Abstract
DNA is a precious molecule. It encodes vital information about cellular content and function. There are only two copies of each chromosome in the cell, and once the sequence is lost no replacement is possible. The irreplaceable nature of the DNA sets it apart from other cellular molecules, and makes it a critical target for age-related deterioration. To prevent DNA damage cells have evolved elaborate DNA repair machinery. Paradoxically, DNA repair can itself be subject to age-related changes and deterioration. In this review we will discuss the changes in efficiency of mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER) and double-strand break (DSB) repair systems during aging, and potential changes in DSB repair pathway usage that occur with age. Mutations in DNA repair genes and premature aging phenotypes they cause have been reviewed extensively elsewhere, therefore the focus of this review is on the comparison of DNA repair mechanisms in young versus old.
EVIDENCE FOR AGE-RELATED CHANGES IN DNA REPAIR FROM THE STUDIES OF SOMATIC MUTATIONS
The prevailing view regarding causes of aging is that aging results from accumulation of somatic damage. Damage to DNA can lead to cell cycle arrest, cell death or mutation. The majority of mutations do not kill the cell, but when accumulated in sufficient numbers may lead to deregulation of transcription patterns (1), reduced fitness and ultimately the aging phenotype.
Accumulation of mutations with age has been studied extensively in mice and humans. The early studies of mutations in the HPRT locus in cultured lymphocytes from young and old individuals have shown accumulation of mutations with age in both humans and mice (2–6). The studies using transgenic mouse models allowed the measurement of the mutation frequency in other mammalian tissues and loci. These assays measure mutation frequency in chromosomally integrated LacZ (7) or LacI (8–10) transgenes, which are rescued in Escherichia coli and analyzed for mutations using beta-galactosidase assay. Using these mice, it was demonstrated that point mutations accumulate with age (9–13) and furthermore, the mutation rate is higher in old animals (10). Not only did mutations accumulate but a characteristic type of mutations, genomic rearrangements, appear in old individuals (11,14–18).
Why does the rate of mutations increase with age and genomic rearrangements appear? Multiple studies have shown a higher load of DNA damage in old organisms (19–23). But why is there more damage? It is tempting to suggest these changes are caused by DNA repair machinery becoming less efficient and more error-prone with age. We will now discuss the studies, which directly measured DNA repair efficiency in young and old.
AGE-RELATED CHANGES IN MISMATCH REPAIR (MMR)
MMR removes mispaired bases resulting from replication errors, recombination between imperfectly matched sequences and deamination of 5-methyl-cytosine. DNA replication past a mismatched base pair would result in a point mutation. The MMR system is also thought to play a role in repair of oxidative damage by mechanisms that are not well understood (24). Several lines of evidence indicate the importance of the MMR system to the aging process. MMR is essential for maintenance of repeated sequences, as mutations in MMR genes are associated with a substantial destabilization of microsatellites (25), and microsatellite instability increases with aging in humans (26–28). The rate of MMR has been analyzed in in vitro aging human T cell clones (29). Cells at different passages were treated with mismatch-inducing agent and mismatch frequency was determined using a modification of the alkaline comet assay. Results showed a decline in MMR with increasing in vitro age. Thus, there is evidence of age-related alterations in MMR; however, more studies are needed which would directly measure MMR capacity in young and old individuals.
AGE-RELATED CHANGES IN BASE EXCISION REPAIR (BER)
Excision repair removes lesions that affect only one DNA strand, which permits excision of the lesion and subsequent use of the complementary strand to fill the gap. BER corrects small DNA alterations that do not distort the overall structure of DNA helix, such as oxidized bases, or incorporation of uracil. Excision repair is critically important for repairing base damage induced by reactive oxygen species. BER is classified into two sub-pathways: short-patch BER; a mechanism whereby only 1 nucleotide is replaced or long-patch BER; a mechanism whereby 2–13 nucleotides are replaced. BER is initiated by DNA glycosylases, which cleave N-glycosylic bond of damaged bases leaving apurinic/apyrimidinic site (AP site). The abasic site is then processed by AP endonuclease (APE1) leaving a single-stranded gap. The gap is filled by DNA polymerase β and ligated by DNA ligase (30,31).
Age-related changes in BER have been intensively studied using several experimental approaches. Measuring the levels and kinetics of AP sites following DNA damage in nuclear DNA (32) showed that senescent human fibroblasts as well as leukocytes from old donors have higher basal level of AP sites than young cells. Upon treatment with H2O2 or MMS the level of AP sites rose faster in the young cells than in the old cells suggesting a deficiency in DNA glycosylase activity (32).
Measurement of oxidized guanine (8-oxoG) kinetics in genomic DNA of young and old mice after exposure to γ-irradiation did not reveal a difference in the rate of 8-oxoG removal; however, the tissues of old animals appeared to accumulate higher levels of 8-oxoG after irradiation (23). Another way of measuring BER is by comparing in vitro incision activity in tissue extracts from young and old animals. Protein extracts are incubated with radiolabeled oligonucleotides containing single base lesions and the appearance of cleavage products is analyzed on a gel (33). A significant decrease in the mitochondrial incision activity of 8-oxoG DNA glycosylase, uracil DNA glycosylase and the endonuclease III homolog was found in the brains of old mice, whereas smaller changes were observed in nuclear incision activity (30,33,34). Similarly, a decline in cleavage activity was observed in mitochondrial, and to a lesser extent nuclear extracts from senescent human fibroblasts (35).
A different in vitro assay measuring the repair of a synthetic DNA substrate containing a single G:U mismatch showed a strong decline in BER activity in brain, liver and germ cell nuclear extracts of old mice (36). Germ cell extracts from old mice were found to contain reduced levels of APE1, and supplementation with purified recombinant enzyme restored the activity to the level of young animals (37). Reduced abundance of DNA polymerase β in brain extracts from mice and rats has been reported (36,38,39).
In addition to altered enzyme activity, the mechanism for age-related decline of BER may lie in altered response to DNA damage. For example, the expression of DNA polymerase β and AP endonuclease was induced by DNA damage in young mice, while aged mice showed a lack of inducibility (40). Furthermore, old mice and senescent human fibroblasts were deficient in the translocation of oxoguanine DNA glycosylase and AP endonuclease into both their nuclei and mitochondria (41,42). An intriguing finding showed that the efficiency of BER may be sequence-specific with the promoters of the genes whose expression is downregulated with age being repaired poorly compared to genes that are not affected by age (43). In summary, multiple evidence indicates that BER undergoes age-related changes, which are likely to contribute to the accumulation of oxidative DNA lesions and mutations with aging.
AGE-RELATED CHANGES IN NUCLEOTIDE EXCISION REPAIR (NER)
NER removes short DNA oligonucleotides containing a damaged base (44). NER recognizes bulky lesions caused by carcinogenic compounds, and covalent linkages between adjacent pyrimidines resulting from UV exposure. NER is further classified into global genome repair (GG–NER) that occurs everywhere in the genome, and transcription-coupled repair (TCR), which removes lesions in the transcribed strand of active genes. NER is a multistep process involving multiple proteins (44). In GG–NER DNA damage is recognized by XPC–HR23B complex, followed by damage verification by XPA. The DNA is unwound by XPB and XPD (ERCC3 and ERCC2) helicases in complex with TFIIH basal transcription initiation factor, and the incision is made in the damaged strand by XPF and XPG. The damaged strand is removed, and repair is completed by DNA polymerase and DNA ligase. In the TCR pathway stalled RNA–PolII on the damaged DNA template is believed to initiate the repair process, which requires the TCR-specific proteins CSB and CSA.
The relationship between NER and aging has long been studied by either evaluating repair during in vitro senescence of human fibroblasts and lymphocytes, or in cells or tissues derived from donors of different age. Studies using in vitro senescence yielded conflicting results. Hart and Setlow (45) reported a reduction in unscheduled DNA synthesis following DNA damage in late passage WI-38 human fibroblasts compared to early passage cells. While no differences in the repair replication were found using the same cell line (46). A more recent study (47) measured the kinetics of the disappearance of cyclobutane pyrimidine dimers (CPDs) from genomic DNA in human fibroblasts and trabecular osteoblasts aged in vitro. In this method (48) cells are treated with UV, genomic DNA is extracted and incubated with T4 endonuclease V, which cleaves the DNA at pyrimidine dimers. Then the DNA is cleaved with restriction enzymes, separated on an alkaline gel and the intensity of the bands corresponding to specific genes is determined by Southern hybridization. Both actively transcribed and inactive genes were analyzed, and no clear differences in the rate of CPD removal were found (47). Removal of CPDs was also examined using in situ assay with CPD antibodies (49). This study showed reduced UV-induced CPD removal in senescent compared to young fibroblasts (49).
Contrasting results between different studies may be explained by the sensitivity of cells to culture conditions such as serum concentration, and differences in cell cycle distribution during treatment (50). Furthermore, replicative aging may vary between cell lines, depending on the cell donor. For example, a decline in NER with increasing passages was documented for lymphocytes derived from two different donors but not in the lymphocytes from the third donor (51).
Studies of NER in cells or tissues from young and old individuals consistently showed a decline of NER capacity with increasing age. Earlier works used unscheduled DNA synthesis (52) or a plasmid reactivation assay as a measure of NER (53–55). In the plasmid reactivation assay a plasmid containing chloramphenicol acetyltransferase (CAT) is irradiated with UV to introduce DNA damage, plasmids are transfected into host cells and percentage of CAT activity relative to undamaged control plasmid is measured. A large study using human peripheral blood lymphocytes from 135 individuals aged 20 to 60 showed a decline of UV damage repair (54,55). The rate of decline was calculated as 0.63% per year, which amounts to ∼25% decrease over a 40-year period (54,55). This analysis was further extended to show similar decline in NER in skin fibroblasts (53). In addition to reduced plasmid reactivation, cells from older donors introduced an increased number of mutations in the transfected plasmid (53). This suggests that not only the repair becomes less efficient with age, but it also makes more errors. A disadvantage of plasmid reactivation assay is that it does not differentiate between different types of repair, and types of photoproducts.
Other assays measured the rate of removal of specific photoproducts. A study using T4 endonuclease V for detection of CPDs in hepatocytes of 6 and 24 months old rats showed that removal of CPDs from two nontranscribed genes was ∼40% lower for cells isolated from old rats than for cells isolated from young animals (56). In contrast, the age-related decline of CPD removal was less apparent in a transcribed gene, where only the rate of CPD removal was slower in old animals, but no difference in the number of CPDs was found after 24 h (56). By a similar assay, repair efficiency of telomeric DNA was reported to be lower in fibroblasts isolated from older human donors (57). Thus, it appears that aging has a greater effect on repair of nontranscribed genes.
Another method of detection of CPDs and (6–4) pyrimidone photoproducts (PP) became available with development of CPD and (6–4) PP-specific antibodies (58). DNA is isolated from cells after UV-irradiation and hybridized with antibodies. In agreement with earlier results this method showed a decrease in removal of CPDs and (6–4) PPs in dermal fibroblasts of older human donors (59). Decline in repair of (6-4) PPs was also shown in round spermatids of 14 months old compared to 2 months old mice (60). The method was further developed to use PP-specific antibodies for immunohistochemistry, which allowed studying the removal of CPDs in situ in human epidermis (61). Skin at the upper arm of young and old volunteers was exposed to UVB, and biopsied at different time points after irradiation. The biopsy material was either used for DNA extraction and probed with CPD antibodies or analyzed by immunohistochemistry. CPDs were removed from epidermis at 4 days after irradiation in the young subjects, and between 7 and 14 days in older subjects (61). Since the process of CPD removal is a combination of NER and epidermal turnover, the CPDs are likely to be removed from aged skin after 7–14 days by turnover of the epidermis. Thus, in situ studies further suggest that NER declines with age. Another in situ study with human volunteers using a postlabeling method based on quantification of photoproducts by HPLC found that photoproducts in UV-irradiated skin are induced at a higher frequency in old individuals (62). The latter study, however, did not detect age-related differences in NER due to large individual variations in the study population (63).
In summary, there is compelling evidence that NER declines with age. Little is known about the mechanism responsible for this decline. Reduced constitutive protein levels of ERCC3, PCNA, RPA, XPA and p53 that participate in NER were reported in older humans (59). Interestingly, treatment with short oligonucleotides that mimic DNA damage signal, stimulated NER in fibroblasts from old donors (64). The oligonucleotide treatment also upregulated the levels of NER proteins (64). Thus, the decline may be caused by lower levels of NER enzymes or by altered induction of DNA damage response.
Recent studies have shown that multiple mutations in the NER genes result in dramatically accelerated aging phenotypes (65–67). The progeroid phenotypes caused by NER defects were associated with characteristic changes of the global transcription patterns. Similar changes were seen in wild type mice in response to stress and during aging. It is tempting to speculate that the decline of NER function that occurs in normal individuals contributes to the onset of aging.
AGE-RELATED CHANGES IN DOUBLE-STRAND BREAK (DSB) REPAIR
A DSB is the most lethal of all DNA lesions. If unrepaired, a DSB leads to loss of chromosome segments and threatens the survival of the cell. Equally detrimental to the organism are misrepaired DSBs that destabilize the genome and lead to genomic rearrangements. Genomic rearrangements become common in aging organisms (11,14–18) ultimately leading to deregulation of transcription (1) and malignancies.
DSBs in DNA are repaired by two major mechanisms: homologous recombination (HR) and nonhomologous end joining (NHEJ). During HR-mediated repair of DSB, the sister chromatid is used as a template to copy the missing information into the broken locus. Repair by HR is mediated by Rad51 protein with the help of other members of Rad52 epistasis group (68,69). Since sister chromatids are identical to each other, DNA damage can be repaired faithfully with no genetic consequence. In contrast, the NHEJ pathway simply fuses two broken ends with little or no regard for sequence homology. NHEJ starts with binding of Ku70/Ku80 heterodimer to the broken DNA ends (70). Ku facilitates recruitment of Artemis-DNA–PKcs complex, which processes the ends to prepare them for ligation (71). Next, the gaps are filled by DNA polymerase of polX family, and covalently joined by XRCC4-DNA ligase IV complex (71). NHEJ is rarely error-free leading to deletions or insertions of filler DNA (72–74). DSBs situated between two direct repeats can also be repaired by single-strand annealing (SSA), a highly mutagenic recombinational mechanism in which the sequence between the repeats is deleted.
The first indication of age-related changes in DSB repair is the exponential increase in the incidence of cancer with age (75), since cancer is associated with genome rearrangements and loss of heterozygosity (LOH). Sequence analysis of genomic rearrangements in the LacZ locus in aged transgenic mice (11) and also in the endogenous HPRT locus in human lymphocytes (76) did not reveal regions of extended homology at the breakpoints, suggesting that the rearrangements may have resulted from erroneous NHEJ.
Despite the tremendous impact of DSB repair on aging process, age-related changes in DSB repair are currently understudied. Early studies using variations of the comet assay documented age-related decline of the efficiency of rejoining of X-ray-induced DNA breaks in normal human lymphocytes (77,78). The methodology, however, did not allow addressing a specific repair mechanism.
Our laboratory has designed a sensitive assay for NHEJ (73). The assay is based on a reporter cassette containing GFP gene interrupted by an intron, containing recognition sites for the rare cutting endonuclease I-SceI. The GFP gene in the original construct is inactive, however, induction of DSBs by I-SceI and subsequent repair restores GFP activity. Since NHEJ takes place within intronic sequences, GFP activity is reconstituted even if repair is accompanied by deletions or insertions. The number of GFP positive cells serves as a measure of NHEJ efficiency, and can be quantified by FACS. In addition the fidelity of repair can be determined by sequencing the repair products. We used this assay to study senescence-related changes of NHEJ in normal human fibroblasts. The efficiency of NHEJ was reduced up to 4.5-fold in presenescent and senescent relative to young cells. Strikingly, we observed that end joining in old cells was associated with extended deletions. These results indicate that end joining becomes less efficient and more error-prone in senescent cells (73).
NHEJ in the brain of young and old rats has been analyzed using in vitro plasmid rejoining assay. In this assay linearized plasmids are incubated with nuclear extract, and plasmid concatamers resulting from NHEJ are quantified on a gel. This assay showed that NHEJ efficiency is reduced in the brains of old rats (79,80). NHEJ is also decreased in the brains of Alzheimer's disease patients compared to normal controls (81).
What is the molecular mechanism responsible for age-related decline of NHEJ? The level of Ku, which is a protein responsible for recognition and binding to DSBs, has been shown to decline markedly in the testis of aging rats (82), and lymphocytes from human donors of increasing age (83). We observed a 50% reduction in Ku70 and Ku80 protein levels in senescent human fibroblasts (84). Furthermore, intracellular distribution of Ku differed between young and senescent cells. In young cells, Ku was distributed throughout the cytoplasm and the nucleus, and translocated to the nucleus in response to γ-irradiation. We hypothesize that the cytoplasm of young cells contains Ku that serves as a reserve that is recruited to the nucleus upon DNA damage. In contrast, in senescent cells, all Ku was localized in the nucleus, so no additional Ku can be brought into the nucleus to repair DNA damage. Moreover, our results suggest that nuclear Ku in senescent cells is unavailable for repair of new lesions (84). Ku may be trapped by unrepairable DNA damage foci (21,22) that accumulate in the nuclei of senescent cells. Ku heterodimer forms a ring that is loaded onto the DNA strand (85), and it is unknown what takes Ku off the DNA. The release of Ku from DNA may be impaired in senescent cells. Alternatively, Ku may remain at the damage sites and shortened telomeres, which cannot be repaired by DNA repair machinery in senescent cells. Impaired nuclear targeting and DNA binding activities of Ku have been reported in peripheral blood mononuclear cells of aged humans (86,87). Thus, decline of NHEJ with age may be caused by reduced availability and altered regulation of Ku.
Disruption of genes involved in DSB repair often leads to premature aging phenotypes with notable examples being Werner syndrome, Ataxia telangiectasia, and mice deficient for Ku80, DNA–PKcs and ERCC1 (88). The exact mechanism by which these mutations cause progeroid phenotype is unknown, and likely involves a combination of disregulation of transcription and an increase of cellular senescence and apoptosis. As DSB repair becomes less efficient during normal aging the same pathways may contribute in a more subtle way into the onset of aged phenotype.
RELATIVE USAGE OF DSB REPAIR PATHWAYS WITH INCREASING AGE
As we discussed above DSBs can be repaired by three distinct pathways: mutagenic NHEJ and SSA, and potentially nonmutagenic HR. Does the usage of DSB repair pathways change with age? This question was addressed in Drosophila male germline (89). The males carried a reporter cassette where DSBs were generated by constitutively expressed I-SceI endonuclease. The cassette allowed to differentiate repair events as NHEJ, SSA or HR between two homologous chromosomes. HR in male germline has increased from 14% in young flies to 60% in old ones, with a corresponding decrease in SSA and NHEJ (89).
Can this finding be generalized to mammalian DNA repair? We did observe 4.5-fold decrease in NHEJ in senescent human fibroblasts (73), however, this was clearly not due to an increase in HR, since HR is virtually absent in replicatively senescent cells (Seluanov,A. and Gorbunova,V., in preparation). Lack of HR in G0-arrested cells is not unexpected, as HR is restricted to G2/M phase of cell cycle when a sister chromatid is available (90,91). Potentially, a homologous chromosome can be used as a template in nondividing cells, however, repair using a homolog is three orders of magnitude less frequent than the use of a sister chromatid in mammalian cells (92,93). This preference is explained by sister chromatids being in close proximity after DNA replication, in contrast to homologs that are not paired in mitosis and may be harder to bring together. Expression of Rad51 protein is cell cycle-regulated (94,95), and Rad51 becomes undetectable when cells enter replicative senescence (Seluanov,A. and Gorbunova,V., unpublished data). Therefore, the major pathway for repair of DSBs in mammalian G1/G0 cells is NHEJ (90,91,96).
While senescent cells do not use HR, the situation is less clear in an aging organism, which still contains dividing cells. However, several studies show that HR is more active in ES cells (97) and in early embryonic development (98), compared to late developmental stages and somatic cells. As the body ages, fewer cells divide, cell cycle slows down and more cells are in G1/G0 stage, therefore we predict that the usage of HR will further decrease. This prediction, of course, needs to be tested in aging mammals.
Another important point is that HR tested by Preston et al. (89) is HR between homologous chromosomes. In mammalian cells mitotic recombination is rare and potentially dangerous process, as it is associated with LOH and cancer. Specific mechanisms controlled by Bloom protein (99), the MMR system (100) and genetic divergence between homologs (101) suppress mitotic recombination in mammalian cells. LOH takes place in precancerous cells and is a major contributor to the tumorogenic process (102). However, LOH and cancer do occur with increased frequency with advancing age (75,103). Thus, the finding of increased HR between homologs in aging flies may be compared to increased frequency of abnormal mitotic recombination events in aging mammals. It is possible that mechanisms suppressing mitotic recombination start to fail in aging cells, or that old individuals contain more premalignant cells that tend to be more recombinogenic. In summary, changes in DSB repair pathway usage may have profound implications for human aging and cancer. Age-related changes in DSB repair pathways have not been studied in mammals, and more research is clearly needed in this area.
CONCLUSIONS
There is sufficient evidence that all pathways of DNA repair, MMR, excision repair and DSB repair, become less efficient with age leading to accumulation of mutations (Figure 1). Much less is known about the causes of this deterioration. Several studies point out age-related decrease in expression of DNA repair enzymes or their activities (36–39,59,82,84). Why do levels of DNA repair enzymes decline with age, and why are all the DNA repair systems affected? Since DNA repair and DNA damage response are tightly controlled processes it is tempting to speculate that DNA damage response becomes less efficient or somewhat disregulated with age. Indeed, alterations in the response of DNA repair proteins to DNA damage have also been reported (40–42,64,84,86,87). Apoptosis, which is another process triggered by DNA damage, was shown to become downregulated in aging (104) and senescence (105), which was further connected to altered p53 activity (105). Even beyond DNA damage response, it is a general notion that old organisms become more sensitive to stress, hence stress responses may be altered. Recently a connection was made between the stress response elicited by DNA damage and insulin/IGF1 signaling (65,106). As stress signaling becomes disregulated with age it may in turn affect DNA repair.
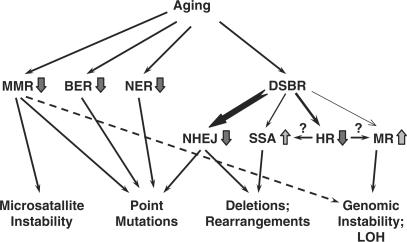
Figure 1.
Age-related changes in DNA repair and their consequences. Inefficient MMR leads to microsatellite instability, point mutations and potentially, to increased frequency of LOH. Decline in efficiency and fidelity of BER and NER leads to point mutations. Less efficient and more error-prone NHEJ results in point mutations and genomic rearrangements. As fewer cells are in G2 stage, the usage of sub-pathways of DSB repair (DSBR) may also change, where precise HR between sister chromatids declines, giving way to more mutagenic SSA and mitotic recombination (MR) with homologous chromosome leading to genomic instability and LOH.
Do the observed changes in DNA repair activities with age contribute to the onset of aging? It may be difficult to infer cause–effect relationship from the studies of normal aging, however, mouse knockout studies can provide answers. Defects in DNA maintenance may lead to accelerated aging. Notable examples are mice deficient in NHEJ or mice deficient in TCR (107,108). It has to be mentioned that not all DNA repair mutants show progeroid phenotypes, and it appears that only mutations in particular pathways affect lifespan. However, as most premature aging syndromes are caused by mutations in DNA repair genes it is reasonable to assume therefore that normal aging is caused, in part by the decline in DNA repair capacity.
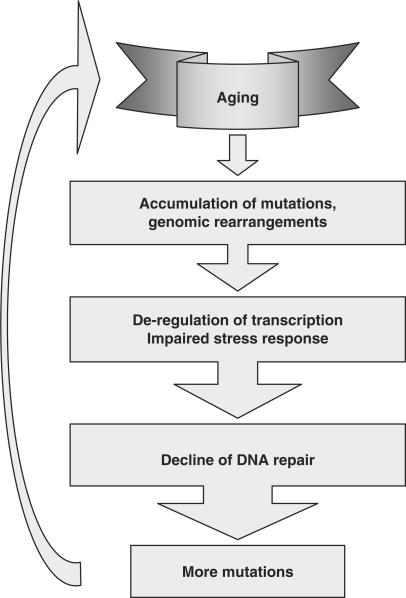
The overall sequence of events can be as follows (Figure 2): spontaneous mutations and rearrangements gradually impair the function of genes involved in stress response and DNA repair. DNA repair becomes less efficient and more error-prone leading to cascading accumulation of DNA damage and mutations, which further exacerbate age-related physiological decline.
Figure 2.
The vicious cycle of aging and genomic instability. Spontaneous mutations and rearrangements accumulate with time leading deregulation of transcription, impaired stress response and diminished function of DNA repair genes. Decline of DNA repair efficiency and fidelity leads to more mutations, and further exacerbates the age-related functional decline.
Accumulation of mutations may contribute to aging by affecting essential genes and disturbing transcription patterns (1). In addition, unrepaired DNA damage may contribute to aging by increasing apoptosis. A shift in the balance between cell death and cell renewal may lead to exhaustion of the stem cell pool, loss of tissue cellularity and functional decline. Accelerated aging due to loss of stem cells has been observed in mice with overactive p53 (109). Mice with mutated DNA polymerase γ lacking the proofreading activity exhibit accelerated aging due to increased apoptosis (109), and remarkably several NER mutants undergo premature aging due to increased apoptosis, which is not associated with an increase in mutation frequency (110). Furthermore, DNA damage and mutation may perturb chromatin structure and cause epigenetic changes.
Why is DNA repair not perfect? Mutations provide material for natural selection and adaptation. Furthermore, organisms in the wild do not live forever and often succumb to predation or accidents long before they accumulate enough mutations to show manifestations of aging. Therefore, there was no advantage in investing into maintenance of perfect DNA repair system. However, tremendous differences in lifespan between animal species, suggest that there is a wide spectrum of how ‘imperfect’ maintenance mechanisms can be. Thus, there is no reason why DNA repair cannot be improved. That said, overexpression of downstream DNA repair enzymes often resulted in toxicity (111), which underscores the importance of coordinated expression of DNA repair factors. However, changes in upstream regulators may be a more promising approach to improve DNA repair. In conclusion, more research is needed to understand the nature and mechanisms of age-related changes in DNA repair. This knowledge may potentially allow treatments to upregulate DNA repair and delay aging and cancer.
ACKNOWLEDGEMENTS
We thank Michael Bozzella for comments on the manuscript. Research in the authors’ laboratory is supported by grants from US National Institute of Health AG027237 (V.G.), American Federation for Aging Research (V.G.), the Komen Foundation (V.G.) and Ellison Medical Foundation (V.G. and A.S.). Funding to pay the Open Access publication charges for this article was provided by US National Institute of Health AG027237.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 2.Finette BA, Sullivan LM, O’Neill JP, Nicklas JA, Vacek PM, Albertini RJ. Determination of hprt mutant frequencies in T-lymphocytes from a healthy pediatric population: statistical comparison between newborn, children and adult mutant frequencies, cloning efficiency and age. Mutat. Res. 1994;308:223–231. doi: 10.1016/0027-5107(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 3.Jones IM, Thomas CB, Tucker B, Thompson CL, Pleshanov P, Vorobtsova I, Moore DHII. Impact of age and environment on somatic mutation at the hprt gene of T lymphocytes in humans. Mutat. Res. 1995;338:129–139. doi: 10.1016/0921-8734(95)00018-2. [DOI] [PubMed] [Google Scholar]
- 4.Dempsey JL, Pfeiffer M, Morley AA. Effect of dietary restriction on in vivo somatic mutation in mice. Mutat. Res. 1993;291:141–145. doi: 10.1016/0165-1161(93)90153-q. [DOI] [PubMed] [Google Scholar]
- 5.Morley AA, Cox S, Holliday R. Human lymphocytes resistant to 6-thioguanine increase with age. Mech. Ageing Dev. 1982;19:21–26. doi: 10.1016/0047-6374(82)90046-x. [DOI] [PubMed] [Google Scholar]
- 6.Morley A. Somatic mutation and aging. Ann. N. Y. Acad. Sci. 1998;854:20–22. doi: 10.1111/j.1749-6632.1998.tb09888.x. [DOI] [PubMed] [Google Scholar]
- 7.Vijg J, Dolle MET, Martus H-J, Boerrigter METI. Transgenic mouse models for studying mutations in vivo: applications in aging research. Mech. Ageing Dev. 1997;98:189–202. doi: 10.1016/s0047-6374(97)00107-3. [DOI] [PubMed] [Google Scholar]
- 8.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Sorge JA, Putman DL, Short JM. Spectra of spontaneous and mutagen-induced mutations in the LacI gene in transgenic mice. Proc. Natl Acad. Sci. USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart GR, Oda Y, de Boer JG, Glickman BW. Mutation frequency and specificity with age in liver, bladder and brain of lacI transgenic mice. Genetics. 2000;154:1291–1300. doi: 10.1093/genetics/154.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart GR, Glickman BW. Through a glass, darkly: reflections of mutation from lacI transgenic mice. Genetics. 2000;155:1359–1367. doi: 10.1093/genetics/155.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolle MET, Giese H, Hopkins CL, Martus H-J, Hausdorf JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat. Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- 12.Dolle MET, Snyder WK, Gossen JA, Lohman PHM, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl Acad. Sci. USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijg J. Somatic mutations and aging: a re-evaluation. Mutat. Res. 2000;447:117–135. doi: 10.1016/s0027-5107(99)00202-x. [DOI] [PubMed] [Google Scholar]
- 14.Vijg J, Dolle MET. Large genome rearrangements as a primary cause of aging. Mech. Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- 15.Curtis H, Crowley C. Chromosome aberrations in the liver cells in relation to the somatic mutation theory of aging. Radiat. Res. 1963;19:337–344. [PubMed] [Google Scholar]
- 16.Ramsey MJ, Moore DH, Briner JF, Lee DA, Olsen LA, Senft JR, Tucker JD. The effects of age and lifestyle factors on the accumulation of cytogenetic damage as measured by chromosome painting. Mutat. Res. 1995;338:95–106. doi: 10.1016/0921-8734(95)00015-x. [DOI] [PubMed] [Google Scholar]
- 17.Tucker JD, Spruill MD, Ramsey MJ, Director AD, Nath J. Frequency of spontaneous chromosome aberrations in mice: effects of age. Mutat. Res. 1999;425:135–141. doi: 10.1016/s0027-5107(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 18.Martin GM, Smith AC, Ketterer DJ, Ogburn CE, Disteche CM. Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Isr. J. Med. Sci. 1985;21:296–301. [PubMed] [Google Scholar]
- 19.Chevanne M, Caldini R, Tombaccini D, Mocali A, Gori G, Paoletti F. Comparative levels of DNA breaks and sensitivity to oxidative stress in aged and senescent human fibroblasts: a distinctive pattern for centenarians. Biogerontology. 2003;4:97–104. doi: 10.1023/a:1023399820770. [DOI] [PubMed] [Google Scholar]
- 20.Singh NP, Ogburn CE, Wolf NS, van Belle G, Martin GM. DNA double-strand breaks in mouse kidney cells with age. Biogerontology. 2001;2:261–270. doi: 10.1023/a:1013262327193. [DOI] [PubMed] [Google Scholar]
- 21.Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 22.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc. Natl Acad. Sci. USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner AM, Turker MS. Oxidative mutagenesis, mismatch repair, and aging. Sci. Aging Knowledge Environ. 2005;2005:re3. doi: 10.1126/sageke.2005.9.re3. [DOI] [PubMed] [Google Scholar]
- 25.Karran P. Microsatellite instability and DNA mismatch repair in human cancer. Semin. Cancer Biol. 1996;7:15–24. doi: 10.1006/scbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 26.Ben Yehuda A, Globerson A, Krichevsky S, Bar On H, Kidron M, Friedlander Y, Friedman G, Ben Yehuda D. Ageing and the mismatch repair system. Mech. Ageing Dev. 2000;121:173–179. doi: 10.1016/s0047-6374(00)00208-6. [DOI] [PubMed] [Google Scholar]
- 27.Neri S, Gardini A, Facchini A, Olivieri F, Franceschi C, Ravaglia G, Mariani E. Mismatch repair system and aging: microsatellite instability in peripheral blood cells from differently aged participants. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:285–292. doi: 10.1093/gerona/60.3.285. [DOI] [PubMed] [Google Scholar]
- 28.Krichevsky S, Pawelec G, Gural A, Effros RB, Globerson A, Yehuda DB, Yehuda AB. Age related microsatellite instability in T cells from healthy individuals. Exp. Gerontol. 2004;39:507–515. doi: 10.1016/j.exger.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Annett K, Duggan O, Freeburn R, Hyland P, Pawelec G, Barnett Y. An investigation of DNA mismatch repair capacity under normal culture conditions and under conditions of supra-physiological challenge in human CD4+T cell clones from donors of different ages. Exp. Gerontol. 2005;40:976–981. doi: 10.1016/j.exger.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Wilson DM, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2006 doi: 10.1016/j.dnarep.2006.10.017. (Amst). [DOI] [PubMed] [Google Scholar]
- 31.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl Acad. Sci. USA. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol. Aging. 2006;27:1129–1136. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen D, Cao G, Hastings T, Feng Y, Pei W, O’Horo C, Chen J. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J. Neurochem. 2002;81:1273–1284. doi: 10.1046/j.1471-4159.2002.00916.x. [DOI] [PubMed] [Google Scholar]
- 35.Shen GP, Galick H, Inoue M, Wallace SS. Decline of nuclear and mitochondrial oxidative base excision repair activity in late passage human diploid fibroblasts. DNA Repair (Amst) 2003;2:673–693. doi: 10.1016/s1568-7864(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 36.Intano GW, Cho EJ, McMahan CA, Walter CA. Age-related base excision repair activity in mouse brain and liver nuclear extracts. J. Gerontol. A Biol. Sci. Med. Sci. 2003;58:205–211. doi: 10.1093/gerona/58.3.b205. [DOI] [PubMed] [Google Scholar]
- 37.Intano GW, McMahan CA, McCarrey JR, Walter RB, McKenna AE, Matsumoto Y, MacInnes MA, Chen DJ, Walter CA. Base excision repair is limited by different proteins in male germ cell nuclear extracts prepared from young and old mice. Mol. Cell. Biol. 2002;22:2410–2418. doi: 10.1128/MCB.22.7.2410-2418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishna TH, Mahipal S, Sudhakar A, Sugimoto H, Kalluri R, Rao KS. Reduced DNA gap repair in aging rat neuronal extracts and its restoration by DNA polymerase beta and DNA-ligase. J. Neurochem. 2005;92:818–823. doi: 10.1111/j.1471-4159.2004.02923.x. [DOI] [PubMed] [Google Scholar]
- 39.Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR. Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat. Res. 2002;500:135–145. doi: 10.1016/s0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabelof DC, Raffoul JJ, Ge Y, Van Remmen H, Matherly LH, Heydari AR. Age-related loss of the DNA repair response following exposure to oxidative stress. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:427–434. doi: 10.1093/gerona/61.5.427. [DOI] [PubMed] [Google Scholar]
- 41.Szczesny B, Bhakat KK, Mitra S, Boldogh I. Age-dependent modulation of DNA repair enzymes by covalent modification and subcellular distribution. Mech. Ageing Dev. 2004;125:755–765. doi: 10.1016/j.mad.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 42.Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc. Natl Acad. Sci. USA. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 44.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 45.Hart RW, Setlow RB. DNA repair in late-passage human cells. Mech. Ageing Dev. 1976;5:67–77. doi: 10.1016/0047-6374(76)90009-9. [DOI] [PubMed] [Google Scholar]
- 46.Painter RB, Clarkson JM, Young BR. Letter: ultraviolet-induced repair replication in aging diploid human cells (WI-38) Radiat. Res. 1973;56:560–564. [PubMed] [Google Scholar]
- 47.Christiansen M, Stevnsner T, Bohr VA, Clark BF, Rattan SI. Gene-specific DNA repair of pyrimidine dimers does not decline during cellular aging in vitro. Exp. Cell Res. 2000;256:308–314. doi: 10.1006/excr.2000.4826. [DOI] [PubMed] [Google Scholar]
- 48.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 49.Boyle J, Kill IR, Parris CN. Heterogeneity of dimer excision in young and senescent human dermal fibroblasts. Aging Cell. 2005;4:247–255. doi: 10.1111/j.1474-9726.2005.00167.x. [DOI] [PubMed] [Google Scholar]
- 50.Al-Baker EA, Oshin M, Hutchison CJ, Kill IR. Analysis of UV-induced damage and repair in young and senescent human dermal fibroblasts using the comet assay. Mech. Ageing Dev. 2005;126:664–672. doi: 10.1016/j.mad.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Annett K, Hyland P, Duggan O, Barnett C, Barnett Y. An investigation of DNA excision repair capacity in human CD4+ T cell clones as a function of age in vitro. Exp. Gerontol. 2004;39:491–498. doi: 10.1016/j.exger.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 52.Vijg J, Mullaart E, Lohman PH, Knook DL. UV-induced unscheduled DNA synthesis in fibroblasts of aging inbred rats. Mutat. Res. 1985;146:197–204. doi: 10.1016/0167-8817(85)90011-2. [DOI] [PubMed] [Google Scholar]
- 53.Moriwaki S-I, Ray S, Tarone RE, Kraemer KH, Grossman L. The effect of donor age on the processing of UV-damaged DNA by cultured human cells: reduced DNA repair capacity and increased mutability. Mutat. Res. 1996;364:117–123. doi: 10.1016/0921-8777(96)00029-8. [DOI] [PubMed] [Google Scholar]
- 54.Grossman L, Wei Q. DNA repair and epidemiology of basal cell carcinoma. Clin. Chem. 1995;41:1854–1863. [PubMed] [Google Scholar]
- 55.Wei Q, Matanoski GM, Farmer ER, Hedayati MA, Grossman L. DNA repair and aging in basal cell carcinoma: a molecular epidemiology study. Proc. Natl Acad. Sci. USA. 1993;90:1614–1618. doi: 10.1073/pnas.90.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Z, Heydari A, Richardson A. Nucleotide excision repair of actively transcribed versus nontranscribed DNA in rat hepatocytes: effect of age and dietary restriction. Exp. Cell Res. 1998;245:228–238. doi: 10.1006/excr.1998.4269. [DOI] [PubMed] [Google Scholar]
- 57.Kruk PA, Rampino NJ, Bohr VA. DNA damage and repair in telomeres: relation to aging. Proc. Natl Acad. Sci. USA. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem. Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 59.Goukassian D, Gad F, Yaar M, Eller MS, Nehal US, Gilchrest BA. Mechanisms and implications of the age-associated decrease in DNA repair capacity. FASEB J. 2000;14:1325–1334. doi: 10.1096/fj.14.10.1325. [DOI] [PubMed] [Google Scholar]
- 60.Xu G, Spivak G, Mitchell DL, Mori T, McCarrey JR, McMahan CA, Walter RB, Hanawalt PC, Walter CA. Nucleotide excision repair activity varies among murine spermatogenic cell types. Biol. Reprod. 2005;73:123–130. doi: 10.1095/biolreprod.104.039123. [DOI] [PubMed] [Google Scholar]
- 61.Yamada M, Udono MU, Hori M, Hirose R, Sato S, Mori T, Nikaido O. Aged human skin removes UVB-induced pyrimidine dimers from the epidermis more slowly than younger adult skin in vivo. Arch. Dermatol. Res. 2006;297:294–302. doi: 10.1007/s00403-005-0618-0. [DOI] [PubMed] [Google Scholar]
- 62.Xu G, Snellman E, Bykov VJ, Jansen CT, Hemminki K. Effect of age on the formation and repair of UV photoproducts in human skin in situ. Mutat. Res. 2000;459:195–202. doi: 10.1016/s0921-8777(99)00069-5. [DOI] [PubMed] [Google Scholar]
- 63.Hemminki K, Snellman E. How fast are UV-dimers repaired in human skin DNA in situ? J. Invest. Dermatol. 2002;119:699. doi: 10.1046/j.1523-1747.2002.00289.x. discussion 700–692. [DOI] [PubMed] [Google Scholar]
- 64.Goukassian DA, Bagheri S, el-Keeb L, Eller MS, Gilchrest BA. DNA oligonucleotide treatment corrects the age-associated decline in DNA repair capacity. FASEB J. 2002;16:754–756. doi: 10.1096/fj.01-0829fje. [DOI] [PubMed] [Google Scholar]
- 65.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 66.van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, et al. Impaired genome maintenance suppresses the growth hormone—insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van de Ven M, Andressoo JO, Holcomb VB, von Lindern M, Jong WM, De Zeeuw CI, Suh Y, Hasty P, Hoeijmakers JH, et al. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2006;2:e192. doi: 10.1371/journal.pgen.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 69.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 70.Lieber MR. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 71.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 72.Gorbunova V, Levy AA. Nonhomologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sargent RG, Branneman MA, Wilson JH. Repair of site-specific double-strand breaks in mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DePinho RA. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- 76.Fuscoe JC, Zimmerman LJ, Harrington-Brock K, Moore MM. Deletion mutations in the hprt gene of T-lymphocytes as a biomarker for genomic rearrangements important in human cancers. Carcinogenesis. 1994;15:1463–1466. doi: 10.1093/carcin/15.7.1463. [DOI] [PubMed] [Google Scholar]
- 77.Mayer PJ, Lange CS, Bradley MO, Nichols WW. Age-dependent decline in rejoining of X-ray-induced DNA double-strand breaks in normal human lymphocytes. Mutat. Res. 1989;219:95–100. doi: 10.1016/0921-8734(89)90019-2. [DOI] [PubMed] [Google Scholar]
- 78.Singh NP, Danner DB, Tice RR, Brant L, Schneider EL. DNA damage and repair with age in individual human lymphocytes. Mutat. Res. 1990;237:123–130. doi: 10.1016/0921-8734(90)90018-m. [DOI] [PubMed] [Google Scholar]
- 79.Ren K, de Ortiz SP. Non-homologous DNA end joining in the mature rat brain. J. Neurochem. 2002;80:949–959. doi: 10.1046/j.0022-3042.2002.00776.x. [DOI] [PubMed] [Google Scholar]
- 80.Vyjayanti VN, Rao KS. DNA double strand break repair in brain: reduced NHEJ activity in aging rat neurons. Neurosci. Lett. 2006;393:18–22. doi: 10.1016/j.neulet.2005.09.053. [DOI] [PubMed] [Google Scholar]
- 81.Shackelford DA. DNA end joining activity is reduced in Alzheimer's disease. Neurobiol. Aging. 2006;27:596–605. doi: 10.1016/j.neurobiolaging.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 82.Um JH, Kim SJ, Kim DW, Ha MY, Jang JH, Chung BS, Kang CD, Kim SH. Tissue-specific changes of DNA repair protein Ku and mtHSP70 in aging rats and their retardation by caloric restriction. Mech. Ageing Dev. 2003;124:967–975. doi: 10.1016/s0047-6374(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 83.Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, Ham YH, Kim TJ, Choi HM, Han GJ, et al. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp. Mol. Med. 2006;38:686–693. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- 84.Seluanov A, Danek J, Hause N, Gorbunova V. Changes in the level and distribution of Ku proteins during cellular senescence. DNA Repair. 2007 doi: 10.1016/j.dnarep.2007.06.010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 86.Doria G, Barattini P, Scarpaci S, Puel A, Guidi L, Frasca D. Role of immune responsiveness and DNA repair capacity genes in ageing. Ageing Res. Rev. 2004;3:143–151. doi: 10.1016/j.arr.2003.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Frasca D, Barattini P, Tirindelli D, Guidi L, Bartoloni C, Errani A, Costanzo M, Tricerri A, Pierelli L, et al. Effect of age on DNA binding of the ku protein in irradiated human peripheral blood mononuclear cells (PBMC) Exp. Gerontol. 1999;34:645–658. doi: 10.1016/s0531-5565(99)00026-1. [DOI] [PubMed] [Google Scholar]
- 88.Gorbunova V, Seluanov A. Making ends meet in old age: DSB repair and aging. Mech. Ageing Dev. 2005;126:621–628. doi: 10.1016/j.mad.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 89.Preston CR, Flores C, Engels WR. Age-dependent usage of double-strand-break repair pathways. Curr. Biol. 2006;16:2009–2015. doi: 10.1016/j.cub.2006.08.058. [DOI] [PubMed] [Google Scholar]
- 90.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 93.Stark JM, Jasin M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yamamoto A, Taki T, Yagi H, Habu T, Yoshida K, Yoshimura Y, Yamamoto K, Matsushiro A, Nishimune Y, et al. Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol. Gen. Genet. 1996;251:1–12. doi: 10.1007/BF02174338. [DOI] [PubMed] [Google Scholar]
- 95.Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat. Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- 96.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 97.Arbones ML, Austin HA, Capon DJ, Greenburg G. Gene targeting in normal somatic cells: inactivation of the interferon-gamma receptor in myoblasts. Nat. Genet. 1994;6:90–97. doi: 10.1038/ng0194-90. [DOI] [PubMed] [Google Scholar]
- 98.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl Acad. Sci. USA. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 100.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 101.Shao C, Stambrook PJ, Tischfield JA. Mitotic recombination is suppressed by chromosomal divergence in hybrids of distantly related mouse strains. Nat. Genet. 2001;28:169–172. doi: 10.1038/88897. [DOI] [PubMed] [Google Scholar]
- 102.Lasko D, Cavenee W, Nordenskjold M. Loss of constitutional heterozygosity in human cancer. Annu. Rev. Genet. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 103.Grist SA, McCarron M, Kutlaca A, Turner DR, Morley AA. In vivo human somatic mutation: frequency and spectrum with age. Mutat. Res. 1992;266:189–196. doi: 10.1016/0027-5107(92)90186-6. [DOI] [PubMed] [Google Scholar]
- 104.Suh Y, Lee KA, Kim WH, Han BG, Vijg J, Park SC. Aging alters the apoptotic response to genotoxic stress. Nat. Med. 2002;8:3–4. doi: 10.1038/nm0102-3. [DOI] [PubMed] [Google Scholar]
- 105.Seluanov A, Gorbunova V, Falcovitz A, Sigal A, Milyavsky M, Zurer I, Shohat G, Goldfinger N, Rotter V. Change of the death pathway in senescent human fibroblasts in response to DNA damage is caused by an inability to stabilize p53. Mol. Cell. Biol. 2001;21:1552–1564. doi: 10.1128/MCB.21.5.1552-1564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, et al. Impaired genome maintenance suppresses the growth hormone–insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2006;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 108.Andressoo JO, Hoeijmakers JH, Mitchell JR. Nucleotide excision repair disorders and the balance between cancer and aging. Cell Cycle. 2006;5:2886–2888. doi: 10.4161/cc.5.24.3565. [DOI] [PubMed] [Google Scholar]
- 109.Gatza C, Moore L, Dumble M, Donehower LA. Tumor suppressor dosage regulates stem cell dynamics during aging. Cell Cycle. 2007;6:52–55. doi: 10.4161/cc.6.1.3667. [DOI] [PubMed] [Google Scholar]
- 110.Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, van der Horst G, Hoeijmakers JH, van Steeg H, et al. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat. Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 111.Frosina G. Overexpression of enzymes that repair endogenous damage to DNA. Eur. J. Biochem. 2000;267:2135–2149. doi: 10.1046/j.1432-1327.2000.01266.x. [DOI] [PubMed] [Google Scholar]