Abstract
Mannitol-1-phosphate (M1P) dehydrogenase (M1PDH; EC 1.1.1.17), an enzyme catalyzing the reduction of Fru-6-phosphate (F6P) to M1P in algal mannitol biosynthesis, was purified to homogeneity from a cell homogenate of the eulittoral red alga Caloglossa continua (Okamura) King et Puttock. The enzyme was a monomer with an apparent molecular mass of 53 kD, as determined by gel filtration and SDS-PAGE, and exhibited an pI of approximately 5.5. The substrate specificity was very high toward F6P and M1P for respective reductive and oxidative reactions. The enzyme was found to be a sulfhydryl-type, because its activity was inhibited by N-ethylmaleimide and p-hydroxymercuribenzoate, and the inhibition by p-hydroxymercuribenzoate was rescued by 2-mercaptoethanol. Some unknown factors in the extract may also have inhibited the activity, because the total activity was greatly increased through the purification procedure. The optimum pH for F6P reduction was changed from 6.0 or lower to 7.2 by the addition of 200 mm NaCl. The reduction of F6P showed strong substrate inhibition above 0.5 mm. However, Km(F6P) of M1PDH was increased eight times by the addition of 200 mm NaCl, whereas Vmax was in a similar range with the avoidance of substrate inhibition by F6P. These results indicate that the enzyme was finely and directly regulated by the salt concentration without the requirement for gene expression. M1PDH can therefore be a key enzyme for regulating mannitol biosynthesis when the alga is stressed by a salinity change.
Mannitol is the most widely occurring sugar alcohol, and its occurrence has been reported in various organisms (Kremer and Kirst, 1982; Stoop and Mooibroek, 1998). The synthesis of mannitol in higher plants is mediated by Man-6-phosphate (Man6P) isomerase, Man6P dehydrogenase, and mannitol-1-phosphatase (M1Pase), which respectively convert Fru-6-phosphate (F6P) to Man6P, Man6P to mannitol-1-phosphate (M1P), and M1P to mannitol. This process in algae is mediated by only two enzymes: M1P dehydrogenase (M1PDH) for the conversion of F6P to M1P and M1Pase for the conversion of M1P to mannitol. Mannitol is known to be involved in osmoregulation, storage and regeneration of reducing power, and service as a compatible solute in both plants (Loescher et al., 1992) and algae (Kremer and Kirst, 1982; Davison and Reed, 1985). The function of mannitol as a compatible solute has been suggested by the evidence that the eulittoral red alga Caloglossa leprieurii accumulated mannitol in response to increasing salinity and that the accumulated mannitol was degraded under decreasing salinity conditions (Mostaert et al., 1995a). The significance of mannitol as an antioxidant has also been shown by trials for creating a mannitol-accumulating transgenic tobacco (Nicotiana tabacum) plant into which the bacterial M1PDH gene, mtlD, was introduced (Shen et al., 1997a, 1997b). However, M1PDH in bacteria usually functions for degradation and not for accumulation in vivo, and mannitol absorbed from the medium is catabolized to F6P via M1P for use as a carbon and energy source by a specific phosphotransferase system. Thus, bacterial M1PDH and the mannitol-catabolizing process are thought not to be affected by a salinity change (Teschner et al., 1990). Although the regulation of mannitol accumulation in plants is well documented (Stoop et al., 1996; Loescher and Everard, 2000), the regulation of mannitol metabolism in algae is less defined. Further studies on the salt-regulatory mechanism for mannitol metabolism would be required for elucidating the salt-tolerant mechanism in algae and also for finding an effective way to improve the salt tolerance of transgenic plants. The importance of M1PDH in salt regulation for mannitol biosynthesis was suggested in our previous report (Iwamoto et al., 2001). We now report the biochemical and enzymological properties of a purified preparation of M1PDH from the eulittoral red alga Caloglossa continua (Okamura) King et Puttock.
RESULTS
Purification and Physical Properties of M1PDH
M1PDH was completely purified from the red alga C. continua as shown in Table I and Figure 1. The F6P-reducing activity of M1PDH was 0.023 and 228.6 μmol min–1 mg–1 protein in the crude extract and in the completely purified preparation, respectively (Table I). The total activity of M1PDH was increased 50 times through the purification procedure from a crude extract to affinity chromatography with Reactive Red 120 agarose. The most effective steps of this procedure for increasing the specific activity were the polyethylene glycol (PEG)/ammonium sulfate (AS) treatment and affinity chromatography. Most of the contaminating proteins removed by the PEG/AS treatment are thought to have been mainly phycobili-proteins from the spectrophotometric properties (data not shown). Reactive Red 120 agarose was selected for affinity chromatography to better separate the enzyme, because its higher ability for the recovery of enzyme activity has been experimentally proved in comparison with other agarose gels such as Reactive Blue 4, Reactive Blue 72, Reactive Brown 10, Cibacron Blue 3GA type 3000-CL, Reactive Green 5, Reactive Green 19, Reactive Yellow 3, and Reactive Yellow 86 (Sigma-Aldrich, St. Louis; data not shown).
Table I.
Purification of M1PDH from the red alga C. continua
The activity was determined by F6P reduction.
| Purification Step | Total Proteins | Total Activity | Specific Activity | |||
|---|---|---|---|---|---|---|
| mg | (%) | μmol min-1 | (—fold) | μmol min-1 mg-1 protein | (—fold) | |
| Crude extracta | 662 | (100) | 15 | (1) | 0.023 | (1) |
| PEG/AS treatmentb | 249 | (37.6) | 278 | (19) | 1.116 | (49) |
| Reactive Red 120 | 6.00 | (0.91) | 751 | (50) | 125.2 | (5,443) |
| BioAssist Q | 0.83 | (0.13) | 219 | (15) | 263.9 | (11,474) |
| Sephacryl S-100 | 0.70 | (0.11) | 160 | (11) | 228.6 | (9,939) |
a Activity was assayed after gel filtration through a Sephadex G-25 column (PD-10 column) equilibrated with buffer A as described in “Materials and Methods.” b Activity was assayed after gel filtration through a Sephadex G-25 column (2.6 × 90 cm) equilibrated with buffer A as described in “Materials and Methods.”
Figure 1.
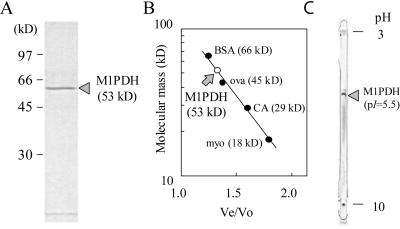
Determination of the molecular masses of the subunit (A) and native form (B), and of the pI (C) of M1PDH purified from the red alga C. continua. A, SDS-PAGE pattern. B, Molecular mass versus ratio of elution volume for the enzyme to the void volume (Ve/Vo) determined by gel filtration through a Sephacryl S-100 column. BSA, Bovine serum albumin; ova, ovalbumin; CA, carbonic anhydrase; myo, myoglobin. C, IEF pattern for the determination of pI.
The apparent homogeneity of the final enzyme preparation was shown by SDS-PAGE (Fig. 1A) and isoelectric focusing (IEF; Fig. 1C). The apparent molecular masses of the subunit and native forms were estimated to be equal at 53 kD by SDS-PAGE and by gel filtration with Sephacryl S-100 (Fig. 1B). The holo-protein of M1PDH from C. continua was therefore concluded to be a monomer whose pI determined by IEF was approximately 5.5 (Fig. 1C).
Catalytic Properties of M1PDH
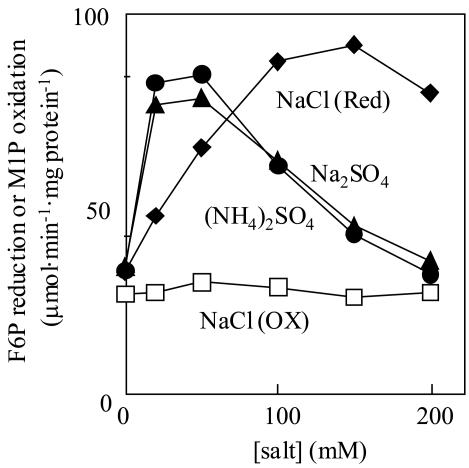
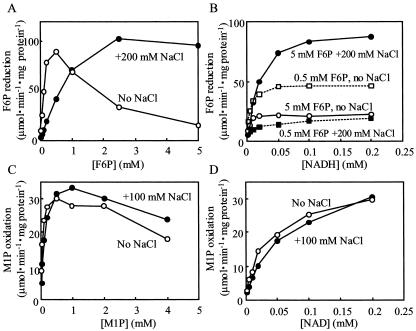
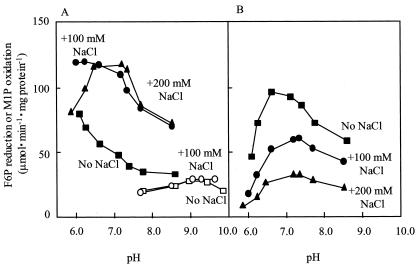
The purified enzyme exhibited very high substrate specificity for both F6P and NADH in the reductive reaction, and for both M1P and NAD in the oxidative reaction. Other compounds such as Fru-1-phosphate, Fru-1,6-bisphosphate, Glc-1-phosphate, Glc-6-phosphate, Man6P, and sorbitol-6-phosphate did not react with any coenzymes such as NADH, NADPH, NAD, and NADP in either reaction (data not shown). The F6P-reducing activity was stimulated approximately three times by such salts as Na2SO4, (NH4)2SO4, and NaCl, the optimum concentrations of those salts being 50, 50, and 150 mm, respectively, but the M1P-oxidizing activity was not affected at all by NaCl (Fig. 2). F6P reduction exhibited a low Km(F6P) value (Table II), although the activity was strongly inhibited by the substrate (F6P) itself when the concentration was above 0.5 mm (Fig. 3A). This inhibition was negated by the addition of 200 mm NaCl (Fig. 3, A and B). At the same time, Km(F6P) was increased by about eight times from 189 to 1,458 μm by the addition of 200 mm NaCl, while Vmax was in a similar range (Table II; Fig. 3A). The Km (NADH) value was also increased by NaCl in the presence of 5 mm F6P (Table II). On the other hand, M1P oxidation did not show such strong substrate inhibition, irrespective of the presence or absence of NaCl (Fig. 3, C and D; Table II). Figure 4 shows that the optimum pH value for F6P reduction was affected by the concentrations of the substrate and NaCl: At 5.0 mm F6P, the optimum pH was below 6.0 in the absence of NaCl, but changed to around 6.3 and 6.5 to 7.4 by the respective addition of 100 and 200 mm NaCl (Fig. 4A). On the other hand, at 0.5 mm F6P, the change in optimum pH was only slight in the neutral range (6.6 without NaCl and 7.4 with 200 mm NaCl; Fig. 4B). The optimum pH value for M1P oxidation was 9.0, and this value was not affected by the NaCl concentration (Fig. 4A).
Figure 2.
Effects of various salts on the activity of M1PDH purified from the red alga C. continua. F6P (3 mm) reduction: •, (NH4)2SO4; ▴, Na2SO4; ♦, NaCl. M1P (1.25 mm) oxidation: □, NaCl.
Table II.
Km and Vmax values for F6P reduction and M1P oxidation of purified M1PDH from the red alga, C. continua
| Substrate 1 | Substrate 2 | NaCl | Km for Substrate 1 | Vmax | |
|---|---|---|---|---|---|
| (mM) | mm | μm | μmol min-1 mg-1 protein | ||
| F6P reduction | |||||
| F6P | NADH | (0.2) | 0 | 188.8 | 128.8 |
| F6P | NADH | (0.2) | 200 | 1458 | 164.4 |
| NADH | F6P | (5.0) | 0 | 3.1 | 22.5 |
| NADH | F6P | (5.0) | 200 | 17.8 | 96.0 |
| NADH | F6P | (0.5) | 0 | 3.5 | 47.5 |
| NADH | F6P | (0.5) | 200 | 5.4 | 15.2 |
| M1P oxidation | |||||
| M1P | NAD | (0.2) | 0 | 48.9 | 33.2 |
| M1P | NAD | (0.2) | 100 | 138.0 | 40.2 |
| NAD | M1P | (0.5) | 0 | 32.0 | 34.0 |
| NAD | M1P | (0.5) | 100 | 56.9 | 38.3 |
Figure 3.
Kinetic analysis of M1PDH purified from the red alga C. continua. A, F6P-reducing activity versus F6P concentration. B, F6P-reducing activity versus NADH concentration. C, M1P-oxidizing activity versus M1P concentration. D, M1P-oxidizing activity versus NAD concentration. ○ and □ indicate assays without NaCl, and • and ▪ indicate assays with NaCl.
Figure 4.
pH profiles of M1PDH purified from the red alga C. continua. The activities were determined without NaCl (▪, F6P reduction; □, M1P oxidation), with 100 mm NaCl (•, F6P reduction; ○, M1P oxidation), or with 200 mm NaCl (▴, F6P reduction). A, 5 mm F6P and 0.5 mm M1P were respectively added as the substrate for the F6P-reducing and M1P-oxidizing reactions. B, F6P (0.5 mm) was added as the substrate for the F6P-reducing reaction. Final pH values are presented on the abscissa. The pH values did not change during the enzyme reaction.
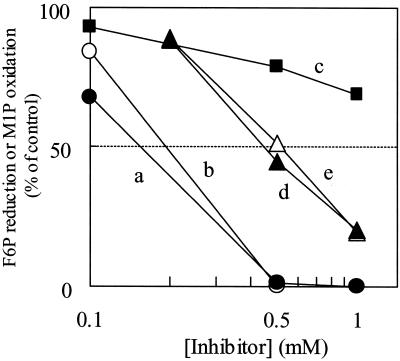
Such inhibitors of the SH enzyme as N-ethylmaleimide (NEM) and p-hydroxymercuribenzoate (pHMB) strongly inhibited the activity of both F6P reduction and M1P oxidation (Fig. 5). The inhibition of F6P reduction by pHMB was released by adding 2.5 mm 2-mercaptoethanol (2-ME), an SH-reducing reagent. The enzyme activities for the reductive and oxidative reactions were also strongly inhibited by 1 mm Zn2+, whereas such other metals as Ca2+, Mg2+, and Mn2+ did not inhibit the activity (Table III; data not shown).
Figure 5.
Effects of various inhibitors on the activity of M1PDH purified from the red alga C. continua. The activity was assayed in the presence of 100 mm NaCl. a, pHMB (•, F6P reduction); b, pHMB (○, M1P oxidation); c, pHMB (▪, F6P reduction) with 2.5 mm 2-ME; d, NEM (▴, F6P reduction); e, NEM (▵, M1P oxidation). The activities of the control were 91.3 and 18.3 μmol min–1 mg–1 protein for F6P reduction and M1P oxidation, respectively.
Table III.
Comparison of M1PDH properties in various organisms
–, No data. NA, No activity. Red, Reductive activity. Ox, Oxidative activity. G6P, Glc-6-phosphate. Gluci6P, glucitol-6-phosphate. S6P, sorbitol-6-phosphate; underlined values indicate testing with a crude or partially purified preparation.
| Molecular
|
Km
|
Optimum pH
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Native
|
Subunit
|
Structure | Red
|
Ox
|
Red
|
Ox
|
Substrate Specificity (Reacted with) | Inhibitor | ||
| F6P | NADH | M1P | NAD | ||||||||
| kD | μm | ||||||||||
| Red alga | |||||||||||
| C. continuaa | 53 | 53 | 1-mer | 147 | 3 | 51 | 26 | 7.2 | 9.0 | Narrow | NEM, pHMB |
| 1,326p | 23p | 133q | 45q | Zn2+ | |||||||
| C. leprieuriib | - | - | - | 1,400 | 20 | 90 | 23 | 7.0 | 8.5 | Wide (G6P) | - |
| Brown alga | |||||||||||
| S. pacificumc | - | - | - | 280 | 13 | 250 | 71 | 6.5 | 10.2 | Wide (G6P, Man6P) | pHMB, Hg2+ ATP, ADP |
| Green alga | |||||||||||
| P. subcordiformisd | - | - | - | 5,500 | - | NA | NA | 7.5 | NA | Narrow | - |
| Fungus | |||||||||||
| A. nigere | 40 | 22 | 2-mer | 540 | 5 | 38 | 83 | - | - | - | - |
| A. parasiticusf | - | - | - | - | - | - | - | - | - | - | Zn2+, Cd2+, Co2+, Ni2+ |
| C. neoformansg | 148 | 36 | 4-mer | 300 | 99 | 55 | 110 | 7.0-7.5 | 9.0-9.5 | Wide (S6P, G6P, EtOH, acetaldehyde) | - |
| S. sclerotiorumh | - | - | - | - | - | - | - | 7.5 | 10.5 | Wide (G6P, Gluci6P) | pHMB, Hg2+, NEM (Ox) |
| Apiconplexa | |||||||||||
| E. tenellai | - | 67 | Hetero-trimero | - | - | - | - | - | - | Wide (NADP, NADPH) | - |
| Bacterium | |||||||||||
| A. aerogenesj | 40 | - | - | 2,100 | 140 | 2,000 | 380 | - | 9.5 | Narrow | - |
| B. thermosphactak | 50 | 50 | 1-mer | 250 | - | 50 | - | 6.5 | 9.5 | - | ATP (Ox), ADP (Ox) |
| E. colil | 47 | 45n | 1-mer | 200 | 10 | 200 | 200 | 7.0 | 9.5 | Narrow | pHMBr |
| S. mutansm | 45 | - | - | 1,660 | 16 | 150 | 66 | 6.5 | 8.5-9.0 | Narrow | ADP |
a Present work. b Karsten et al. (1997a). c Ikawa et al. (1972). d Richter and Kirst (1987). e Kiser and Niehaus (1981). f Forema n and Niehaus (1985). g Suvarna et al. (2000). h Wang and Le Tourneau (1972). iSchmatz (1997). jLiss et al. (1962). k Singh and Rogers (1993). l Novontny et al. (1984). mBrown and Bowles (1977). nTeschner et al. (1990). oOne catalytic protein and two 35-kD 14-3-3 proteins. p Assayed in the presence of 200 mm NaCl. q Assayed in the presence of 100 mm NaCl. r Wolff and Kaplan (1955).
DISCUSSION
This report describes the homogeneous purification of M1PDH in a photosynthetic organism. The characterization of algal M1PDH has only previously been performed with crude or partially purified preparations from the prasinophycean alga Platymonas subcordiformis (Richter and Kirst, 1987), the brown alga Spatoglossum pacificum (Ikawa et al., 1972), and the red alga C. leprieurii (Karsten et al., 1997a, 1997b; Table III). Effective and complete purification was possible by introducing the PEG/AS treatment and affinity chromatography with Reactive Red 120 agarose. Surprisingly, the total enzyme activity was respectively stimulated approximately 20 and three times by the PEG/AS treatment and by affinity chromatography, whereas the protein concentration was greatly reduced (Table I). These results suggest that an unknown inhibitory factor was contained in the crude extract, and that activation was achieved by removing this factor during the purification procedure. This factor would not have been present in the free form but completely bound to the enzyme, because the purified M1PDH activity was not inhibited by the addition of a crude extract and the inhibitory factor was not removed by passage through a Sephadex G-25 desalting column to remove those substances with low molecular mass (data not shown). The removal of some regulatory subunit or modification of protein during the purification procedure is also considered to be a possible reason. Further study on the activation mechanism is required to elucidate this.
Caloglossa spp. M1PDH was found to be monomer like that of bacteria, although the molecular masses are different and the monomeric structure is unique among eukaryotic organisms (see Table III). A comparison of the complete coding sequences of the M1PDH gene in the database (AF175685, C. neoformans, MPD1; AF055716, E. tenella, M1PDH) indicated no homology at all among the MPD1, M1PDH, and bacterial mtlD genes, whereas mtlDs showed high homology. Hence, the unique aspects of C. continua M1PDH, including its structure, might have developed during the evolution of eukaryotes. Purified M1PDH from C. continua exhibited high specificity to F6P and NADH for its reducing activity and to M1P and NAD for its oxidizing activity (Table II). Such narrow substrate specificity of M1PDH has also been reported for other organisms (see Table III). The wider substrate specificity previously reported (e.g. Karsten et al., 1997a) might possibly have been due to contaminants, because the assay was performed with crude extracts. The inhibition by such sulfhydryl reagents as pHMB and NEM indicates that M1PDH can be considered as an SH enzyme (see Table III).
The uniqueness of M1PDH from C. continua is evident from the results that Km(F6P) and the optimum pH value for the F6P-reducing activity, but not for the M1P-oxidizing activity, were affected by the concentrations of both NaCl and F6P (Figs. 2, 3, 4). Although the activation of M1PDH by NaCl has been reported for the prasinophyte P. subcordiformis (Richter and Kirst, 1987), no report has been found about its effect on Km(F6P) or the optimum pH value. Sulfate was found to be more effective than chloride for the activation of M1PDH in C. continua (Fig. 2). This property may be advantageous for a quick response when fronds are submerged in seawater, because seawater generally contains 28 mm SO42–, which is enough to activate M1PDH. However, data on the internal concentration of SO42– in salt-stressed Caloglossa spp. are required to prove this speculation. The activation of the mannitol-synthesizing enzyme by Cl– possibly plays an important role in the accumulation of mannitol in vivo in the natural environment, because Cl– is known to increase its concentration by being transported into the cell depending on the concentration gradient under hypersaline conditions (Mostaert et al., 1995b). The sum of the concentrations of K+, Na+, and Cl– was reportedly increased from approximately 140 mm (94 mm K+, 8 mm Na+, and 38 mm Cl–) to 350 mm (156 mm K+, 17 mm Na+, and 177 mm Cl–) when the freshwater-acclimatized red alga C. leprieurii was transferred to seawater whose salinity was increased from 0.2‰ to 35‰ (Mostaert et al., 1995b). Although no data on the cytosolic levels of ions and mannitol in C. continua are available yet, data from other Caloglossa spp. can be used, because the algae in this genus generally accumulate mannitol and survive under various salt concentrations (Karsten et al., 1992). Therefore, the results obtained for C. leprieurii strongly suggest that the red alga C. continua also accumulates such ions in the cells under hypersaline stress, regulates the osmotic pressure by their accumulation, and then stimulates mannitol biosynthesis. In the archeabacterium Haloarcula marismortui, a molecular mechanism for increasing the stability and activity of an enzyme under high-salt conditions has been shown to create a hydration sphere to protect the enzyme from aggregation in malate dehydrogenase (Dym et al., 1995). An acidic residue such as Glu is considered to stabilize the folded native protein conformation by the participation of an unusual number of salt bridges. Because M1PDH in C. continua was found to be an acidic protein with low pI (approximately 5.5), a similar mechanism for stabilization might also be expected in this enzyme. On the other hand, when the salt stress is released, the concentration of Cl– in vivo can be expected to decrease (Mostaert et al., 1995b), and that of F6P would be increased by the mannitol-catabolizing pathway with mannitol dehydrogenase and hexokinase (Karsten et al., 1997a, 1997b). Under these conditions the F6P-reducing activity would be limited, and carbon flow toward mannitol biosynthesis might be reduced. Such regulation of mannitol metabolism at the enzyme level may permit the rapid response to a change in the environmental salt concentration.
Regulation of the activity of Caloglossa spp. M1PDH would be attained by changes in the optimum pH value and kinetic parameters of the enzyme. The optimum pH value for the F6P-reducing activity in C. continua was changed to around 7, which would be generally expected for the cytoplasmic pH level, by the addition of NaCl (Fig. 4). Because M1PDH is a cytoplasmic enzyme (Karsten et al., 1997a), the enzyme will be activated in vivo by such a pH change. As for the kinetic parameters, Figure 3A and Table II indicate that the activity for F6P reduction in the absence of NaCl was a maximum at 0.5 mm F6P and was strongly inhibited by the substrate above this concentration. The addition of NaCl increased the Km(F6P) value for F6P reduction by approximately eight times, showing a great decrease in the affinity for F6P, and in contrast, the substrate inhibition disappeared and the Vmax value was increased slightly, although the mechanism is unknown. In the absence of NaCl, the enzyme was changed to a form with high affinity for F6P, although this form was strongly sensitive to a high concentration of F6P to produce substrate inhibition. This kind of change in kinetic properties cannot be explained by a single mechanism for inhibition, but might have been influenced by a complex mechanism involving a conformational change of the enzyme molecule in the absence or presence of NaCl. Further analysis is needed to elucidate the salt regulatory mechanism at the molecular level.
A supply of carbon and reducing power are both essential for active mannitol biosynthesis. Consequently, carbon supplied from photosynthesis in the light and the degradation of storage compounds (e.g. floridian starch) by respiration in the dark can be expected to stimulate mannitol biosynthesis in this alga. It has been reported that starch degradation and photosynthetic organic matter production were respectively stimulated in the dark and light under hypersaline conditions in the green alga Dunaliella sp., which accumulates glycerol in response to high-salinity stress (Ginzburg, 1987). Speculating on a similar regulatory concept, the F6P concentration might be increased in the mannitol-producing red alga under hypersaline conditions by the degradation of floridian starch in the dark and by photosynthetic production of metabolites in the light. Because F6P is metabolically at a branch point along the mannitol pathway involving glycolysis and reductive and oxidative pentose phosphate cycles, the biosynthesis of mannitol via M1P could also be controlled by the supply of F6P. F6P could therefore function as an important control factor for mannitol metabolism, in addition to the regulation of M1PDH activity by NaCl. The significance of M1PDH to mannitol biosynthesis in Caloglossa spp. has been suggested by Iwamoto et al. (2001), and our results for the regulatory properties of M1PDH by the F6P and NaCl concentrations clearly support this idea. We have shown in this present study the regulation of mannitol biosynthesis by the direct regulation of M1PDH by salt. This system may enable a rapid response to an environmental salinity change without any time lag that would be necessary for either the de novo synthesis of the enzyme via gene expression or for a physiological and structural change at the cellular level.
MATERIALS AND METHODS
Algal Material
Thalli of the red alga Caloglossa continua (Okamura) King et Puttock were obtained from the Kidogawa River estuary in Chiba, Japan. The alga grows by adhering to the revetment where it is submerged in water, but is periodically exposed by the tidal effect. After collection, the samples were kept at a low temperature and transported to the laboratory. Any epibiota were removed by hand, and microscopic contaminants were removed by washing in filtered seawater. The resulting samples were stored at –80°C until being used.
Purification of M1PDH
All purification steps were carried out at 4°C, and column chromatography was performed with an FPLC system (Amersham Biosciences, Piscataway, NJ). The thalli (100 g) were homogenized with an automated mortar and pestle (Ishikawa Kogyo, Tokyo) for 1 h with a small amount of quartz sand in an extraction medium (300 mL) containing 500 mm HEPES-KOH (pH 7.0), 1 mm benzamidine-HCl, 10 mm 6-aminocapronic acid, 5 mm MgCl2, 2 mm Na2-EDTA, 4 mm 2-ME, and 1 mm phenylmethylsulfonyl fluoride. At the last stage of the extraction procedure, 3 g of polyvinylpyrolidone K-90 was added. After filtering through four layers of nylon mesh, the resulting homogenate was centrifuged at 20,000g for 30 min to obtain a crude extract. Unless otherwise mentioned, the conditions for other centrifuging operations were the same as those just described.
The aqueous two-phase partitioning method described by Nakamura and Ikawa (1993) was used with some modifications. A 50% (w/v) solution of PEG 6000 dissolved in an extraction medium was added to the crude extract while gently stirring to give a final concentration of 12.5% (w/v). Powdered solid AS was slowly added to the mixture to a 12% (w/v) concentration while constantly stirring. The mixture was then centrifuged, the upper phase of the supernatant being discarded via an aspirator, and the lower phase being collected as the PEG/AS fraction. This fraction was applied at a flow rate of 0.5 mL min–1 to a column of Sephadex G-25 (2.6 × 90 cm, Amersham Biosciences) that had been equilibrated with 50 mm HEPES-KOH (pH 7.0) containing 5 mm MgCl2, 2 mm Na2-EDTA, 4 mm 2-ME, and 20% (v/v) glycerol (buffer A). The proteins were eluted with the same buffer and at the same flow rate. The fractions (5 mL) exhibiting M1PDH activity were combined and then applied to a column of Reactive Red 120 agarose (2.6 × 8 cm, 40-mL bed volume; Sigma-Aldrich) that had been equilibrated with buffer A at a flow rate of 0.25 mL min–1. After washing the column with 50 mm Bis-Tris-propane-HCl (pH 7.0) containing 5 mm MgCl2, 2 mm Na2-EDTA, 4 mm 2-ME, and 20% (v/v) glycerol (buffer B), the proteins were eluted with buffer B containing 0.5 mm NADH at a flow rate of 1 mL min–1. The fractions (5 mL) containing M1PDH activity were then applied to a TSKgel BioAssist Q column (Tosoh Co., Tokyo) that had been equilibrated with buffer A. Proteins bound to the column were eluted with 50 mL of buffer A with a linearly ascending gradient of NaCl from 0 to 0.5 m at a flow rate of 0.25 mL min–1. The resulting active fractions of M1PDH were applied to a column of HiPrep 26/60 Sephacryl S-100 HR (Amersham Biosciences) that had been equilibrated with buffer A and then eluted with the same buffer at a flow rate of 1 mL min–1. The obtained active fraction was stored as a purified enzyme preparation at –80°C.
Assays
The M1PDH assay was performed with a spectrophotometer (UV/VIS 2200, Shimadzu, Kyoto) at 25°C according to the method of Ikawa et al. (1972) with some modifications. Unless otherwise mentioned, the F6P-reducing activity was measured by monitoring the disappearance of NADH at 340 nm in a 1-mL reaction mixture containing 50 mm HEPES-KOH (pH 7.0), 3 mm Na2-EDTA, 3 mm F6P, 0.2 mm NADH, 100 mm NaCl (buffer B), and 50 μL of the enzyme preparation. The reaction was initiated by adding F6P. The M1P-oxidizing activity was measured by monitoring the NADH formation in a 1-mL reaction mixture containing 50 mm Bis-Tris-propane-HCl (pH 9.0), 0.5 mm M1P, 0.5 mm NAD (buffer C), and 50 μL of the enzyme preparation. The reaction was initiated by adding M1P. To test the effects of various salts, the concentration of NaCl and the pH value (Figs. 2, 3, 4; Table II) for both reactions, buffer A in the enzyme preparation was replaced via gel filtration through a column of PD-10 (Amersham Biosciences) by 10 mm HEPES-KOH (pH 7.0) or 10 mm Bis-Tris-propane (pH 9.0) containing 5 mm MgCl2 and 2 mm Na2-EDTA. The resulting enzyme preparations were used for the F6P-reducing or M1P-oxidizing assay by respectively mixing with buffer B or C that contained 5 mm Bis-Tris-propane instead of 50 mm HEPES-KOH and NaCl or 50 mm Bis-Tris-propane. The pH value and the concentration of NaCl had previously been set before the enzyme assay. Measurement of the pH value in the reaction mixtures before and after the reactions clearly indicated that this pH value remained constant throughout the reaction. To test the effects of various metal ions, MgCl2 and EDTA were removed from the enzyme preparation by replacing with the appropriate buffer without the respective components via gel filtration through a column of PD-10 (Amersham Biosciences). When determining the substrate specificity, both activities were assayed in the presence of 100 mm NaCl.
The kinetic parameters of Km and Vmax were calculated by a Hanes plot analysis of the experimental data (not shown). The protein concentration was determined by a protein assay kit (Bio-Rad Protein Assay, Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as the standard.
Electrophoresis
SDS-PAGE was performed with 10% (w/v) acrylamide gel according to the method of Laemmli (1970), and protein transfer from the gel on to a PVDF membrane was performed as previously described (Iwamoto et al., 2001). The molecular mass markers used were phosphorylase b (97 kD), bovine serum albumin (66 kD), ovalbumin (45 kD), and carbonic anhydrase (30 kD). IEF of purified M1PDH was carried out by using IPGphore (Amersham Biosciences) with precast gel (Immobiline DryStrip 11 cm, pH 3–10 L, Amersham Biosciences) according to the manufacturer's instructions. The running conditions were 500 volts·hour (Vh) at 500 V, 1,000 Vh at 1,000 V, 2,000 Vh at 2,000 V, 4,000 Vh at 4,000 V, and 8,000 Vh at 8,000 V, after the gel had been incubated with 200 μL of a swelling solution containing 6 m urea, 1.5% (w/v) CHAPS, 0.4% (w/v) Pharmalyte (3–10, Amersham Biosciences), and 1 μg of purified M1PDH for 12 h at 20°C. Proteins on the resulting gel were stained with Coomassie Brilliant Blue. The pI value was determined according to the standard curve provided by Amersham Biosciences.
Chemicals
M1P was prepared as a barium salt by the method of Seegmiller and Horecker (1951). All other chemicals, including gels for affinity chromatography, were purchased from Sigma-Aldrich or Wako Pure Chemicals (Osaka).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026906.
This work was supported in part by The Salt Science Foundation (Tokyo, Japan; grant-in-aid no. 0124).
References
- Brown AT, Bowles RD (1977) Polyol metabolism by a caries-conducive Streptococcus: purification and properties of a nicotinamide adenine dinucleotide-dependent mannitol-1-phosphate dehydrogenase. Infect Immun 16: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison IR, Reed RH (1985) The physiological significance of mannitol accumulation in brown algae: the role of mannitol as a compatible cytoplasmic solute. Phycologia 24: 449–457 [Google Scholar]
- Dym O, Mevarech M, Sussman JL (1995) Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science 267: 1344–1346 [DOI] [PubMed] [Google Scholar]
- Foreman JE, Niehaus WGJ (1985) Zn2+-induced cooperativity of mannitol-1-phosphate dehydrogenase from Aspergillus parasiticus. J Biol Chem 260: 10019–10022 [PubMed] [Google Scholar]
- Ginzburg M (1987) Dunaliella: a green alga adapted to salt. Adv Bot Res 14: 93–181 [Google Scholar]
- Ikawa T, Watanabe T, Nisizawa K (1972) Enzymes involved in the last steps of the biosynthesis of mannitol in brown algae. Plant Cell Physiol 13: 1017–1029 [Google Scholar]
- Iwamoto K, Kawanobe H, Shiraiwa Y, Ikawa T (2001) Purification and characterization of mannitol-l-phosphatase in the red alga Caloglossa continua (Ceramiales, Rhodophyta). Mar Biotechnol 3: 493–500 [DOI] [PubMed] [Google Scholar]
- Karsten U, Barrow KD, Nixdorf O, West JA, King RJ (1997a) Characterization of mannitol metabolism in the mangrove red alga Caloglossa leprieurii (Montagne) J Agardh. Planta 201: 173–178 [Google Scholar]
- Karsten U, Barrow KD, West JA, King RJ (1997b) Mannitol metabolism in the intertidal mangrove red alga Caloglossa leprieurii: salinity effects on enzymatic activity. Phycologia 36: 150–156 [Google Scholar]
- Karsten U, West JA, Mostaert AS, King RJ, Barrow KD, Kirst GO (1992) Mannitol in the red algal genus Caloglossa (Harvey) J Agardh. J Plant Physiol 140: 292–297 [Google Scholar]
- Kiser RC, Niehaus WGJ (1981) Purification and kinetic characterization of mannitol-1-phosphate dehydrogenase from Aspergillus niger. Arch Biochem Biophys 211: 613–621 [DOI] [PubMed] [Google Scholar]
- Kremer BP, Kirst GO (1982) Biosynthesis of photosynthates and taxonomy of algae. Z Naturforsch 37: 761–771 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Liss M, Horwitz SB, Kaplan NO (1962) d-Mannitol 1-phosphate dehydrogenase and d-sorbitol 6-phosphate dehydrogenase in Aerobacter aerogenes. J Biol Chem 237: 1342–1350 [PubMed] [Google Scholar]
- Loescher WH, Everard JD (2000) Regulation of sugar alcohol biosynthesis. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 275–299
- Loescher WH, Tyson RH, Everard JD, Redgwell RJ, Bieleski RL (1992) Mannitol synthesis in higher plants: evidence for the role and characterization of a NADPH-dependent mannose 6-phosphate reductase. Plant Physiol 98: 1396–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostaert AS, Karsten U, King RJ (1995a) Physiological responses of Caloglossa leprieurii (Ceramiales, Rhodophyta) to salinity stress. Phycol Res 43: 215–222 [Google Scholar]
- Mostaert AS, Karsten U, King RJ (1995b) Inorganic ions and mannitol in the red alga Caloglossa leprieurii (Ceramiales, Rhodophyta): response to salinity change. Phycologia 34: 501–507 [Google Scholar]
- Nakamura Y, Ikawa T (1993) Purification and properties of NADH: nitrate reductase from the red alga Porphyra yezoensis. Plant Cell Physiol 34: 1239–1249 [Google Scholar]
- Novotny MJ, Reizer J, Esch F, Saier MHJ (1984) Purification and properties of d-mannitol-1-phosphate dehydrogenase and d-glucitol-6-phosphate dehydrogenase from Escherichia coli. J Bacteriol 159: 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DFE, Kirst GO (1987) d-Mannitol dehydrogenase and d-mannitol-1-phosphate dehydrogenase in Platymonas subcordiformis: some characteristics and their role in osmotic adaptation. Planta 170: 528–534 [DOI] [PubMed] [Google Scholar]
- Schmatz DM (1997) The mannitol cycle in Eimeria. Parasitology 114: S81–S89 [PubMed] [Google Scholar]
- Seegmiller JE, Horecker BL (1951) The synthesis of glucose-6-phosphate and 6-phosphogluconate. J Biol Chem 192: 175–180 [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ (1997a) Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol 113: 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Jensen RG, Bohnert HJ (1997b) Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol 115: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Rogers PJ (1993) Isolation and characterization of mannitol-1-phosphate dehydrogenase from Brochothrix thermosphacta. J Gen Appl Microbiol 39: 327–337 [Google Scholar]
- Stoop JMH, Mooibroek H (1998) Cloning and characterization of NADP-mannitol dehydrogenase cDNA from the button mushroom, Agaricus bisporus, and its expression in response to NaCl stress. Appl Environ Microbiol 64: 4689–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop JMH, Williamson JD, Pharr DM (1996) Mannitol metabolism in plants: a method for coping with stress. Trend Plant Sci 1: 139–144 [Google Scholar]
- Suvarna K, Bartiss A, Wong B (2000) Mannitol-1-phosphate dehydrogenase from Cryptococcus neoformans is a zinc-containing long-chain alcohol/polyol dehydrogenase. Microbiology 146: 2705–2713 [DOI] [PubMed] [Google Scholar]
- Teschner W, Serre MC, Garel JR (1990) Enzymatic properties, renaturation and metabolic role of mannitol-1-phosphate dehydrogenase from Escherichia coli. Biochimie 72: 33–40 [DOI] [PubMed] [Google Scholar]
- Wang SYC, Le Tourneau D (1972) Manniol biosynthesis in Sclerotinia sclerotiorum. Arch Mikrobiol 81: 91–99 [DOI] [PubMed] [Google Scholar]
- Wolff JB, Kaplan NO (1955) Hexose phosphate and hexose reductase: A. d-Mannitol-1-phosphate dehydrogenase from E. coli. Method Enzymol 1: 346–348 [Google Scholar]