Abstract
The Rho GTPase Cdc42 regulates adhesion, migration, and homing, as well as cell cycle progression, of hematopoietic stem cells, but its role in multilineage blood development remains unclear. We report here that inducible deletion of cdc42 in cdc42-floxed mouse bone marrow by the interferon-responsive, Mx1-Cre–mediated excision led to myeloid and erythroid developmental defects. Cdc42 deletion affected the number of early myeloid progenitors while suppressing erythroid differentiation. Cdc42-deficient mice developed a fatal myeloproliferative disorder manifested by significant leukocytosis with neutrophilia, myeloid hyperproliferation, and myeloid cell infiltration into distal organs. Concurrently, Cdc42 deficiency caused anemia and splenomegaly accompanied with decreased bone marrow erythroid burst-forming units (BFU-Es) and colony-forming units-erythroid (CFU-Es) activities and reduced immature erythroid progenitors, suggesting that Cdc42 deficiency causes a block in the early stage of erythropoiesis. Cdc42 activity is responsive to stimulation by SCF, IL3, SDF-1α, and fibronectin. The increased myelopoiesis and decreased erythropoiesis of the knockout mice are associated with an altered gene transcription program in hematopoietic progenitors, including up-regulation of promyeloid genes such as PU.1, C/EBP1α, and Gfi-1 in the common myeloid progenitors and granulocyte-macrophage progenitors and down-regulation of proerythroid gene such as GATA-2 in the megakaryocyte-erythroid progenitors. Thus, Cdc42 is an essential regulator of the balance between myelopoiesis and erythropoiesis.
Introduction
Hematopoiesis is a highly orchestrated process involving multilineage blood cell development. In a classical view, the earliest cells in the hematopoietic hierarchy are the hematopoietic stem cells (HSCs). The production of mature blood cells requires the sequential proliferation and differentiation of HSCs through a successive series of increasingly lineage-restricted intermediate progenitors including the common lymphoid progenitors (CLPs), the common myeloid progenitors (CMPs), the granulocyte-macrophage progenitors (GMPs), and the megakaryocyte-erythroid progenitors (MEPs).1,2 Deregulation in hematopoiesis can result in a number of blood dysplasias or malignancies including aplastic anemia, myeloproliferative disease (MPD), and leukemia. Elucidating the molecular mechanisms governing hematopoiesis during the process of lineage commitment and lineage-specific expansion is thus fundamental for developing improved treatment of hematologic abnormalities
Although hematopoietic lineage specification and homeostasis have drawn intensive research efforts, the complex molecular network that critically regulates specific progenitor lineage commitment remains poorly understood. An increasing body of work suggests that differentiation of HSCs and progenitors into various blood lineages is controlled by multiple extrinsic and intrinsic factors.3–5 In particular, the maturation program of various lineages appears to be tightly regulated by a closely interrelated and mutually dependent transcription factor network.6–9 Ectopic expression or gene knockout studies of specific transcription factors have provided convincing evidence that blood cell lineage fate, such as myeloid, erythroid, or lymphoid commitment, can be switched by altered expression patterns of transcription factors including PU.1, C/EBPα, Gfi-1, and/or GATA2.10
Cdc42 is a ubiquitously expressed member of the Rho GTPase family. It acts as an intracellular signal transducer in cells by cycling between the active, GTP-bound form and the inactive, GDP-bound form in response to diverse stimuli. In mammalian cells, Cdc42 has been shown to be a critical regulator of a broad range of cellular functions including actin cytoskeletal organization, gene transcription, cell proliferation, and differentiation.11,12 In mature blood cells, Cdc42 has been found to regulate macrophage chemotaxis,13 monocyte transmigration,14 lymphocyte polarization and directional migration,15,16 and neutrophil migration.17 Recently, by conditional gene targeting in mice our group has shown that Cdc42 is required for the homing, lodging, and engraftment, as well as the maintenance of a quiescent cell cycle state, of long-term HSCs in the bone marrow microenvironment.18
To define the role of Cdc42 in hematopoiesis, particularly its contribution to multilineage development of HSCs and early progenitors, in the present studies we have examined the effects of inducible, bone marrow–targeted deletion of Cdc42 on erythropoiesis and myelopoiesis. Our results reveal that Cdc42 deficiency causes hematopoietic cell-intrinsic defects leading to profound blood phenotypes. Cdc42 deletion alters the frequency and the number of primitive IL7R−Lin−Sca-1+c-Kit+ (LSK), IL7R−Lin−c-Kit+Sca-1− (LK), CMP, GMP, and MEP populations in the bone marrow and disrupts the normal blood cell distribution in peripheral blood, bone marrow, liver, and spleen. In particular, Cdc42 deletion inhibits early steps of erythropoiesis and promotes myelopoiesis, leading to MPD in the mice. These phenotypes are associated with altered expression profile of a number of key transcription factors that are capable of determining erythroid and myeloid cell fate in various hematopoietic progenitor populations. The results provide the evidence that Cdc42 critically controls myeloid and erythroid homeostasis at the progenitor cell level.
Materials and methods
Mice
Mx1-Cre;cdc42flox/flox mice were generated as previously described and maintained on a 129/C57BL/6 mixed background in pathogen-free conditions.18 Induction of Mx1-Cre expression was carried out by intraperitoneally injecting 8- to 12-week-old mice with 3 doses of sterile polyinosinic-polycytidylic acid (polyI:C; Amersham, Arlington Heights, IL) every other day at 10 μg/per gram body weight. The time after polyI:C induction was counted from the first day of injection (day 0) as days after induction (DPI). Total bone marrow cells were obtained from 6 bones of each mouse (2 femurs, 2 tibias, and 2 iliac crests).
Hematologic analysis and flow cytometry
Adult wild-type (WT) and Cdc42−/− (KO) mice were anesthetized and bled retro-orbitally into an EDTA-coated tube. Complete blood counts were performed using an automated hematology analyzer (Hemavet 850; Drew Scientific, Oxford, CT). Blood smears were stained using Wright-Giemsa. Bone marrow, spleen, lung, and liver tissues were dissected, fixed in formalin, sectioned and mounted on slides, and stained with hematoxylin-eosin.
Fluorescence activated cell sorting (FACS) analysis was performed on peripheral blood, bone marrow, thymocytes, and splenocytes. In brief, single-cell suspension was prepared and stained with primary and secondary antibodies if necessary. Peripheral blood cells were incubated in red blood cell lysis buffer (BD Pharmingen, San Diego, CA) for 10 minutes at room temperature. The cells were washed twice in PBS and resuspended in PBS/0.1% bovine serum albumin (BSA), divided into aliquots, and placed in tubes; antibodies were added and incubated on ice. Lineage markers (biotin-CD3, -CD4, -CD8, –Mac-1, –Gr-1, -Ter119, and -B220) and other antibodies (Sca-1, c-Kit, IL7Rα, and CD34) were purchased from BD Biosciences (San Jose, CA) and CD16/32 from e-Bioscience (San Diego, CA).
For 6-color HSC and hematopoietic progenitor analysis and sorting, color compensation samples were produced by singly staining bone marrow cells with one antibody of each of 6 fluorochrome or with a combination of cells stained by a method called Fluorescence Minus One (FMO), which was performed by sequentially adding fluorescent-labeled antibodies to the staining cocktail.19 For example, one FMO sample contained only B220-biotin-strepavidin-PerCP, Ter119-biotin-strepavidin-PerCP, CD3-biotin-strepavidin-PerCP, CD4-biotin-strepavidin-PerCP, CD3-biotin-strepavidin-PerCP, Mac1-biotin-strepavidin-PerCP, and Gr1-biotin-strepavidin-PerCP antibodies (Lin-PerCP). Other FMO samples contained Lin-PerCP plus IL7Rα-PeCy5 or its Pe-Cy5 isotype. Additional FMO samples contained Lin-PerCP and IL7Rα-PeCy5 with the addition of Sca1-PE; one other FMO sample contained Lin-PerCP, IL7Rα-PeCy5, and Sca1-PE with the addition of c-Kit-APC. One FMO sample contained Lin-PerCP, IL7Rα-PeCy5, Sca1-PE, and c-Kit-APC with the addition of CD16/32-PeCy7. The final assay sample contained Lin-PerCP, Scal-PE, IL7Rα-PeCy5, CD16/32-PeCy7, c-Kit-APC, and CD34-FITC. FACS analysis and sorting were performed using a FACS Canto Flow Cytometer (Becton Dickinson, Lincoln Park, NJ) or a FACS DiVa Flow Cytometer (Becton Dickinson).
Erythroid progenitor assays
For colony-forming units-erythroid (CFU-Es) assay, 100 000 bone marrow cells were plated per milliliter serum-free methylcellulose (M3134; StemCell Technologies, Vancouver, BC) supplemented with 10% FBS, 100 ng/mL rmSCF (PeproTech, Rocky Hill, NJ), and 4 U/mL human erythropoietin (hEpo; Amgen, Thousand Oaks, CA). For erythroid burst-forming units (BFU-Es) assay, 25 000 fresh bone marrow cells were plated in 1 mL methylcellulose (M3134; StemCell Technologies) containing 10% FBS, 4 U/mL hEPO, 100 ng/mL rrSCF, 100 ng/mL G-CSF, and 20 ng/mL IL3 (PeproTech). Colonies were scored on day 2 (CFU-Es) or day 10 (BFU-Es) following staining with benzidine dihydrochloride (Sigma, St Louis, MO).
Bone marrow transplantation
To generate bone marrow–reconstituted mice, a total of 2 × 106 donor bone marrow cells from Mx1-Cre;cdc42flox/flox and Mx1-Cre;cdc42+/+ donor mice (CD45.2+) were injected into the tail vein of the lethally irradiated recipient B6.SJL/BoyJ (CD45.1+) mice. For reciprocal transplantation, a total of 2 × 106 donor bone marrow cells from WT B6.SJL/BoyJ (CD45.1+) mice were injected into Mx1-Cre;cdc42f/f or Mx1-Cre;cdc42+/+ recipients. For competitive repopulation, 1 × 106 donor bone marrow cells were mixed with B6.SJL/BoyJ WT bone marrow cells at 1:1 ratio. Eight weeks after transplantation, the mice were treated with polyI:C to induce Cdc42 deletion. Chimerisms of the peripheral blood before and after polyI:C induction were analyzed by flow cytometry at various times.
Proliferation and survival assays
For assessment of proliferative status of bone marrow cells, mice received single intraperitoneal injection of BrdU (Sigma) at 100 mg/kg body weight. The mice were killed 2 hours later, and bone marrow cells and splenocytes were collected and stained for surface markers, and then fixed and stained with APC-conjugated anti-BrdU antibody using the Cytofix/Cytoperm Kit (BD Biosciences) according to the manufacturer's instructions.
For survival assays, the apoptotic cell population was determined by annexin V and 7-AAD staining (Apoptosis Detection Kit; BD Biosciences) and analyzed by flow cytometry as described.20 The numbers are mean plus or minus standard deviation from 3 mice in each group. Dot plots are representative of 3 independently performed experiments.
Quantitative real-time RT-PCR
RNA was isolated from sorted HSCs and progenitors (derived from pooled bone marrow of 3-7 mice; 2 independent groups) using the RNeasy Micro Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol including DNase treatment. cDNA was generated using the Sensiscript RT Kit (Qiagen). The mRNA levels of selected genes in the sorted population were measured by real time reverse-transcription–polymerase chain reaction (RT-PCR) on an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA) using SYBR Green PCR Master Mix reagent (Applied Biosystems). To generate standard curves, cDNA from wild-type Lin−Kit+ bone marrow cells was used as template in a 10-fold dilution series (0- to 1000-fold). Sample cDNA was used without dilution. Relative expression was calculated using the standard curve. Primers were purchased from SuperArray Bioscience (Frederick, MD).
Cc42 activity effector domain pull-down assay
The relative level of Cdc42-GTP in the low-density bone marrow cells was examined by an effector domain GST-PAK1 pull-down protocol as previously described.20 Briefly, the isolated WT low-density bone marrow cells were starved in serum/cytokine-free IMDM overnight, and challenged with SCF (100 ng/mL), SDF-1α (100 ng/mL), IL3 (20 ng/mL), or a mixed cocktail containing TPO (100 ng/mL), SCF (100 ng/mL), IL3 (20 ng/mL), and EPO (4 unit/mL) for 15 minutes prior to the effector domain pulldowns. Separately, the cells adhered to a fibronectin fragment (CH296)–coated surface for 2 hours were subjected to a similar GST-PAK1 pull-down assay compared with the cells in suspension.
Statistical analysis
Student t test and analysis of variance were used for statistical analysis. Differences were considered significant when P was less than .05. In all cases, * represents P less than .05; **, P less than .01; and ***, P less than .001.
Results
Inducible deletion of Cdc42 from mouse bone marrow results in lethality and multilineage hematopoiesis defects
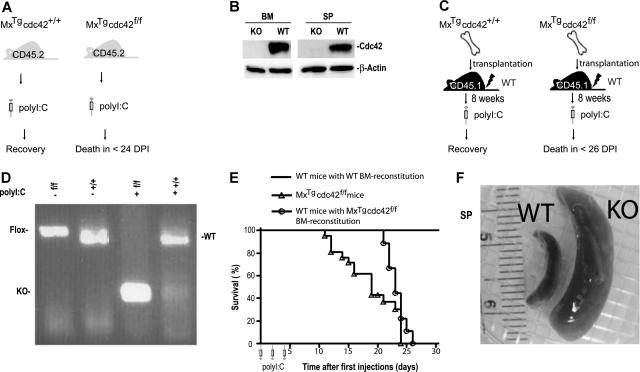
To determine whether Cdc42 is required for hematopoietic homeostasis, mice carrying the conditional cdc42flox allele were crossed with mice harboring the interferon-inducible Mx1-Cre transgene22 to produce the MxTg;cdc42f/f mice and their MxTg;cdc42+/+ littermate controls (WT).18 Deletion of the floxed exon 2 that encodes the nucleotide-binding sequences of Cdc42 GTPase domain was achieved by intraperitoneal administration of 3 doses of polyI:C at 6 to 8 weeks after birth (Figure 1A). Cdc42 protein as well as cdc42 gene were effectively removed from the bone marrow and spleen of KO mice as detected by anti-Cdc42 Western blotting (Figure 1B) and PCR genotyping (data not shown). The Cdc42-deficient mice died between 12 and 26 days after the initial polyI:C treatment, while the control mice did not show any detectable effects (Figure 1E).
Figure 1.
cdc42 deletion in the bone marrow causes lethality and splenomegaly. (A) A schematic representation of the experimental design to delete cdc42 in primary MxTgcdc42f/f mice by polyI:C induction. (B) Cdc42 protein was absent in bone marrow and spleen from polyI:C-induced MxTgcdc42f/f mice as detected by Western blotting. (C) Experimental procedures for bone marrow transplantation into lethally irradiated recipient mice. Control (MxTgcdc42+/+) or mutant (MxTgcdc42f/f) donor bone marrow cells of the CD45.2+ genotype were transplanted into lethally irradiated CD45.1+ recipients. After 8 weeks, the recipients were treated with 3 doses of polyI:C in 2-day intervals. (D) The floxed cdc42 allele was effectively deleted from the transplanted MxTgcdc42f/f bone marrow cells after the polyI:C treatment as detected by PCR genotyping. (E) Viability of the cdc42 knockout primary mice and the bone marrow transplant recipient mice is depicted by the Kaplan-Meier survival curve after polyI:C treatment. Primary MxTgcdc42f/f (f/f) mice, n = 16; WT mice with f/f bone marrow reconstitution, n = 6; mice with WT bone marrow reconstitution, n = 12. (F) Representative gross anatomy of spleens from polyI:C-treated MxTgcdc42+/+ (WT) and MxTgcdc42f/f (KO) mice.
To ensure that Cdc42 deletion occurs in a tissue-specific manner, we generated bone marrow–reconstituted mice that produced CD45.2+, MxTg;cdc42+/+, or MxTg;cdc42f/f blood cells by transplantation of bone marrow cells of the respective genotypes into lethally irradiated CD45.1+ WT recipients (Figure 1C). Flow cytometry analysis showed that greater than 95% of peripheral blood cells of the recipients were of the CD45.2+ genotype 8 weeks after transplantation (data not shown). After treatment with 3 doses of polyI:C, the bone marrow cells from reconstituted MxTg;cdc42f/f recipient mice showed effective Cdc42 gene deletion as assayed by PCR genotyping (Figure 1D) or by anti-Cdc42 Western blotting (data not shown). Consistent with the lethality phenotype observed in the MxTg;cdc42f/f primary mice (Figure 1A,E), reconstituted WT mice engrafted with MxTg;cdc42f/f bone marrow cells died between 22 and 24 days after the initial polyI:C injection, while mice receiving MxTg;cdc42+/+ bone marrow transplant did not show any detectable effect (Figure 1E). Thus, Cdc42 deletion from bone marrow cells results in lethality.
Complete blood count analysis of the primary mice and the transplant recipients showed that the KO animals displayed severe leukocytosis, neutrophilia, and eosinophilia, as well as reduced hemoglobin level (Table 1). The lymphocyte and platelet counts in peripheral blood did not change significantly (Table 1). The spleen size of the KO mice increased up to 10-fold compared with that of WT after the polyI:C induction (Figure 1F). These observations indicate that Cdc42 deletion from bone marrow cells causes multilineage hematopoietic defects and concomitant splenomegaly.
Table 1.
Altered hematopoiesis in Cdc42-deficient mice
| Parameters | MxCreTgcdc42+/+, n = 14 | MxCreTgcdc42f/f, n = 15 | CD45.1+ recipients reconstituted with MxCreTgcdc42+/+ bone marrow cells, n = 10 | CD45.1+ recipients reconstituted with MxCreTgcdc42f/f bone marrow cells, n = 10 |
|---|---|---|---|---|
| WBC, ×109/L | 4.3 ± 2.5 | 34.9 ± 10.0* ↑ | 12.0 ± 1.4 | 52.7 ± 11.6* ↑ |
| Neutrophils, ×109/L | 1.5 ± 0.8 | 29.3 ± 9.4* ↑ | 3.8 ± 0.7 | 41.1 ± 7.3* ↑ |
| Lymphocytes, ×109/L | 2.4 ± 1.9 | 3.5 ± 1.4 | 6.6 ± 1.9 | 7.8 ± 3.4 |
| Monocytes, ×109/L | 0.23 ± 0.16 | 0.67 ± 0.44† ↑ | 0.45 ± 0.19 | 1.49 ± 0.43† ↑ |
| Eosinophils, ×109/L | 0.09 ± 0.1 | 1.2 ± 0.7* ↑ | 0.14 ± 0.18 | 2.06 ± 0.33† ↑ |
| RBC, ×1012/L | 7.8 ± 0.9 | 7.2 ± 1.8 | 3.1 ± 4.4 | 4.3 ± 0.5 |
| Hb, g/L | 13.0 ± 2.0 | 10.9 ± 2.6‡ ↓ | 12.8 ± 1.5 | 8.2 ± 2.3‡ ↓ |
| HCT, % | 42.9 ± 4.8 | 38.5 ± 7.9‡ ↓ | 36.2 ± 3.4 | 24.6 ± 6.5‡ ↓ |
| RDW | 19.5 ± 1.0 | 26.2 ± 7.3† ↑ | 22.0 ± 0.1 | 29.0 ± 5.1‡ ↑ |
| Platelets, ×109/L | 903 ± 577 | 740 ± 202 | 455 ± 380 | 586 ± 442 |
Complete blood counts from primary Mx-CreTgcdc42f/f mice, WT mice engrafted with Mx-CreTgcdc42f/f bone marrow cells, and the corresponding control mice were performed at 14 to 19 DPI with polyI:C as described in Figure 1A,C. Hematologic measurements were performed on a Hemavet 850 Hematology Analyzer. The data are means plus or minus SD.
WBC indicates white blood cells; RBCs, red blood cells; Hb, hemoglobin; HCT, hematocrit; RDW, RBC distribution width; ↑, significant increase; and ↓, significant decrease.
P < .001.
P < .01.
P < .05.
Effect of Cdc42 deficiency on hematopoietic progenitor populations in the bone marrow
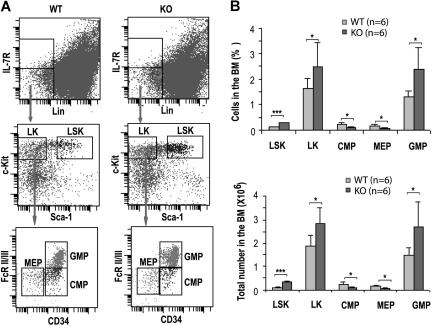
We have previously shown that loss of Cdc42 causes decreased long-term HSC and increased short-term HSC frequencies in the bone marrow but does not detectably affect the total bone marrow cellularity.18 To dissect the effect of Cdc42 deletion on progenitor populations, we immunophenotypically analyzed the bone marrow cells of KO mice at 14 DPI for the percentage and absolute numbers of IL7Rα−Lin−Sca-1+c-Kit+ (LSK), IL7Rα−Lin−c-Kit+Sca-1− (LK), CMP (IL7Rα−Lin−Sca-1−c-Kit+CD34+FcRII/III−),2 GMP (Lin−Sca-1−IL7Rα−c-Kit+CD34+FcRII/III+) and MEP (Lin−Sca-1−IL7Rα−c-Kit+CD34−FcRII/III−), populations.
Deletion of cdc42 resulted in a significant increase of LSK and LK progenitors (Figure 2). However, CMP frequency reduced by approximately 50%, whereas the frequency and number of GMP increased by approximately 1.8-fold compared with the respective WT compartments (Figure 2). In parallel, the frequency and number of MEPs decreased by approximately 2-fold in Cdc42-deficient mice (Figure 2). These data suggest that Cdc42 deletion leads to increased common primitive progenitors with a selective expansion of GMPs and a suppression of the MEPs, although a partial blockage of the development from primitive progenitors (ie, LK population) to CMPs is also apparent.
Figure 2.
Cdc42 deletion affected hematopoietic progenitor populations. (A) Immunophenotypic analysis of various progenitors in WT and KO bone marrow. Representative FACS staining profiles of the progenitor populations, including LSKs, LKs, CMPs, MEPs, and GMPs, from the respective bone marrow of WT or KO mice at 14 DPI are shown. The gating is described in “Hematologic analysis and flow cytometry.” (B) The numbers and percentages of LSK, LK, CMP, MEP, and GMP populations in the bone marrow of WT and KO mice are calculated according to the phenotype determined by flow cytometry shown in panel A. Values were derived from 6 mice of each genotype. *P < .05; **P < .01; and ***P < .001.
Loss of Cdc42 results in myeloproliferative disorder (MPD)
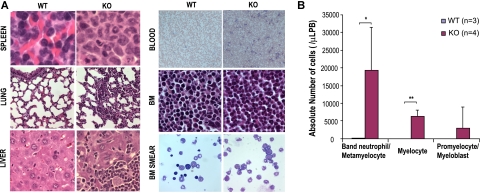
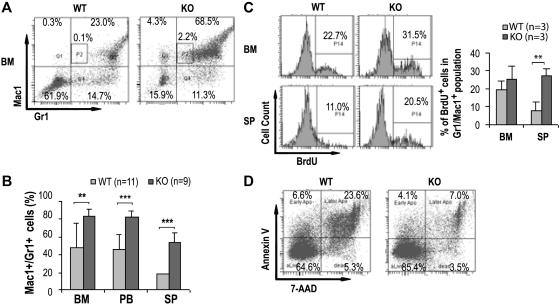
Since a significant increase in GMP population in the bone marrow with marked leukocytosis, neutrophilia, and eosinophilia was noted in the Cdc42-deficient mice, we next evaluated myelopoiesis in the mice in more detail. Histopathological analysis of KO mice revealed that bone marrow, spleen, liver, lung, and peripheral blood were massively infiltrated with myeloid cells of various degrees of maturation (Figure 3A). An examination of the blood smears confirmed increased numbers of monocytes and neutrophils and further revealed the presence of significant numbers of immature myeloid precursors including myelocytes, metamyelocytes, and band neutrophils (Figure 3B). These pathological and morphologic results were further confirmed by FACS analysis of the bone marrow, spleen, and peripheral blood that demonstrated a significantly higher percentage of Gr1+ and/or Mac1+ (Gr1+/Mac1+) cells present in KO mice (KO:WT = 83.3% ± 8.9%:47.8% ± 27.2%, P < .01, in the bone marrow; KO:WT = 83.0% ± 6.2%:45.7% ± 17.0%, P < .001, in peripheral blood; and KO:WT = 54.2% ± 10.3%:18.3% ± 7.0%, P < .001, in spleen) (Figure 4A-B). In addition, increased numbers of Mac-1+, Gr-1lo cells (P2 gating in Figure 4A) that likely represent immature monocytic population were readily detected in the KO bone marrow but not in WT bone marrow (KO:WT = 2.3% ± 0.9%:0.1% ± 0.02%, P < .05.).
Figure 3.
Cdc42 deletion causes myeloid cell infiltration to various organs and accumulation of myeloid precursors in peripheral blood. (A) Cdc42-deficient mice show myeloid expansion in spleen, liver, lung, and bone marrow, as well as peripheral blood. Histologic analyses of spleen, lung, liver, peripheral blood smears, bone marrow, and bone marrow smear from WT or KO mice were carried out. Sections were stained with hematoxylin and eosin (magnifications: ×600 for spleen, lung, liver, bone marrow, and bone marrow smear; ×200 for blood). Slides were viewed with a Nikon Eclipse 50i microscope (Nikon, Tokyo, Japan) using Plan Apo and Plan Fluor lenses at 20×/0.75 PH, 60×/0.85 PH, and 100×/1.30 oil and Micromount mounting medium (Surgical Medical Instruments, Grayslake, IL). Images were acquired using a Spot camera model Insight 4 (Diagnostic Instruments, Irvine, CA) and were processed with Spot software version 4.0.9 (Diagnostic Instruments). (B) Quantification of differential counts of myeloid precursors in peripheral blood. Myelocytes (> 500) at various stages of differentiation were counted by visual inspection of multiple, randomly selected fields under a microscope. *P < .05; **P < .01; and ***P < .001.
Figure 4.
Cdc42 deletion promotes expansion of myeloid cells. (A) Representative FACS analysis of bone marrow cells by Gr1/Mac1 staining of the indicated genotypes at 14 DPI. P2 gating represents the Mac-1+, Gr-1lo population of immature monocytic cells. The numbers shown indicate the percentage of cells within each gate. (B) The frequencies of Gr1+/Mac1+ cells from bone marrow, peripheral blood, and spleens determined by flow cytometry are shown. (C) Mice were injected with one dose of BrdU 2 hours prior to being killed. Representative histograms (left) and statistical quantification (right) of BrdU incorporation determined by flow cytometry of the Gr1+/Mac1+ bone marrow cells and splenocytes are shown. (D) Representative FACS analysis of Gr1+/Mac1+ cells from WT and KO bone marrow by annexin V/7-AAD staining shows increased survival of the KO cells. Numbers indicate percentage of cells within each gate.
The hyperexpansion of myeloid cells could be due to increased proliferation and/or increased survival of the cells. We next measured BrdU incorporation and performed annexin V and 7-AAD staining of the primary Gr1+/Mac1+ cells to ascertain the effects of Cdc42 deletion. KO mice and WT controls were killed 2 hours after a single injection of BrdU, and the percentages of bone marrow and spleen cells that had incorporated the label were determined by flow cytometry. The fraction of BrdU-positive Gr1+/Mac1+ cells was higher in the spleen (KO:WT = 27.2% ± 4.2%:8.1% ± 4.4%, P < .01, Figure 4C) and substantially greater in the bone marrows of KO mice (KO:WT = 25.3% ± 7.5%:19.2% ± 5.1%, P = .35, Figure 4C). The percentage of freshly isolated cells stained positive by annexin V was lower in KO Gr1+/Mac1+ bone marrow cell population compared with control mice (KO:WT = 7.1% ± 3.6%:23.6% ± 6.6%, P < .05, Figure 4D). Similarly, Gr1+/Mac1+ cells in KO spleen also displayed greatly enhanced survival (data not shown).
The Cdc42-deficient mice died 2 to 3 weeks after Cdc42 deletion (Figure 1E), showing increased myeloid expansion in spleen and bone marrow along with increased white blood counts (Figures 3,4; Table 1). These mice also displayed massive infiltration of myeloid and myeloid precursor cells to the peripheral organs including the lung (Figure 2). In addition, anemia and progressive splenomegaly were evident (Table 1; Figure 1F). The combined phenotypes conform to the criteria of murine myeloproliferative disorder (MPD) according to criteria defined by the “Bethesda proposal.”23,24 Thus, Cdc42 deficiency causes MPD in mice.
Cdc42 deletion causes impaired erythroid lineage development
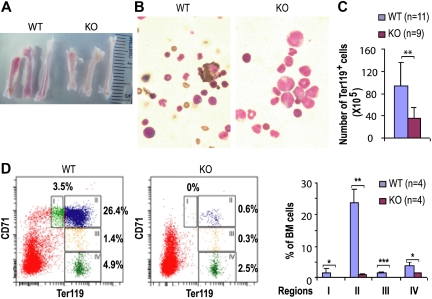
Two weeks after Cdc42 deletion, primary or the bone marrow–reconstituted KO mice began to show signs of anemia with reduced hemoglobin and hematocrits compared with wild-type mice (Table 1). KO mice surviving beyond 21 days after the initial dose of polyI:C induction displayed severe anemia. Bone marrow of the KO mice appeared pale (Figure 5A) and showed significant reduction in benzidine-positive cells (Figure 5B), indicating reduced erythroid cells in the KO (Figure 5B). Consistent with these observations, the frequency and total number of Ter119+ cells, which include early proerythroblast to mature erythrocytes, in the KO bone marrow were significantly reduced compared with that of WT at 14 DPI (KO:WT = 35.7% ± 9.1%:93.6% ± 41.4%, P < .01, Figure 5C).
Figure 5.
Cdc42 deletion causes defective erythropoiesis. (A) Gross anatomy of bones from WT and KO mice. (B) Visualization of erythroid cells by benzidine staining (magnification × 1000). Slides were viewed with a Nikon Optiphot-2 microscope (Nikon, Melville, NY) using Plan Apo lens at 100×/1.25 oil and Cytoseal 60 mounting medium (Richard-Allan Scientific, Kalamazoo, MI). Images were acquired using a Spot camera model Insight 4 (Diagnostic Instruments) and were processed with Spot software version 4.0.9 (Diagnostic Instruments). (C) The total numbers of Ter119-positive erythroid cells in bone marrow were assessed at 14 DPI. (D) Flow cytometry analysis of CD71 and TER119 expression of freshly harvested bone marrow cells at 19 DPI, with percentages of erythroid cells at different developmental stages shown as in regions I to IV.
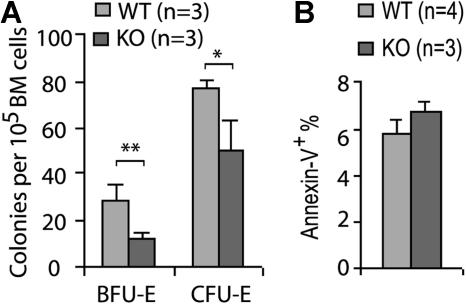
We next analyzed the effect of Cdc42 deletion on erythroid differentiation by flow cytometry using a Ter119 and CD71 surface marker staining protocol described previously.25 This analysis revealed a paucity of the KO erythroid progenitors in various developmental stages at 19 DPI (Figure 5D). In addition to the reduced frequency of late erythroblasts (regions III-IV, Ter119hiCD71med and Ter119hiCD71lo), the FACS analysis showed a more than 20-fold reduction of basophilic erythroblasts (region II, Ter119hiCD71hi) and more than 10-fold reduction of the proerythroblast population (region I, Ter119medCD71hi), suggesting an erythroid differentiation block in a stage earlier than proerythroblast. Consistent with the defective genotypic distribution of erythroid progenitors, Cdc42 deficiency was associated with a significantly reduced erythroid BFU-E and CFU-E activities of bone marrow cells (Figure 6A), indicating that the erythropoiesis blockage may occur functionally at an early developmental stage.
Figure 6.
Cdc42 deletion decreases erythroid progenitor activity. (A) BFU-E and CFU-E activities in the WT or KO bone marrow cells were assayed as described in “Erythroid progenitor assays.” (B) The relative apoptotic erythroid cells stained positive for annexin V and Ter119 were analyzed by FACS in freshly harvested bone marrow cells. *P < .05; **P < .01; and ***P < .001.
To test whether the decrease in erythropoietic populations was due to reduced erythroid cell survival or proliferation activity, we examined the freshly harvested bone marrows and spleens from the KO mice by annexin V and BrdU labeling. The percentage of annexin V+ cells in the Ter119+ population was comparable between KO and WT cells (Figure 6B), and no detectable difference in BrdU incorporation was observed between the 2 genotypes (data not shown). These data suggest that excessive apoptosis or suppressed proliferation in the erythroblast stages does not play a major role in the erythropoiesis defect of the KO animals. Therefore, it appears that Cdc42 deletion affects the erythroid differentiation program in a stage prior to proerythoblasts and likely as early as the MEP stage.
Cdc42 deletion alters the expression profile of key transcription factors in the progenitor populations
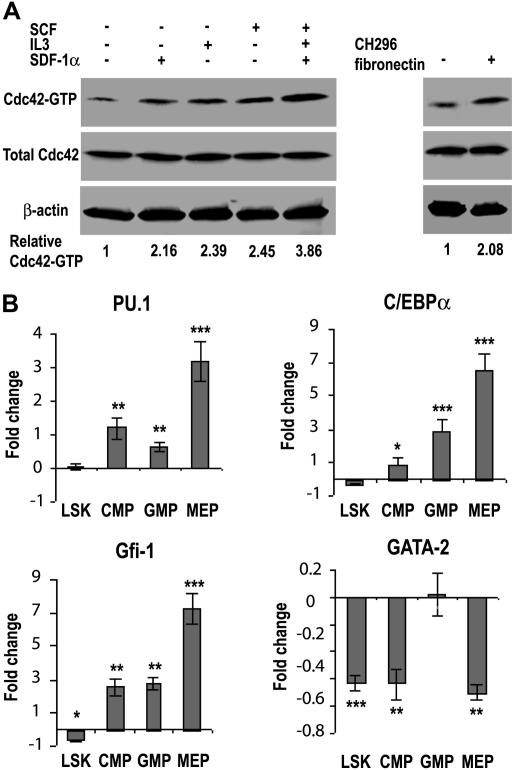
To begin to dissect the signaling mechanisms involved in the Cdc42-regulated hematopoietic process, we first examined what stimuli in the bone marrow could activate Cdc42 activity. As shown in Figure 7A, Cdc42-GTP level in the isolated low-density bone marrow cells was elevated significantly in response to stimulation by the cytokine SCF and IL3, the chemokine SDF-1α, or the adhesion molecule fibronectin, as well as by a mixed cytokine cocktail containing SCF, IL3, and TPO. These data suggest that Cdc42 can be regulated by diverse cytokines, chemokines, and adhesion molecules in the BM to impact on the HSC differentiation hierarchy.
Figure 7.
Cdc42 deletion results in altered transcription factor network in progenitors. (A) The relative Cdc42-GTP level in the WT low-density bone marrow cells under various stimulatory conditions was determined by the GST-PAK1 effector domain pull-down assay as described in “Cdc42 activity effector domain pull-down assay.” (B) Changes in gene transcription of purified LSK, CMP, GMP, and MEP cells from WT and KO bone marrow were measured by quantitative real-time PCR. The relative transcript levels of each cell population were internally normalized to that of GAPDH, and the results are expressed as (= KO/WT − 1). Each cell population was pooled from at least 4 mice and values are representative of at least 2 independent experiments performed in triplicate. *P < .05; **P < .01; and ***P < .001.
Cdc42 is known to regulate gene transcription, whereas lineage commitment of hematopoietic progenitors is driven by the expression pattern of a tightly controlled transcription factor network. To examine if Cdc42 gene targeting may alter the transcriptional network that impacts on myelopoiesis and erythropoiesis, we measured the mRNA levels of several candidate transcription factors (ie, PU.1, C/EBPα, GATA2, and Gfi-1) whose expressions could determine the fate of myeloid and/or erythroid cell development.26–29 The mRNA was isolated from FACS-purified LSK, CMP, GMP, and MEP populations for real-time RT-PCR analysis. As shown in Figure 7B, although there was no detectable expression difference in the LSK population, PU.1 mRNA levels increased by 1.2-fold and 0.6-fold in the CMP and GMP populations, respectively, after Cdc42 deletion. Significantly, PU.1 expression increased by 3.2-fold in the Cdc42−/− MEP population, consistent with a possible role of Cdc42 in regulating PU.1 and the observed suppression of erythropoiesis. Previous studies have demonstrated that overexpression of PU.1 in erythroid cells is sufficient to inhibit erythroid differentiation.30–33 In parallel, the levels of C/EBPα mRNA were significantly increased in Cdc42-deficient GMP and MEP populations by 2.9- and 6.4-fold, respectively, supporting a role of C/EBPα in promoting myelopoiesis24,25 and possibly, the development of MPD in Cdc42-deficient mice. Cdc42 deficiency was also associated with a significant increase in the expression of Gfi-1 by 2.5-, 2.8-, and 7.3-fold in the CMP, GMP, and MEP populations, respectively. Since Gfi-1 has been shown to regulate the proliferation and survival of early myeloid cells,34 such changes in Gfi-1 expression could also contribute to the observed myeloid expansion. The mRNA levels of GATA2, an erythroid lineage determinant at early stem/progenitor population,35–37 were reduced by approximately 2-fold in Cdc42-decifient LSK, CMP, and MEP populations (Figure 7B), a finding that suggests a possible involvement of Gata2 down-regulation in the early erythropoiesis blockage of the KO mice.21,38 Collectively, these results reveal that Cdc42 deletion has a profound effect on the transcription program of various progenitors. The pattern of the transcription factor profile appears to favor myeloid lineage selection while suppressing erythroid lineage differentiation.
Discussion
In previous studies using a Cdc42GAP knockout, Cdc42 gain-of-activity mouse model, we have shown that constitutively increased Cdc42-GTP species can lead to defective hematopoietic stem/progenitor survival, adhesion, and actin cytoskeleton that may contribute to an engraftment defect in hematopoietic stem/progenitors.20 Cdc42GAP−/− mice also displayed mild anemia and deficiencies in erythroid progenitors. While this animal model provides valuable information for a possible involvement of Cdc42 activity in hematopoiesis, there are inherent weaknesses of the gain-of-activity approach that complicate interpretation of the data as they relate to the physiologic role of Cdc42. For example, this animal model cannot reveal the requirement of Cdc42 in hematopoiesis nor rebut a valid criticism that Cdc42GAP knockout may also bring about Cdc42-independent effects.
To understand the physiological role of Cdc42, we have recently generated a Cdc42-conditional knockout mouse model that uses the loxP/Cre technology.18 Consistent with previous observations, the Cdc42 knockout primary mouse embryonic fibroblasts showed defective filopodia formation, defective migration, and impaired gene transcription that were readily rescued by adding back WT Cdc42,39 indicating that the conditional knockout model allows studies of Cdc42-specific function. By tissue-specific targeting in the hematopoietic stem cell compartment, we found that Cdc42 is required for the localization of HSCs in the proper bone marrow microenvironment and regulates the quiescent state of HSCs.18 The regulation of HSC quiescent state maintenance and bone marrow microenvironment interactions by Cdc42 is mediated at least in part by Cdc42's effects on the expression of adhesion molecules such as β1-integrin and N-cadherin and cell cycle regulators such as p21Cip1 and c-Myc. In the present studies, we have carried out detailed characterizations of hematopoiesis in the cdc42 gene-targeted mice and demonstrate that Cdc42 plays a critical role in maintaining hematopoietic homeostasis in vivo by regulating multilineage blood cell development. Specifically, we show that Cdc42 activity can be subjected to regulation by multiple stimuli in the bone marrow. Cdc42 deficiency in the bone marrow alters the frequency and distribution of primitive progenitors, leading to a decrease in the MEP population and an increase in the GMP population. As a consequence of the developmental defects in the myeloid and erythroid lineages that are likely due to the dysregulated expression profiles of transcription factor networks including PU.1, C/EBPα, GATA-2, and Gfi-1, the cdc42 KO mice display severe leukocytosis, anemia, and myeloproliferative disorder. These results define the stages of the involvement of Cdc42 in hematopoiesis and implicate an important role for Cdc42 in controlling hematopoietic progenitor differentiation.
Cdc42 deficiency significantly affected erythroid maturation. Immunophenotypic analysis indicates that Cdc42 is important for MEP production and plays a role in the subsequent development of early erythroid progenitors. Consistent with this, the capability to generate erythroid progenitors in Cdc42−/− bone marrow cells was significantly impaired. Interestingly, Cdc42 deficiency did not have an obvious effect on the megakaryocytic lineage development (data not shown) despite the inhibitory effect on the erythroid lineage. How Cdc42 elicits the erythroid-specific function and what specific signaling pathways are involved in this process require future investigation.
In contrast to the pronounced inhibitory effect on the development of the erythroid lineage, excision of cdc42 led to a massive expansion of myeloid cells at multiple stages of differentiation as judged by several criteria, including the presence of leukocytosis, neutrophilia, and eosinophilia, and MPD. Mechanistically it appears that Cdc42 could impact on myeloid development in at least 2 ways: by regulating the proliferation and survival of the myeloid cells and by modulating differentiation toward the myeloid lineage.
Because Cdc42 can be regulated by diverse cytokines, chemokines, and adhesion molecules in the bone marrow and Cdc42 knockout affects the LK, CMP, MEP, and GMP populations differentially, it is possible that Cdc42 uses distinct signaling cascades to impact on multiple steps of myeloid and erythroid lineage determination. One established mechanism underlying myeloid and erythroid lineage commitment and balance is through the concerted action of a transcription factor network. Our data showed that cdc42 deletion causes deregulated expression of a number of key transcription factors that control myelopoiesis and erythropoiesis. These data suggest that Cdc42 operates as a critical regulator of myeloid and erythroid lineage choice at the primitive progenitor level by maintaining the proper expression and balance of the transcription factors. The important questions these results raise at the same time include how Cdc42 exerts its regulatory function on the transcription factor network and which effect of lineage determination is primary among Cdc42-regulated pathways. Given Cdc42 is known to play a key role in regulating HSC adhesion, spreading, and migration, a related issue to be addressed is how the Cdc42-mediated adhesion and/or migration of HSCs to the bone marrow stroma/endothelial cells regulates the differentiation and proliferation state of HSCs.
The MPD phenotype associated with cdc42 deletion suggests that Cdc42 deficiency may contribute to transformation by a loss of proliferative control at the stem/progenitor stage or in a committed myeloid lineage. Since Cdc42-deficient hematopoietic stem cells are unable to engraft in recipient mice,18 analysis of the MPD by bone marrow transplantation is not possible. It remains a possibility that Cdc42 loss-of-function can serve as a second hit to corporate with other leukemogenic events for the development of leukemia.
Overall, the current work directly implicates Cdc42 in myeloid and erythroid cell development. Cdc42 regulates myeloid and erythroid lineage commitment starting at as early as the primitive progenitor stage. A detailed analysis of the molecular pathways regulated by Cdc42 in the subsequent differentiation is warranted to understand the upstream and downstream signals controlled by Cdc42 in the lineage specifications.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 HL085362 and R01 CA105117.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.Y. performed research, contributed vital new reagents or analytical tools, analyzed data, and wrote the paper; L.W. performed research and analyzed data; T.K., X.S., and S.P. performed research; J.A.C. performed research and analyzed data; J.M. and D.A.W. analyzed data; Y. Z. designed research, contributed vital new reagents or analytical tools, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yi Zheng, Division of Experimental Hematology, Children's Hospital Research Foundation, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: yi.zheng@chmcc.org.
References
- 1.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 3.Metcalf D. Stem cells, pre-progenitor cells and lineage-committed cells: are our dogmas correct? Ann N Y Acad Sci. 1999;872:289–303. doi: 10.1111/j.1749-6632.1999.tb08473.x. discussion 303–304. [DOI] [PubMed] [Google Scholar]
- 4.Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E, Segre JA. Stem cells: a new lease on life. Cell. 2000;100:143–155. doi: 10.1016/s0092-8674(00)81691-8. [DOI] [PubMed] [Google Scholar]
- 6.Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- 7.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 8.Munugalavadla V, Dore LC, Tan BL, et al. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro LH, Look AT. Transcriptional regulation in myeloid cell differentiation. Curr Opin Hematol. 1995;2:3–11. doi: 10.1097/00062752-199502010-00002. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki H, Mizuno S, Arinobu Y, et al. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 12.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 13.Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber KS, Klickstein LB, Weber PC, Weber C. Chemokine-induced monocyte transmigration requires cdc42-mediated cytoskeletal changes. Eur J Immunol. 1998;28:2245–2251. doi: 10.1002/(SICI)1521-4141(199807)28:07<2245::AID-IMMU2245>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 15.del Pozo MA, Vicente-Manzanares M, Tejedor R, Serrador JM, Sanchez-Madrid F. Rho GTPases control migration and polarization of adhesion molecules and cytoskeletal ERM components in T lymphocytes. Eur J Immunol. 1999;29:3609–3620. doi: 10.1002/(SICI)1521-4141(199911)29:11<3609::AID-IMMU3609>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 16.Westerberg L, Greicius G, Snapper SB, Aspenstrom P, Severinson E. Cdc42, Rac1, and the Wiskott-Aldrich syndrome protein are involved in the cytoskeletal regulation of B lymphocytes. Blood. 2001;98:1086–1094. doi: 10.1182/blood.v98.4.1086. [DOI] [PubMed] [Google Scholar]
- 17.Szczur K, Xu H, Atkinson S, Zheng Y, Filippi MD. Rho GTPase CDC42 regulates directionality and random movement via distinct MAPK pathways in neutrophils. Blood. 2006;108:4205–4213. doi: 10.1182/blood-2006-03-013789. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Wang L, Geiger H, Cancelas JA, Mo J, Zheng Y. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci U S A. 2007;104:5091–5096. doi: 10.1073/pnas.0610819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Yang L, Filippi MD, Williams DA, Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harigae H, Okitsu Y, Yokoyama H, et al. Induction of erythroid-specific genes by overexpression of GATA-2 in K562 cells. Int J Hematol. 2006;84:38–42. doi: 10.1532/IJH97.06020. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 23.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 24.Van Etten RA, Shannon KM. Focus on myeloproliferative diseases and myelodysplastic syndromes. Cancer Cell. 2004;6:547–552. doi: 10.1016/j.ccr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 26.Fisher RC, Scott EW. Role of PU. 1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 27.Reddy VA, Iwama A, Iotzova G, et al. Granulocyte inducer C/EBPalpha inactivates the myeloid master regulator PU. 1: possible role in lineage commitment decisions. Blood. 2002;100:483–490. doi: 10.1182/blood.v100.2.483. [DOI] [PubMed] [Google Scholar]
- 28.Nagamura-Inoue T, Tamura T, Ozato K. Transcription factors that regulate growth and differentiation of myeloid cells. Int Rev Immunol. 2001;20:83–105. doi: 10.3109/08830180109056724. [DOI] [PubMed] [Google Scholar]
- 29.Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005;33:131–143. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 31.Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU. 1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89:1383–1393. [PubMed] [Google Scholar]
- 32.McIvor Z, Hein S, Fiegler H, et al. Transient expression of PU. 1 commits multipotent progenitors to a myeloid fate whereas continued expression favors macrophage over granulocyte differentiation. Exp Hematol. 2003;31:39–47. doi: 10.1016/s0301-472x(02)01017-2. [DOI] [PubMed] [Google Scholar]
- 33.DeKoter RP, Walsh JC, Singh H. PU. 1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 1998;17:4456–4468. doi: 10.1093/emboj/17.15.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhuang D, Qiu Y, Kogan SC, Dong F. Increased CCAAT enhancer-binding protein epsilon (C/EBPepsilon) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J Biol Chem. 2006;281:10745–10751. doi: 10.1074/jbc.M510924200. [DOI] [PubMed] [Google Scholar]
- 35.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 36.Walsh JC, DeKoter RP, Lee HJ, et al. Cooperative and antagonistic interplay between PU. 1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi S, Shimizu R, Suwabe N, et al. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96:910–916. [PubMed] [Google Scholar]
- 39.Yang L, Wang L, Zheng Y. Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia formation, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol Biol Cell. 2006;17:4675–4685. doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]