1007/BF02175596. [DOI] [PubMed] [Google Scholar]
- Murata T., Goshima F., Daikoku T., Inagaki-Ohara K., Takakuwa H., Kato K., Nishiyama Y. Mitochondrial distribution and function in herpes simplex virus-infected cells. J. Gen. Virol. 2000;81:401–406. doi: 10.1099/0022-1317-81-2-401. [DOI] [PubMed] [Google Scholar]
- Murti K.G., Goorha R. Interaction of frog virus-3 with the cytoskeleton. I. Altered organization of microtubules, intermediate filaments, and microfilaments. J. Cell Biol. 1983;96:1248–1257. doi: 10.1083/jcb.96.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédellec P., Vicart P., Laurent-Winter C., Martinat C., Prévost M.C., Brahic M. Interaction of Theiler's virus with intermediate filaments of infected cells. J. Virol. 1998;72:9553–9560. doi: 10.1128/jvi.72.12.9553-9560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A., Moreau V., Ashman K., Reckmann I., González C., Way M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I., Brookes S.M., Hyatt A.D., Windsor M., Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 1998;72:2373–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson C.M., Hollinshead M., Smith G.L. The vaccinia A27L protein is needed for microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- Sharpe A.H., Chen L.B., Fields B.N. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology. 1982;120:399–411. doi: 10.1016/0042-6822(82)90040-x. [DOI] [PubMed] [Google Scholar]
- Sodeik B., Ebersold M.W., Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M., Nakano M.Y., Keller S., Boucke K., Stidwill R.P., Geber U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdeira M.L., Bernardes C., Cruz B., Geraldes A. Entry of African swine fever virus into Vero cells and uncoating. Vet. Microbiol. 1998;60:131–140. doi: 10.1016/s0378-1135(98)00152-7. [DOI] [PubMed] [Google Scholar]
- Wigley W.C., Fabunmi R.P., Goo Lee M., Marino C.R., Muallem S., DeMartino G.N., Thomas P.J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez R.J., Rodríguez J.M., Nogal M.L., Yuste L., Enríquez C., Rodriguez J.F., Viñuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




