Abstract
The proximal 5′-flanking region of the human platelet-derived growth factor A (PDGF-A) promoter contains one nuclease hypersensitive element (NHE) that is critical for PDGF-A gene transcription. On the basis of circular dichroism (CD) and electrophoretic mobility shift assay (EMSA), we have shown that the guanine-rich (G-rich) strand of the DNA in this region can form stable intramolecular parallel G-quadruplexes under physiological conditions. A Taq polymerase stop assay has shown that the G-rich strand of the NHE can form two major G-quadruplex structures, which are in dynamic equilibrium and differentially stabilized by three G-quadruplex-interactive drugs. One major parallel G-quadruplex structure of the G-rich strand DNA of NHE was identified by CD and dimethyl sulfate (DMS) footprinting. Surprisingly, CD spectroscopy shows a stable parallel G-quadruplex structure formed within the duplex DNA of the NHE at temperatures up to 100°C. This structure has been characterized by DMS footprinting in the double-stranded DNA of the NHE. In transfection experiments, 10 μM TMPyP4 reduced the activity of the basal promoter of PDGF-A ∼40%, relative to the control. On the basis of these results, we have established that ligand-mediated stabilization of G-quadruplex structures within the PDGF-A NHE can silence PDGF-A expression.
INTRODUCTION
Platelet-derived growth factor (PDGF), which was originally isolated from human platelets, is composed of four polypeptide chains designated A, B, C and D. These polypeptide chains make up five biologically active dimeric PDGF isoforms: PDGF-AA, -AB, -BB, -CC and -DD, which exert their effects through two tyrosine kinase receptors, PDGFR-α and PDGFR-β (1–3). PDGF-A and PDGF-B are considered to be major mitogens for a variety of mesenchymal cell types. They play critical roles in normal embryonic development, cellular differentiation and wound healing (1–4). The overactivity of the PDGFs has been implicated in the pathogenesis of a number of diseases characterized by excessive cell growth, including cancer, atherosclerosis and various fibrotic conditions (3,5,6). Significantly augmented expression of the PDGF-A gene has been observed in a number of human tumor cell lines and tissues, particularly in gliomas, sarcomas and ostoastrocytomas (6–10). Abnormal PDGF-A expression is linked to accelerated malignant cell growth through autocrine and paracrine pathways (1,6,11). For these reasons, PDGF signaling pathways are important targets for anticancer therapeutics; specifically, inhibitors of PDGF-A gene expression could be valuable in the treatment of some cancers.
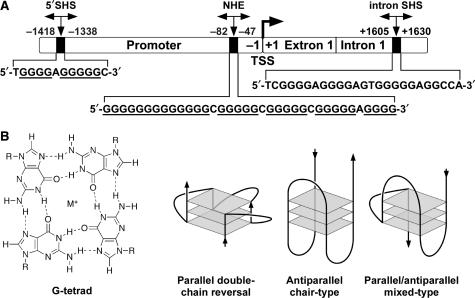
Transcription of the PDGF-A gene is controlled by several positive and negative regulatory elements located in the promoter region and the 5′-distal upstream region of the gene (Figure 1) (12–14). An important, highly guanine- and cytosine-rich region located at nucleotides –120 to –33 within the PDGF-A promoter accounts for 80% of basal promoter activity. This GC-rich region was originally identified by its sensitivity to the single-strand-specific endonuclease S1 nuclease and has been termed the nuclease hypersensitive element (NHE) (12–14). PDGF-A is also subject to transcriptional repression by two discrete S1 nuclease hypersensitive (SHS) elements. One is located within the 5′-distal region (–1418 to –1338) of the promoter (5′SHS), while the other is located within the first intron (+1605 to +1630, intron SHS) (14–17). Multiple transcription factors bind to and regulate PDGF-A genes through the NHE region, such as Sp1, Egr-1, Wilms’ tumor protein (WT1), GC factor 2 (GCF2), nuclear factor 1-X (NF1-X) and the metastasis suppressor protein NM23 (17–23). This region is structurally dynamic and capable of adopting non-B-DNA conformations (24). The importance of the unwound paranemic structure of NHE in PDGF-A promoter activity was confirmed by demonstrating that a single-stranded DNA oligomer, derived from the guanine-rich strand (–74 to –51), inhibits the promoter activity of PDGF-A (24). It is believed that the equilibrium between the B-form DNA, the single-stranded DNA, and intramolecular secondary DNA structures is instrumental in the determination of the binding of transcription factors to the NHE region of PDGF-A (NHEPDGF-A). However, the molecular details of the unwinding of secondary structures in the NHEPDGF-A are poorly understood.
Figure 1.
(A) Promoter structure of PDGF-A and locations of the important NHEs. The sequences of the 5′SHS, the G-rich strand of the NHE and the intron SHS are shown. TSS = transcriptional start site. (B) Structure of a G-tetrad and examples of the folding patterns of known intramolecular G-quadruplexes.
Tandem repeats of the guanine-tract (G-tract) sequence adopt specific intramolecular G-quadruplex structures in the telomeres and promoters of several important genes, such as c-MYC, KRAS, Bcl-2, c-Kit and VEGF (25–29). The basic building block, the G-tetrad, and examples of folding patterns are shown in Figure 1B. The involvement of G-quadruplex structures in cellular processes, such as transcriptional control of c-MYC expression, has stimulated the development of anticancer drugs that are selective for G-quadruplexes. The G-rich strand of NHEPDGF-A contains five contiguous guanine runs, each separated by one base. The G-tract motif in NHEPDGF-A has some similarity to the G-tract motif in the NHE region of c-MYC (NHEc-MYC). However, in NHEc-MYC the minimum number of guanines in any run is three rather than four, and NHEPDGF-A contains a very long run of 13 contiguous guanines at the 5′-end (see sequences in Figure 2A) (25,30). Since the G-rich strand of NHEc-MYC is known to form G-quadruplex structures, by analogy the G-rich strand of NHEPDGF-A has the potential to form G-quadruplex structures under physiological conditions. Consequently, these G-quadruplex structures may play an important role in modulating the expression of PDGF-A. For these reasons, this work examines the in vitro formation and properties of G-quadruplex structures within the G-rich strand of NHEPDGF-A, including its transcriptional modulation by TMPyP4.
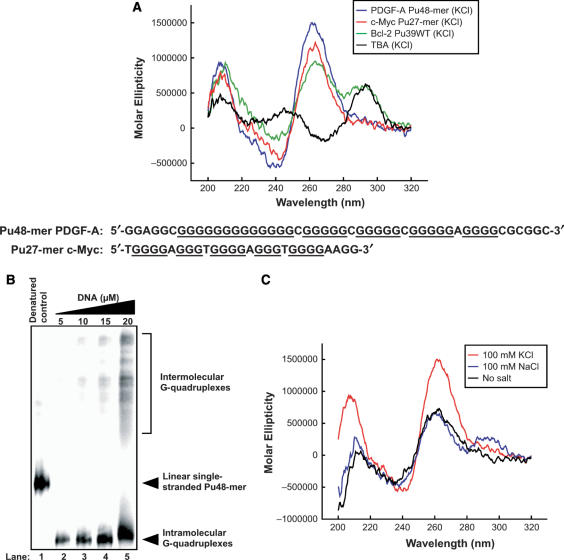
Figure 2.
(A) Comparative CD spectra of three known G-quadruplex-forming sequences with the PDGF-A Pu48-mer. Blue line = PDGF-A Pu48-mer (25 mM KCl), red line = c-Myc Pu27-mer (parallel G-quadruplex in 25 mM KCl), green line = Bcl-2 Pu39WT (mixed parallel/antiparallel G-quadruplex in 100 mM KCl), black line = TBA (antiparallel G-quadruplex in 100 mM KCl). All CD data were obtained with a 5 μM strand concentration at 25°C. Comparison of sequences of these two DNA oligomers is shown under the CD spectra. (B) Native gel electrophoresis indicating the formation of intramolecular G-quadruplex structures by Pu48-mer. The fast mobility major band represents the intramolecular G-quadruplex and the slow mobility bands represent the higher-order intermolecular G-quadruplexes. (C) Effect of alkali metals on the Pu48-mer CD spectra. Effect of KCl (red line) and NaCl (blue line) on the ellipticity signal compared to the signal in the absence of salt (black line). All CD data were obtained with a 5 μM strand concentration at 25°C.
MATERIALS AND METHODS
Oligodeoxynucleotides, enzymes, materials and drugs
DNA oligomers (sequences shown in Table 1) were obtained from Sigma Genosys and Biosearch Technologies, Inc. All DNA oligomers were PAGE-purified and dissolved in double-distilled water before use. The extinction coefficients that the manufacturers provided for a given DNA sequence were calculated using the nearest-neighbor method (31). When the DNA oligomers fold into a G-quadruplex, the base stacking results in hyperchromicity. Thus, the extinction coefficients of DNA oligomers that can form DNA secondary structures need to be corrected for the hyperchromicity. To address this issue, we used UV/vis thermal denaturation techniques to determine the high-order structural hyperchromicity of the G-quadruplex-forming DNA oligomers (32). The UV experiments were carried out on a Cary 300 Bio UV-visible spectrophotometer. Cells with optical path lengths of 10 mm were used, and the temperature of the cell holder was controlled by an external circulating water bath (Varian). The UV melting experiments were done by collecting spectra every five degrees as the sample heated up from 20 to 100°C, and monitoring the absorption change at λmax. The plateau at the top of the melting curve (usually 20°C above the respective Tm) was extrapolated back to 20°C to obtain the hyperchromicity. The extinction coefficient for the G-quadruplex-forming DNA oligomer was corrected by using the following equation:  260(Q) =
260(Q) =  260(N)/(1 + (%hyperchromicity/100)).
260(N)/(1 + (%hyperchromicity/100)).  260(Q) represents the extinction coefficient for the G-quadruplex-forming DNA oligomer, and
260(Q) represents the extinction coefficient for the G-quadruplex-forming DNA oligomer, and  260(N) represents the extinction coefficient for the non-structured single-stranded DNA oligomers. The corrected
260(N) represents the extinction coefficient for the non-structured single-stranded DNA oligomers. The corrected  260(Q) values for select DNA oligomers in our study are as follows: PDGF-A Pu48, 533680 M−1 cm−1; PDGF-A Pu60, 672353 M−1 cm−1; PDGF-A Pu90, 1000303 M−1 cm−1; PA-5W, 379701 M−1 cm−1; PA-Mut1, 360524 M−1 cm−1; PA-Mut2, 369612 M−1 cm−1; c-MYC Pu27, 354938 M−1 cm−1; TBA, 151770 M−1 cm−1 and Bcl-2 Pu38WT, 453766 M−1 cm−1. Acrylamide:bisacrylamide (29:1) solution and ammonium persulfate were purchased from Bio-Rad, and N,N,N′,N′-tetramethylethylenediamine was purchased from Fisher. T4 polynucleotide kinase and Taq DNA polymerase were purchased from Promega. [γ-32P]ATP was purchased from NEN Dupont. TMPyP2, TMPyP4 and Se2SAP were synthesized in our laboratory. Telomestatin was kindly provided by Dr Kazuo Shin-ya (University of Tokyo, Japan). Stock solutions of telomestatin and Se2SAP (10 mM) were made using DMSO (10%) and double-distilled water, respectively. Further dilutions to working concentrations were made with double-distilled water immediately prior to use. Plasmid pGL3-basic, pRL-TK and dual luciferase assay kits were purchased from Promega.
260(Q) values for select DNA oligomers in our study are as follows: PDGF-A Pu48, 533680 M−1 cm−1; PDGF-A Pu60, 672353 M−1 cm−1; PDGF-A Pu90, 1000303 M−1 cm−1; PA-5W, 379701 M−1 cm−1; PA-Mut1, 360524 M−1 cm−1; PA-Mut2, 369612 M−1 cm−1; c-MYC Pu27, 354938 M−1 cm−1; TBA, 151770 M−1 cm−1 and Bcl-2 Pu38WT, 453766 M−1 cm−1. Acrylamide:bisacrylamide (29:1) solution and ammonium persulfate were purchased from Bio-Rad, and N,N,N′,N′-tetramethylethylenediamine was purchased from Fisher. T4 polynucleotide kinase and Taq DNA polymerase were purchased from Promega. [γ-32P]ATP was purchased from NEN Dupont. TMPyP2, TMPyP4 and Se2SAP were synthesized in our laboratory. Telomestatin was kindly provided by Dr Kazuo Shin-ya (University of Tokyo, Japan). Stock solutions of telomestatin and Se2SAP (10 mM) were made using DMSO (10%) and double-distilled water, respectively. Further dilutions to working concentrations were made with double-distilled water immediately prior to use. Plasmid pGL3-basic, pRL-TK and dual luciferase assay kits were purchased from Promega.
Table 1.
Oligodeoxynucleotides used in this study
| DNA oligomer | Sequence (5′ → 3′) |
|---|---|
| PDGF-A Pu48 | GGAGGC GGGGG GGGGG GGGCG GGGGC GGGGG CGGGG GAGGG GCGCG GC |
| PDGF-A Pu60 | CCGGG GAGGC GGGGG GGGGG GGGCG GGGGC GGGGG CGGGG GAGGG GCGCG GCGGC GGCGG |
| PDGF-A Py60 | CCGCC GCCGC CGCGC CCCTC CCCCG CCCCC GCCCC CGCCC CCCCC CCCCC GCCTC CCCGG |
| PDGF-A Pu90 | TGCGG GTCCC AGGCC CGGAA TCCGG GGAGG CGGGG GGGGG GGGGC GGGGG CGGGG GCGGG GGAGG GGCGC GGCGG CGGCG GCTAT AACCC |
| Templatea | CGGG GAGGC GGGGG GGGGG GGGCG GGGGC GGGGG CGGGG GAGGG GCGCG GTTAG TCAGA CCTCG ACGCA TTTGC TTAGA GTCGA |
| Primerb | TCGAC TCTAA GCAAA TGCGT CGAG |
| c-MYC Pu27 | TGGGG AGGGT GGGGA GGGT GGGG AAGG |
| TBA | GGTT GGTG TGGT TGG |
| Bcl-2 Pu39WT | AGGGG CGGGC GCGGG AGGAA GGGGG CGGGA GCGGGG CTG |
| PA-5W | AGGC GGGGG GGGGG GGGCG GGGGC GGGGG CGG |
| PA-Mut1 | AGGC GGGGG GGGGG GGGCT GGGGC TGGGG CGG |
| PA-Mut2 | AGGC GGGGG GGGGG GGGCA GGGGC AGGGG CGG |
aTemplate used in Taq DNA polymerase stop assay.
bPrimer used in Taq DNA polymerase stop assay.
Circular dichroism spectroscopy
CD spectra were recorded on a Jasco-810 spectropolarimeter (Easton, MD, USA) using a quartz cell of 1 mm optical path length and an instrument scanning speed of 100 nm/min, with a response time of 1 s, over a wavelength range of 200–330 nm. All DNA samples were dissolved and diluted in Tris–HCl buffer (50 mM, pH 7.6), and where appropriate, the samples also contained different concentrations of KCl and/or NaCl. The DNA strand concentrations were 5 μM, and the CD data represent four averaged scans taken at an experimental temperature (25–110°C). All CD spectra are baseline-corrected for signal contributions due to the buffer.
Labeling and purification of oligodeoxyribonucleotides
The DNA oligomers were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase for 1 h at 37°C. The labeling reaction was inactivated by heating the samples at 90°C for 8 min after the addition of 1.5 μl of 0.5 M EDTA. The 5′-end labeled DNA was then purified using a Bio-Spin 6 chromatography column (Bio-Rad). The labeled DNA was further purified by running a 12% denatured polyacrylamide gel that contained 8.0 M urea.
Preparation of intramolecular G-quadruplex structures by EMSA
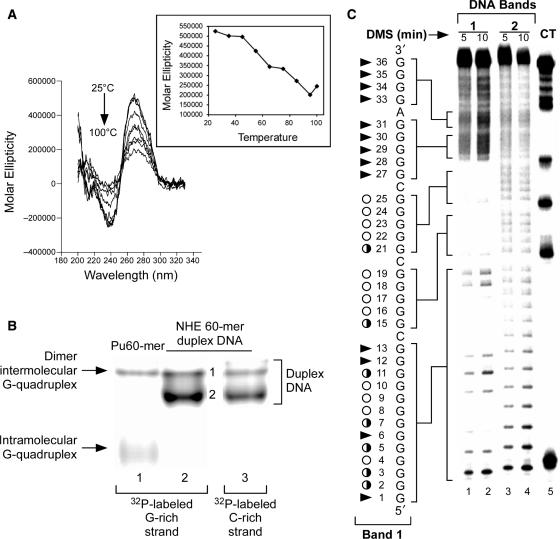
The 5′-end 32P-labeled single-stranded DNA oligomer was incubated in Tris–HCl buffer (50 mM, pH 7.6) containing different concentrations of KCl and/or NaCl for 1 h at room temperature. The denatured marker DNA oligomers were prepared separately before the EMSA. The control DNA oligomers were heated at 95°C for 15 min. They were then immediately put on ice and cooled to 4°C before loading the gel. The secondary structures of the DNA oligomers were obtained by heating the stock solutions at 90°C for 10 min and then slowly cooling to room temperature over 4 h. The monomolecular G-quadruplex structure of the G-rich strand of the NHE was isolated by non-denatured gel electrophoresis in the presence of 10 mM NaCl and KCl in Tris-Borate-EDTA buffer. Electrophoresis of the DNA was conducted at 150 V. The different mobility shift DNA bands were cut from the non-denatured gel and soaked in double-distilled water. The double-stranded 60-mer NHE was obtained first by annealing equal amounts of 60-mer G-rich strand (Pu60-mer, 5′-end 32P-labeled) and C-rich strand (Py60-mer) in annealing buffer (10 mM Tris–HCl, pH 7.5, 10 mM NaCl), heating the mixtures at 75°C for 15 min, and then slowly cooling the DNA sample to room temperature. The major double-stranded DNA band was isolated from a 12% native polyacrylamide gel. For inducing the formation of G-quadruplexes in the duplex DNA, the double-stranded 60-mer NHE was incubated in different concentrations of KCl buffers at room temperature for at least 1 h, then heated at 65°C and allowed to slowly cool to room temperature over 4 h.
Taq polymerase stop assay
The DNA primer 5′-d(TCGACTCTAAGCAAATGCGTCGAG)-3′ was 5′-end-labeled with 32P as described above. The labeled primer was annealed to the DNA template (sequence shown in Table 1) by using the aforementioned protocol. The DNA complex formed by annealing the primer to the template was purified using gel electrophoresis on a 12% native polyacrylamide gel. The purified DNA was then diluted to a concentration of 0.2 nM and mixed with the reaction buffer (10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 1.5 μg/μl BSA) and 0.1 mM dNTPs. Where appropriate, KCl, NaCl and/or drugs were also added. The reaction mixtures were incubated for 1 h at room temperature, allowing the alkali metal ions or drugs to stabilize the G-quadruplexes. Taq DNA polymerase was then added, and samples were incubated for 30 min at 60°C for polymerase extension. The polymerase extension reaction was stopped by adding 2× stop buffer (10 mM EDTA, 10 mM NaOH, 0.1% xylene cyanole, 0.1% bromophenol blue in formamide solution), and the samples were analyzed on a 16% denatured polyacrylamide gel (33).
DMS footprinting
Each band of interest was excised from the EMSA and soaked in 1× Tris-Borate-EDTA buffer containing 10 mM NaCl and KCl. The samples were then vigorously agitated at room temperature for 3 h in water. The solutions were filtered and 50 000 c.p.m. per reaction of DNA solution was further diluted with 0.1× TE (10 mM Tris, 1 mM EDTA, pH 7.5) to a total volume of 90 μl per reaction. Following the addition of 1 μl calf thymus DNA (0.1 μg/μl), the reaction mixture was incubated with 1 μl of 25% dimethyl sulfate solution (DMS:ethanol; 1:4, vol/vol) for 5 or 10 min. Each reaction was stopped by adding 18 μl of stop buffer (3 M β-mercaptoethanol:water:NaOAc; 1:6:7, vol/vol). After ethanol precipitation and washing, the piperidine cleavage reaction was performed by heating the samples to 90°C for 30 min in 30 μl piperidine. The samples were separated on a 20% denatured polyacrylamide gel and visualized on a phosphorimager (Storm 820). DMS analysis of random unstructured control DNA oligomer was performed in the same way by using heat-denatured (quick cooling on ice) 5′-end-labeled oligomer (25).
Imaging and quantitation
The dried gel was exposed on a phosphor screen. Imaging and quantitation were performed using a phosphorimager (Storm 820) and ImageQuant 5.1 software from Amersham Biosciences.
Plasmid construction
The DNA sequence of the –222 to +119 region of the PDGF-A promoter, relative to the transcriptional start site (12,13), was obtained by PCR-amplifying human genome DNA extracted from HPDE-6 cells with the primer pair (5′-GGGGCTTTGATGGATTTAGC-3′ and 5′-CCTGAGGGCGGCGCAAGGCCG-3′). This fragment was subcloned into the multiple cloning sites region, between HindIII and ScaI, of the pGL3-basic vector (Promega), which is a promoter-less plasmid containing the firefly luciferase reporter gene. The resultant clone pA361 was characterized by DNA sequencing, and the correct orientation of the insert was verified by restriction enzyme analysis.
Cell culture, transient transfection and dual luciferase assay
Human pancreatic cancer MIA PaCa-2 cells were cultured in Dulbecco's Modified Eagle's Medium, containing 100 U/ml penicillin, 100 μg/ml streptomycin, 200 mM L-glutamine and 10% FBS (Mediated Inc.). Cells were maintained in a logarithmic phase of growth. For transfection experiments, cells were seeded in 6-well dishes with a concentration of 2 × 105 cells per well and treated with varying concentrations of TMPyP4 or TMPyP2 (up to 50 μM). Each well was co-transfected with 2 μg of pA361 and 25 ng of pRL-TK with TransFectin (BioRad). pRL-TK (Promega), containing a renilla luciferase gene driven by the herpes simplex virus thymidine kinase promoter, was used as a control for transfection efficiency. The expression of firefly luciferase, with respect to that of renilla luciferase, was determined by dual luciferase assay (Promega), 24 h after transfection (as described by the manufacturer). Cell lysate (20 μl) was mixed with 100 μl of reconstituted luciferase assay reagent, and light output was measured for 12 s with a FB12 Luminometer (Berthold).
RESULTS
The G-rich strand of the PDGF-A NHE forms intramolecular G-quadruplex structures
Since signature CD spectra have been determined for several well-defined G-quadruplex structures in solution, comparative CD analysis can provide primary evidence for the existence of G-quadruplex structures in an unknown DNA sequence and also provide insight into the topology of G-quadruplexes (34–44). To determine the potential presence of G-quadruplex structures within the G-rich strand of NHEPDGF-A, we used CD to analyze the purine strand Pu48-mer, which represents the whole core sequence of the G-rich strand of NHEPDGF-A (–40 to –87 in the PDGF-A promoter). Figure 2A (blue line) shows that Pu48-mer exhibited a CD spectrum characterized by a maximum positive ellipticity at 266 nm, a negative band at 240 nm and a minor positive band at 212 nm. This spectrum is similar to that of the guanine-rich DNA oligomer Pu27-mer of NHEc-MYC (red line in Figure 2A), which forms intramolecular parallel G-quadruplex structures that have been well characterized by NMR, CD and DMS footprinting studies (29,38,45). To further identify the topology of the G-quadruplex structures formed in Pu48-mer, we compared the CD spectrum of Pu48-mer to two other DNA sequences, the thrombin binding aptamer (TBA) (black line in Figure 2A) and the Bcl-2 Pu39WT (green line in Figure 2A). The chair-type antiparallel G-quadruplex structure of TBA has been proposed on the basis of NMR and X-ray crystallography (46), and it has a signature CD spectrum characterized by a positive maximum band around 290 nm, a negative band at 265 nm and a smaller positive band at 246 nm. The hybrid parallel/antiparallel G-quadruplex structure of Bcl-2 Pu39WT was solved by NMR (47). The CD spectrum of this hybrid G-quadruplex structure shows a major positive band at ∼ 265 nm with a negative band at 240 nm, which is similar to the parallel G-quadruplex. However, it has a unique pronounced shoulder absorption between 280 and 300 nm, which is not observed in the CD spectrum of the parallel G-quadruplex (27). The CD spectrum of the Pu48-mer is quite different from those of Bcl-2 Pu39WT and TBA, and it resembles most closely that of the c-MYC Pu27-mer. Collectively, these data suggest that the major G-quadruplex structure in the Pu48-mer is a parallel structure.
DNA oligomers folding into intramolecular G-quadruplex structures migrate faster than non-structured single-stranded DNA and the corresponding intermolecular dimers in a native gel (48,49). Thus, it is possible to isolate the intramolecular G-quadruplex structure from the other DNA structures by native gel electrophoresis. To determine whether the G-quadruplexes formed by Pu48-mer in our CD studies are intra- or intermolecular, we analyzed the electrophoretic mobilities of different concentrations of Pu48-mer in the presence of 50 mM KCl. When the DNA concentration was 15 μM or lower (Figure 2B, lanes 2–4), the major DNA species of Pu48-mer migrated in an 8% native polyacrylamide gel much faster than the corresponding denatured Pu48-mer that represents the random linear DNA structure (Figure 2B, lane 1 as control). These results suggest that the Pu48-mer folds into a compact intramolecular G-quadruplex at low DNA concentrations. However, when the DNA concentration was 20 μM, more than 40% of the Pu48-mer was found as a slowly migrating species, which is presumed to represent higher-order intermolecular DNA complexes (Figure 2B, lane 5). Since the concentration of the DNA oligomer used in our CD studies is 5 μM, it is reasonable to assume that the G-quadruplexes formed in these conditions are intramolecular.
K+ facilitates the formation of intramolecular G-quadruplexes in the G-rich strand of NHEPDGF-A
To examine the effects of the alkali metal ions (K+ and Na+) on the formation of G-quadruplexes in the G-rich strand of NHEPDGF-A, we monitored the CD spectra of Pu48-mer in the presence of either 100 mM KCl or 100 mM NaCl. These CD spectra showed similar parallel G-quadruplex characteristics. However, a dramatic increase in the elliptic intensity was observed with the sample that was pre-incubated with 100 mM KCl (Figure 3A, red line), while 100 mM NaCl only slightly increased the magnitude of the CD signal around 280–300 nm (Figure 2C, blue line) as compared to the spectrum of DNA in the Tris–HCl buffer alone (Figure 2C, black line). This indicates that K+ or Na+ can increase the formation of the PDGF-A Pu48-mer G-quadruplex, but a more significant effect occurs with K+. K+ strongly increases the formation of the parallel Pu48-mer G-quadruplex structure, while Na+ appears to induce the formation of a hybrid parallel/antiparallel Pu48-mer G-quadruplex structure. Further evidence that K+ stabilizes the Pu48-mer G-quadruplex structure was obtained by comparing the melting curve of Pu48-mer in the absence or presence of KCl. We monitored the CD spectra of Pu48-mer as a function of temperature and then plotted the ellipticity at 266 nm versus temperature to generate a melting curve. The full melting curve of Pu48-mer in Tris–HCl (50 mM, pH 7.2) showed a Tm of 75°C (data not shown). This high Tm suggests that intramolecular Pu48-mer G-quadruplex structures are very stable. Surprisingly, in the presence of 25 mM KCl, the Pu48-mer still showed a detectable parallel G-quadruplex CD signal at 110°C. Because of the high-temperature limit of our CD instrument, we could not obtain a full melting curve for the Pu48-mer in 25 mM KCl. This result implies that K+ can significantly stabilize the Pu48-mer G-quadruplex and increase its Tm.
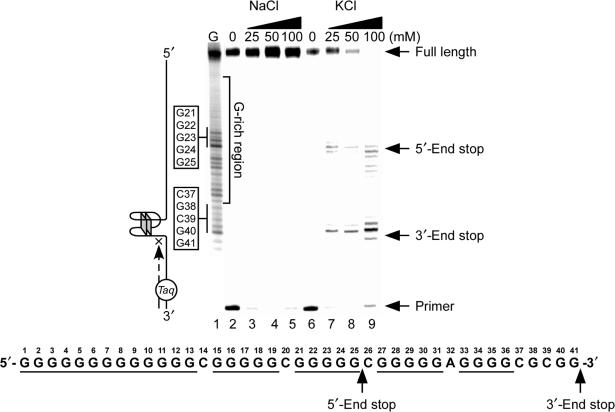
Figure 3.
Effect of K+ and Na+ on the formation of the NHEPDGF-A G-quadruplex in a Taq polymerase stop assay. Two stop products are designated as the 5′-end product and the 3′-end stop product. The corresponding arrest sites are indicated on the core G-tract sequence.
The specific stabilizing effect of K+ on the formation of G-quadruplexes in the NHEPDGF-A was confirmed by Taq polymerase stop assay. This assay is a useful tool to investigate the capability of G-rich DNA sequences to form stable G-quadruplex structures in vitro, based on the assumption that a stable DNA secondary structure in the DNA template is able to stop primer extension by the DNA polymerase (33). A cassette template DNA containing the Pu48-mer of NHEPDGF-A was annealed with a labeled primer, which subsequently was extended with Taq polymerase in the presence of increasing concentrations of KCl or NaCl. A full-length product was synthesized in the presence of NaCl (Figure 3, lanes 3–5); however, upon replacement of NaCl by KCl to the reaction, two major premature primer extension products were produced and designated as the 5′-end stop product and the 3′-end stop product (Figure 3, lanes 7–9). Both of these arrest products increased in a K+-dependent manner but were not affected by Na+. Thus, the K+-specific block to DNA synthesis is not due to general effects of alkali ions. Collectively, these results suggest that the G-rich strand of NHEPDGF-A has the capacity to form two separate intramolecular parallel G-quadruplex structures that are preferentially stabilized by K+.
Formation of multiple G-quadruplex structures in the G-rich strand of NHEPDGF-A
The G-rich strand of NHEPDGF-A is composed of five G-tracts and has the potential to form multiple G-quadruplexes structures, based on the assumption that each G-tract has an equal possibility of participating in the formation of G-quadruplexes. The Taq polymerase stop assay in Figure 3 showed that two significant stop products are formed within the NHEPDGF-A. This indicates that at least two major, stable intramolecular G-quadruplex structures are formed by different G-tracts in the G-rich strand of NHEPDGF-A. The Taq polymerase arrest sites for these two major stop products were determined by sequencing the template DNA. The arrested site for the 3′-end stop product corresponded to G41 in the PDGF-A Pu48-mer (core sequence of NHEPDGF-A, as shown under the gel in Figure 3). The arrest product at G41 suggest that bases G37–G41 must form a stable cap structure on the 3′-end of the G-quadruplex. The arrested site for the 5′-end stop product corresponded to G25 in the Pu48-mer (Figure 3). This indicates that the three consecutive runs of guanines at the 5′-end of Pu48-mer (G1–G25) form the G-quadruplex structure of the 5′-end stop product. One structural characteristic of particular interest is that the 13-mer G-tract must provide the two runs of presumably four guanines at the 5′-end of the G-quadruplex.
Native polyacrylamide gel electrophoresis of Pu48-mer, which was pre-incubated with 100 mM KCl, shows two fast migrating bands (Figure 4A, bands 1 and 2) that move faster than the denatured unstructured linear DNA species (band 3). These high-mobility species are presumed to represent compact intramolecular G-quadruplex structures. The close mobility and broadness of two intramolecular G-quadruplexes (bands 1 and 2) indicate that the Pu48-mer may form multiple similarly structured intramolecular G-quadruplexes.
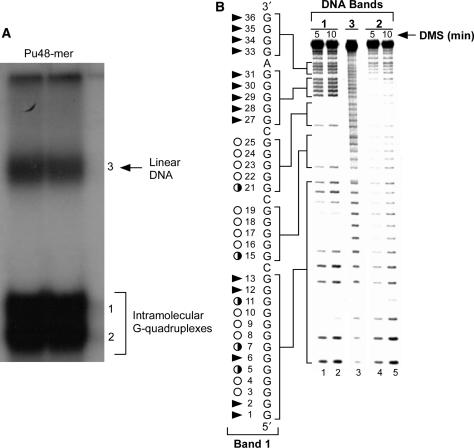
Figure 4.
The EMSA (A) and DMS footprinting (B) of intramolecular G-quadruplex structures of PDGF-A Pu48-mer. In (A), high-mobility bands 1 and 2 represent compact intramolecular G-quadruplex structures, and band 3 represents linear DNA, which was fully denatured by heating. In (B), DMS footprinting of band 1 (lanes 1 and 2), band 2 (lanes 4 and 5) and band 3 (lane 3) from (A) is shown. Open circles indicate the guanines that are fully protected, partially open circles indicate the guanines that are partially protected and arrowheads indicate the guanines that are cleaved.
To further probe the structures of the two intramolecular G-quadruplexes, a DMS footprinting experiment was carried out. Since N7 of guanine in a G-tetrad is involved in Hoogsteen bonding and is inaccessible to alkylation by DMS, we can identify those guanine bases engaged in the formation of a G-quadruplex structure. The DMS methylation pattern produced by band 3 was consistent with a linear single-stranded DNA (Figure 4B, lane 3). For bands 1 and 2 (Figure 4B, lanes 1 and 2 and lanes 3 and 4), the DMS protection patterns suggested the formation of intramolecular G-quadruplexes. As shown in Figure 4B (lanes 1 and 2), full DMS protection occurred within the four runs of guanines (G3–G4, G8–G10, G16–G19 and G22–G25) of band 1. For band 1, the DMS protection of the two four-guanine G-tracts (G16–G19, G22–G25) at the 3′-end of the NHEPDGF-A is clear, while the remaining two G-tracts within the run of 13 guanines at the 5′-end of the NHEPDGF-A are less well defined. Within the long run of 13 guanines there are two runs of guanines that are clearly protected: a run of two guanines (G3–G4) and three guanines (G8–G10). Partial protection of the intervening run of guanines (G5 and G7) occurs between these two fully protected regions, and likewise G11 is partially protected. The first guanine involved in the formation of the G-tetrad at the 3′-end of band 1 is G25. This corresponds to the arrest site for the 5′-end stop product in the Taq polymerase stop assay (Figure 3). For band 2 (lanes 4 and 5), most guanines within the two 5′-runs of guanine (G1–G13 and G15–G19), with the exception of G1 and G2, were partially protected from DMS methylation, with only two runs of guanines (G22–G25 and G27–G30) showed full protection (Figure 4B, lanes 4 and 5). These somewhat ill-defined N7 methylation protection patterns do not provide straightforward insight into the specific G-quadruplex structures for the DNA species in bands 1 and 2. Most probably these ill-defined bands result from mixtures of multiple intramolecular G-quadruplexes existing in dynamic equilibrium (see Discussion Section).
Formation of a G-quadruplex within the double-stranded DNA of NHEPDGF-A
To date, all well-characterized G-quadruplex structures have been determined by studying single-stranded DNA in vitro (25–30,35,36,41,42). To investigate the potential for G-quadruplex formation within the duplex DNA of NHEPDGF-A, we used CD to study a 90 base pair duplex DNA, derived from the –24 to –113 region of the PDGF-A promoter, that contained about 20 base pairs flanking each side of the NHE core sequence. Significantly, the CD spectrum of the NHE 90 bp double-stranded DNA exhibited an almost identical CD spectrum to the parallel G-quadruplex formed in the single-stranded DNA (Figure 2A, blue line), which is characterized by a positive ellipticity maximum at 266 nm and a negative minimum at 240 nm (Figure 5A). This CD spectrum is substantially different from the CD signal of B-form duplex DNA, which typically has an ellipticity maximum absorption around 275–280 nm and zero ellipticity at 258 nm. In addition, the CD bands of B-form DNA are much weaker (34,50,51). This CD profile of the NHE 90 bp double-stranded DNA indicates that a dominant intramolecular parallel G-quadruplex is formed within the duplex DNA of NHEPDGF-A. To determine the stability of this G-quadruplex structure, we monitored the CD spectra of the NHE 90 bp double-stranded DNA over a 25–100°C temperature range. Notably, the DNA sample still generated very strong parallel G-quadruplex CD signals, even above 100°C (Figure 5A). This indicates that the G-quadruplex structure formed in the NHE duplex DNA is very stable.
Figure 5.
(A) Effect of temperature (25–100°C) on the CD spectra of 90 bp duplex DNA containing NHEPDGF-A. At 100°C, 90 bp duplex DNA of NHEPDGF-A still generated a strong G-quadruplex CD signal. (B) Non-denatured gel analysis of 60-mer G-rich single-stranded DNA of NHEPDGF-A (lane 1) and 60 bp double-stranded DNA of NHEPDGF-A (lanes 2 and 3). In lanes 1 and 2, the G-rich strand was 5′-end-radiolabeled with 32P, and the C-rich strand was 5′-end-radiolabeled with 32P in lane 3. (C) DMS footprinting for G-rich strands of 60 bp duplex DNA band 1 (lanes 1 and 2) and band 2 (lanes 3 and 4). Lane 5 shows the CT sequencing on the G-rich strand of the 60 bp duplex DNA of NHEPDGF-A. Open circles indicate the guanines that are fully protected, partially open circles indicate the guanines that are partially protected, and arrowheads indicate the guanines that are cleaved.
To identify the G-quadruplex structure formed in the NHE duplex DNA, a 60 bp double-stranded DNA of NHE (–33 to –92 in the PDGF-A promoter), which was shown to have better resolution for DMS footprinting than the 90 bp double-stranded DNA, was used. The CD spectrum of purified NHE 60-mer double-stranded DNA was consistent with that of the NHE 90 bp double-stranded DNA (data not shown). EMSA was performed to isolate the duplex DNA species containing the G-quadruplex structure, which was 5′-end 32P-labeled in the G-rich and C-rich strands. Two bands (Figure 5B, bands 1 and 2) were observed in both duplex DNA samples (lanes 2 and 3) pre-incubated with 100 mM KCl. These bands were duplex DNA species, since they both migrated differently to the single-stranded intramolecular G-quadruplex species. The duplex DNA species containing the partially unwound region was less compact than the linear duplex DNA and as expected showed slower migration in EMSA. Thus band 1 corresponds to the duplex DNA species containing the G-quadruplex structure, while band 2 corresponds to the fully linear duplex DNA. By coincidence, this band migrates at the same point as the presumed dimer intermolecular species from the Pu60-mer. However, it cannot correspond to this species since the NHE 60 bp duplex DNA labeled on the C-rich strand also gives this species. Bands 1 and 2 were both extracted and subjected to DMS footprinting. The full DMS cleavage pattern of band 2 confirmed that it was a linear duplex DNA (Figure 5C, lanes 3 and 4). A distinct DMS protection pattern among three runs of guanines (G3–G11, G16–G19 and G22–G25) was observed for the G-rich strand of band 1 (Figure 5C, lanes 1 and 2). Although it is not quite as well defined as the DMS footprinting patterns of the G-quadruplex species obtained from single-stranded DNA (see Figure 4B, lanes 1 and 2), the DMS footprinting pattern of band 1 corresponds very closely to that of the single-stranded species associated with the 5′-end product seen in the Taq polymerase stop assay.
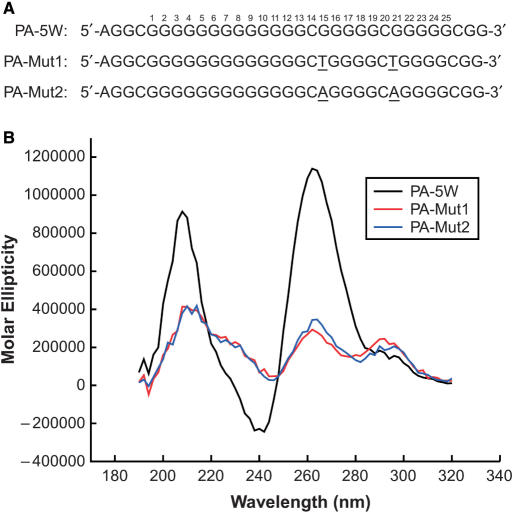
Mutation of guanines in the loops leads to dramatic destabilization of the PDGF-A G-quadruplexes
A full mutational analysis of the three consecutive 5′-runs of guanines provided further insight into the sequence requirements for stability of the NHEPDGF-A G-quadruplexes. In accordance with the DMS footprinting studies on the predicted mixture of loop isomers (see Discussion Section), 12 sets of mutants, in which the individual loop isomers were selected for, were compared by the relative intensity of the CD at 285 nM (data not shown). In all cases, the mutant sequences that were predicted to form defined loop isomers showed lower stability. Surprisingly, all these guanine mutant isomers showed a significant decrease in parallel G-quadruplex signals and an increase in antiparallel signals. This shows that these mutants predominantly form antiparallel or hybrid parallel/antiparallel G-quadruplex structures, which is different from the biologically relevant NHEPDGF-A G-quadruplex. Among these guanine mutant isomers, the most surprising results originated from two dual mutants, PA-Mut1 and PA-Mut2 (Figure 6A). These two mutant oligomers were designed to provide dual G-to-T or G-to-A base mutations at G15 and G21 in the core sequence of NHEPDGF-A. PA-Mut1 and PA-Mut2 were chosen because of the identified cleavage sites shown in the DMS footprinting of the PDGF-A Pu48 and the 60 bp duplex of NHEPDGF-A (Figures 4B and 5C). Therefore, G15 and G21 are not required for G-tetrad formation, and it was predicted that a mutation at these positions would not affect the stability or folding of the G-quadruplex in the NHEPDGF-A. Unexpectedly, PA-Mut1 and PA-Mut2 both showed a significant decrease in ellipticity at 212 and 266 nm and an increase in the antiparallel shoulder absorption between 280 and 300 nm, compared with the wild-type sequence of the three consecutive 5′-runs of guanines of NHEPDGF-A (PA-5W) (Figure 6B). Substitution of other loop-position guanines with other bases also resulted in the same changes in CD.
Figure 6.
Sequences (A) and comparative CD spectra (B) of the wild-type sequence of the three 5′-end runs of guanines and the G15/G21 double mutation sequences in the presence of 100 mM KCl. Black line = wild-type sequence PA-5W, red line = double G-to-T mutation sequence PA-Mut1 and blue line = double G-to-A mutation sequence PA-Mut2. All CD data were obtained with a 5 μM strand concentration at 25°C.
Stabilization of the G-quadruplexes in the NHEPDGF-A with small molecules
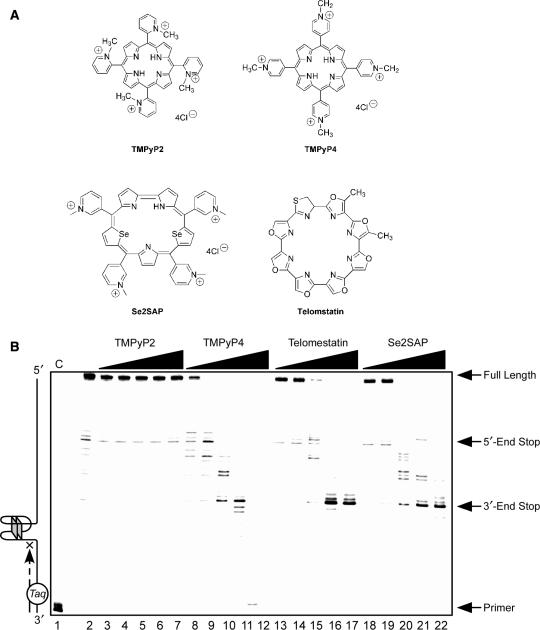
One important goal of our studies is to identify small molecules with the ability to specifically modulate PDGF-A gene expression through stabilization or modification of the biologically relevant G-quadruplex structures in the NHEPDGF-A. To gain further insight into the selective action of G-quadruplex-interactive agents with the NHEPDGF-A sequence, three well-established G-quadruplex-interactive agents, TMPyP4, telomestatin and Se2SAP (Figure 7A), were evaluated by a Taq polymerase stop assay with a DNA template containing the whole core sequence of the G-rich strand of NHEPDGF-A. TMPyP2 (Figure 7A), a positional isomer of TMPyP4 that lacks the ability to insert or stack into the G-tetrad structures (52), served as the negative control compound in our studies. Since there was a considerable amount of 5′-end stop products formed in the absence of drugs and KCl (Figure 7B, lane 2), we used an elevated primer extension temperature (60°C) to partially destabilize the arresting G-quadruplex structure and thereby permit a larger window for drug stabilization of the G-quadruplex structures. The pausing at the 5′-end stop product in the absence of G-quadruplex-interactive ligands and KCl at a high temperature of 60°C is another indication that the G-quadruplex structure in the 5′-end product is more stable than other possible G-quadruplex structures formed by the G-rich strand of NHEPDGF-A (Figure 7B, lane 2). This suggests that the 5′-end G-quadruplex is the primary G-quadruplex structure formed in NHEPDGF-A, corresponding to the one formed in the NHE duplex DNA.
Figure 7.
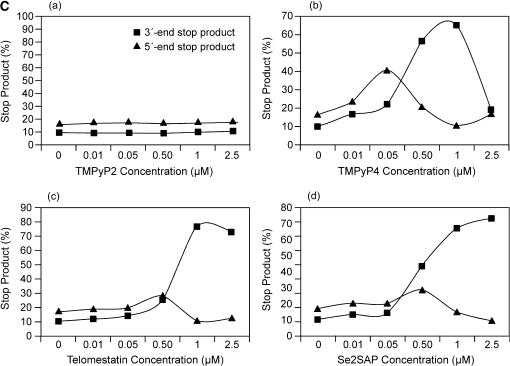
(A) Structures of the G-quadruplex-interactive compounds TMPyP4, telomestatin and Se2SAP, and the control compound TMPyP2. (B) The Taq polymerase stop assay was used to compare the stabilization of the NHEPDGF-A G-quadruplex by TMPyP2 (lanes 3–7), TMPyP4 (lanes 8–12), telomestatin (lanes 13–17) and Se2SAP (lanes 18–22) by using increasing concentrations of drugs (0.01, 0.05, 0.5, 1 and 2.5 μM) at 60°C. Lane 1 is control, and lane 2 is without drug. (C) The ratios of the major arrest products of each sample to the total product were plotted against drug concentrations.
The results in Figure 7B (lanes 3–7) and Figure 7C (a) show that TMPyP2 had a negligible effect on the formation of polymerase stop arrest products. When the concentration of drugs was 0.05 μM and lower (Figure 7B, lanes 8 and 9), TMPyP4 significantly increased stabilization of the 5′-end stop products in a concentration-dependent manner, but telomestatin and Se2SAP only marginally increased the formation of 5′-end stop products (lanes 13, 14, 19 and 20 in Figure 7B). In contrast, when the concentration of all three drugs was 0.5 μM and higher, decreased formation of the 5′-end stop products was observed for all three G-quadruplex-interactive drugs, because of the accumulation of the 3′-end stop product. TMPyP4 achieved the maximum stabilizing effect on the 5′-end stop products at a concentration of 0.05 μM [Figure 7B, lane 9, and C (b)]. The Taq polymerase stop assay also showed that these three G-quadruplex-interactive drugs induced formation of the 3′-end stop products at high drug concentrations (≥0.5 μM). A concentration-dependent increase in stabilization of the 3′-end stop products was also observed for all three drugs. However, telomestatin and Se2SAP were more selective at stabilizing the 3′-end stop product; both achieved the best stabilizing effect on the 3′-end G-quadruplex at a concentration of 1 μM [Figure 7B, lane 21, and C (c and d)]. In addition, at drug concentrations of 1 and 2.5 μM, TMPyP4 exhibited preferential binding for duplex DNA over the G-quadruplex structures (lanes 11 and 12 in Figure 7B).
TMPyP4 selectively inhibits the promoter activity of the NHEPDGF-A
In light of our results demonstrating that the duplex DNA of NHEPDGF-A can form a G-quadruplex structure, we sought to determine if stabilization of this G-quadruplex structure affects PDGF-A transcription in vivo. Therefore, we constructed pGL3-Basic derivative (pA361) in which the human genomic DNA fragment containing the proximal promoter region of PDGF-A (–218 to +142, relative to the transcriptional start site) was inserted upstream of the firefly luciferase reporter gene. When transfected into MIA PaCa-2 human pancreatic cancer cells, pA361 drove luciferase expression into these cells. Thus, the PDGF-A proximal promoter, which contains the complete NHEPDGF-A core sequence, can drive transcription in vivo. Furthermore, RT-PCR studies demonstrated significantly higher PDGF-A expression in MIA PaCa-2 cells than in the normal pancreatic cell line HPDE-6 (data not shown).
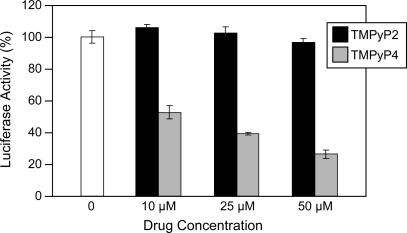
Since TMPyP4 at low concentrations selectively stabilized the biologically relevant G-quadruplex structure (the 5′-end stop product) formed by duplex NHEPDGF-A DNA in vitro, we directly assessed its specific biological effects on PDGF-A transcription. MIA PaCa-2 cells were co-transfected with plasmids pA361 and pRL-TK as an internal control and treated with varying concentrations of TMPyP4 or TMPyP2. We measured the expression of firefly luciferase relative to the renilla luciferase of pRL-TK by dual luciferase assay, 24 h after transfection. At all concentrations used, TMPyP4 reduced the expression of firefly luciferase activity by inhibiting the promoter activity of NHEPDGF-A, whereas TMPyP2 had no significant effect on the firefly luciferase activity (Figure 8). We found that 10 μM TMPyP4 could reduce the expression of firefly luciferase to ∼50% of the control, indicating that the stabilization of the G-quadruplex has a strong repression effect on promoter activity of NHEPDGF-A. This data suggests that the stable G-quadruplex structure formed locally in the human PDGF-A promoter at NHE is likely to behave as a transcription repression in vivo.
Figure 8.
Dual luciferase assay to determine the effect of TMPyP4 and TMPyP2 on the transcriptional activity of PDGF-A basal promoter containing the NHE. The comparative firefly luciferase expressions (firefly/renilla) of TMPyP2 and TMPyP4 are shown in the histograms. The values are the average of three independent experiments. Error bars are ± SE.
DISCUSSION
Up-regulation of PDGF activity has been implicated in the pathogenesis of a number of serious diseases, including cancer, as well as other disorders characterized by excessive cell growth, such as atherosclerosis and various fibrotic conditions (3,5,6), as described in the Introduction Section. One strategy to block PDGF signaling pathways is to target the PDGF receptors. Employment of this strategy led to the discovery of selective PDGF antagonists (5,53,54). For example, the PDGF receptor kinase inhibitor STI-571 (Gleevec) has been approved for clinical use for the treatment of certain tumors (54–56). Because the PDGF-A polypeptide is short in length, it has not been thought of as a druggable anticancer target. However, another viable strategy for targeting PDGF signaling pathways in tumors is to decrease PDGF-A expression by inhibiting its promoter activity. Transcription of PDGF-A is under the strict control of NHEPDGF-A in the promoter. The unwound paranemic NHEPDGF-A structure plays an important role in regulating transcription factors that bind to the PDGF-A promoter. The G-rich strand of NHEPDGF-A, which contains five G-tracts with each G-tract separated from the neighboring G-tract by a single intervening base, is anticipated to form G-quadruplex structures under physiological conditions. Therefore, the NHEPDGF-A G-quadruplex structure is potentially involved in transcriptional control of PDGF-A expression, perhaps as the silencer element. Consequently, the search for compounds that have selectivity for binding to the NHEPDGF-A G-quadruplex and that inhibit PDGF-A gene expression might well be valuable in the treatment of some cancers and other disorders, as described earlier.
In the present study, we demonstrate that the G-rich strand of NHEPDGF-A, located within the proximal promoter region of PDGF-A, is able to form two stable intramolecular parallel G-quadruplex structures (3′-end and 5′-end G-quadruplexes) that are potentially involved in transcriptional regulation. The formation of these G-quadruplex structures is K+-dependent. The DMS footprinting patterns of the G-quadruplexes formed by PDGF-A Pu48 and Pu60 imply that these structures must contain at least four G-tetrads, and this is supported by the CD data in Figure 2A, in which the molar ellipticity is greater for the PDGF-A G-quadruplex than for the c-Myc G-quadruplex. Significantly, an intramolecular G-quadruplex that has a similar folding pattern to the 5′-end G-quadruplex found in single-stranded DNA of NHEPDGF-A can be formed within the duplex DNA of NHEPDGF-A. In light of the protocol for formation of the duplex containing the G-quadruplex, the G-quadruplex obtained within the duplex DNA of NHEPDGF-A probably forms prior to the formation of the duplex species. Thus, the species formed during the annealing is a result of hybridization of the single-stranded G-rich strand already containing the G-quadruplex with the single-stranded C-rich strand. This is unlikely to be the same mechanism for the formation of the G-quadruplex structure within the duplex DNA in in vivo situations where superhelicity is involved (see subsequently). Nevertheless, this is the first time that we have been able to isolate the duplex DNA containing a G-quadruplex structure in vitro. This unique structure is likely to represent the dominant biologically relevant G-quadruplex structure formed in the PDGF-A promoter. It has been proposed that superhelicity of genomic DNA may play an important role in determining the presence of such an interconversion (57,58). The functional analysis of the PDGF-A promoter revealed that the NHEPDGF-A adopted a non-B-DNA conformation and was hypersensitive to the S1 nuclease in the supercoiled plasmids (13–16). This provides further evidence that the formation of a G-quadruplex in the NHEPDGF-A can compete with the superhelical duplex conformation in vivo. The implication of the formation of a stable G-quadruplex in duplex DNA is that the C-rich strand forms a single-stranded DNA loop, which may be in equilibrium with an i-motif structure.
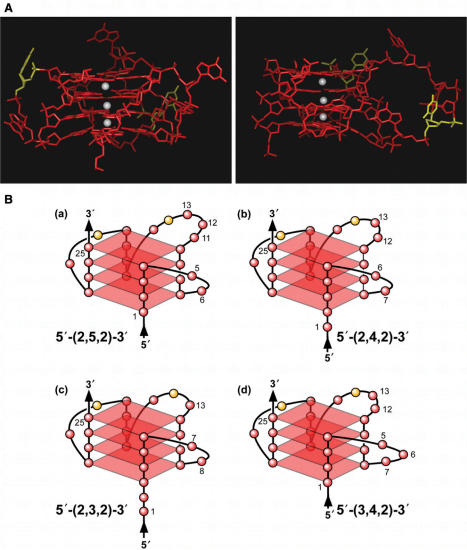
The folding pattern of the biologically relevant G-quadruplex structure formed by the three consecutive G-tracts (G1–G25) at the 5′-end of NHEPDGF-A duplex DNA most likely represents a mixture of different loop isomers. Of particular interest, the 13-mer G-tract (G1–G13) must provide the two runs of guanines at the 5′-end. The flanking regions of NHEPDGF-A are also made up of polypurine and polypyrimidine elements, which may have a significant impact on the propensity to form the G-quadruplex in the G-rich strand of NHEPDGF-A double-stranded DNA. The overall folding patterns of the parallel G-quadruplex structure in NHEPDGF-A duplex DNA has been defined by its characteristic CD spectra, which demonstrates that it has a unique double-chain reversal structure. Previously, examples of G-quadruplexes consisting of double-chain reversals in promoter regions have been confined to three tetrads with a single base in the internal loop. This is the first example of a G-quadruplex with four tetrads and a two-base internal loop. The DMS footprinting pattern of the G-quadruplex in NHEPDGF-A duplex DNA reveals that the 3′-face of this G-quadruplex structure has two runs of four guanines, which are separated by a two-base (CG) internal loop. A molecular model of the four G-tetrads in the NHEPDGF-A G-quadruplex structure shows that a two-base internal loop is the minimum required to bridge two runs of four guanines (Figure 9A). The opposite 5′-face of this G-quadruplex structure is constructed from the run of 13 contiguous guanines, in which the DMS footprinting pattern predicts a mixture of at least four double-chain reversal loop isomers (Figure 9B). Loop isomers ‘a’, ‘b’, and ‘c’ in Figure 9B have a two-base internal loop on the 5′-face dictated by the demonstrated DMS cleavages of G11, G12 and G13 (‘a’ isomer), G12 and G13 (‘b’ isomer) and G1 and G2 (‘c’ isomer) in the core sequence of NHEPDGF-A (see Figure 4B). Finally, the cleavage of G5, G6 and G7 predicts loop isomer ‘d’. By analogy with the c-Myc parallel G-quadruplex structure, in which the double-chain reversal loops on the opposite face are sufficient to stabilize a third intervening loop size of six bases, the double-chain reversals in the NHEPDGF-A G-quadruplex can stabilize a third internal loop of up to five bases (as predicted in loop isomer ‘a’ in Figure 9B) (38). Two views of a molecular model of loop isomer ‘a’ having the predicted folding pattern are shown in Figure 9A.
Figure 9.
(A) Model of the biologically relevant NHEPDGF-A G-quadruplex [loop isomer 5′-(2,5,2)-3′], which contains two 2-base double-chain reversal loops and one 5-base intervening loop (guanines = red, cytosines = yellow and K+ ions = white). For clarity, hydrogen atoms have been removed. In the left panel, the two 2-base double-chain reversal loops are shown on each side of model, and in the right panel, the model has been rotated to show the 5-base intervening loop on the right side of model. (B) Proposed folding patterns of the four different loop isomers formed in the core sequence of NHEPDGF-A. Guanines = red, cytosines = yellow.
A surprising finding illustrated by the CD results shown in Figure 6B for two dual mutant oligomers is that base changes, even in potential loop positions, result in destabilization and apparent dramatic changes in the folding patterns. This is in contrast to the c-Myc G-quadruplex, where elimination of the redundancy in loop isomers by judicial G-to-T mutations does not dramatically change the stability or folding patterns of the parallel-stranded G-quadruplex (38). The effects of G-to-N mutations in the NHEPDGF-A G-quadruplexes are not limited to the run of 13 contiguous guanines, but also occur when the 3′ terminal guanines in the two runs of 5 guanines (G15 and G21) are mutated to thymines or adenines. On the basis of DMS cleavage results, these guanines are predicted to be predominantly in the loops, implying that the dynamic equilibrium between different loop isomers is an important feature of both stability and formation of the parallel-stranded species. This phenomenon may be uniquely linked to the 13 contiguous guanines in this promoter sequence.
An important aim of our research program is to identify small molecules that can selectively modulate PDGF-A transcription by stabilizing or modifying the unique G-quadruplex structures formed by NHEPDGF-A. Therefore, we examined the affinities of three well-known G-quadruplex-interactive drugs (TMPyP4, Se2SAP and telomestatin) for the NHEPDGF-A G-quadruplex. The results of our Taq polymerase stop assays revealed that these three drugs preferentially interact with and stabilize the 5′-end NHEPDGF-A G-quadruplex, which has a similar folding pattern to the biological G-quadruplexes formed in the NHEPDGF-A duplex DNA at low concentrations. TMPyP4 exhibited stronger selectivity than either Se2SAP or telomestatin for stabilization of the 5′-end stop product. Thus, TMPyP4 appeared to be an ideal candidate for modulating PDGF-A transcription. Our data confirms that, in cells, TMPyP4 can specifically downregulate PDGF-A promoter activity by interacting with the NHEPDGF-A G-quadruplex. Because TMPyP4, and not TMPyP2, inhibits PDGF-A proximal promoter activity, this is strong evidence that G-quadruplexes exist in vivo and that NHEPDGF-A G-quadruplex structures act as transcriptional silencers for PDGF-A.
Finally, the relationship of the 5′SHS and intron SHS elements to the dynamic equilibrium of the G-quadruplex and associated C-strand structures remains to be determined. The complementarity of the 5′SHS and the 5′-end of the intron SHS is suggestive of the interaction of these elements. Experiments addressing this question are in progress.
ACKNOWLEDGEMENTS
This research has been supported by grants from the National Institutes of Health (CA94166) and the Arizona Biomedical Research Commission (9006). We thank John Fitch, Michael Cusanovich and members of the Cusanovich lab (University of Arizona) for providing equipment and technical support for oligonucleotide extinction coefficient calculation. We are grateful to David Bishop for preparing, proofreading and editing the final version of the manuscript and figures. Funding to pay the Open Access publication charges for this article was provided by NIH CA94166.
Conflict of interest statement. None declared.
REFERENCES
- 1.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 2.Heldin CH, Eriksson U, Ostman A. New members of the platelet-derived growth factor family of mitogens. Arch. Biochem. Biophys. 2002;398:284–290. doi: 10.1006/abbi.2001.2707. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez RH, Kantarjian HM, Cortes JE. Biology of platelet-derived growth factor and its involvement in disease. Mayo Clin. Proc. 2006;81:1241–1257. doi: 10.4065/81.9.1241. [DOI] [PubMed] [Google Scholar]
- 4.Betsholtz C. Biology of platelet-derived growth factors in development. Birth Defects Res. C Embryo. Today. 2003;69:272–285. doi: 10.1002/bdrc.10030. [DOI] [PubMed] [Google Scholar]
- 5.Ostman A, Heldin CH. Involvement of platelet-derived growth factor in disease: development of specific antagonists. Adv. Cancer Res. 2001;80:1–38. doi: 10.1016/s0065-230x(01)80010-5. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Ustach C, Kim HR. Platelet-derived growth factor signaling and human cancer. J. Biochem. Mol. Biol. 2003;36:49–59. doi: 10.5483/bmbrep.2003.36.1.049. [DOI] [PubMed] [Google Scholar]
- 7.Westermark B, Heldin CH, Nister M. Platelet-derived growth factor in human glioma. Glia. 1995;15:257–263. doi: 10.1002/glia.440150307. [DOI] [PubMed] [Google Scholar]
- 8.Sulzbacher I, Birner P, Trieb K, Traxler M, Lang S, Chott A. Expression of platelet-derived growth factor-AA is associated with tumor progression in osteosarcoma. Mod. Pathol. 2003;16:66–71. doi: 10.1097/01.MP.0000043522.76788.0A. [DOI] [PubMed] [Google Scholar]
- 9.Afrakhte M, Nister M, Ostman A, Westermark B, Paulsson Y. Production of cell-associated PDGF-AA by a human sarcoma cell line: evidence for a latent autocrine effect. Int. J. Cancer. 1996;68:802–809. doi: 10.1002/(SICI)1097-0215(19961211)68:6<802::AID-IJC19>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Guha A, Dashner K, Black PM, Wagner JA, Stiles CD. Expression of PDGF and PDGF receptors in human astrocytoma operation specimens supports the existence of an autocrine loop. Int. J. Cancer. 1995;60:168–173. doi: 10.1002/ijc.2910600206. [DOI] [PubMed] [Google Scholar]
- 11.Sulzbacher I, Träxler M, Mosberger I, Lang S, Chott A. Platelet-derived growth factor-AA and -α receptor expression suggests an autocrine and/or paracrine loop in osteosarcoma. Mod. Pathol. 2000;13:632–637. doi: 10.1038/modpathol.3880109. [DOI] [PubMed] [Google Scholar]
- 12.Takimoto Y, Wang ZY, Kobler K, Deuel TF. Promoter region of the human platelet-derived growth factor A-chain gene. Proc. Natl Acad. Sci. USA. 1991;88:1686–1690. doi: 10.1073/pnas.88.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin X, Wang Z, Gu L, Deuel TF. Functional analysis of the human platelet-derived growth factor A-chain promoter region. J. Biol. Chem. 1992;267:25614–25619. [PubMed] [Google Scholar]
- 14.Kaetzel D.M.Jr, Maul RS, Liu B, Bonthron D, Fenstermaker RA, Coyne DW. Platelet-derived growth factor A-chain gene transcription is mediated by positive and negative regulatory regions in the promoter. Biochem. J. 1994;301:321–327. doi: 10.1042/bj3010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Maul RS, Kaetzel D.M., Jr. Repression of platelet-derived growth factor A-chain gene transcription by an upstream silencer element. J. Biol. Chem. 1996;271:26281–26290. doi: 10.1074/jbc.271.42.26281. [DOI] [PubMed] [Google Scholar]
- 16.Wang ZY, Deuel TF. S1-nuclease-sensitive structures contribute to transcriptional regulation of the human PDGF A-chain gene. Prog. Nucleic Acid Res. Mol. Biol. 1996;55:227–244. doi: 10.1016/s0079-6603(08)60195-6. [DOI] [PubMed] [Google Scholar]
- 17.Kaetzel DM. Transcription of the platelet-derived growth factor A-chain gene. Cytokine Growth Factor Rev. 2003;14:427–446. doi: 10.1016/s1359-6101(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 18.Silverman ES, Khachigian LM, Lindner V, Williams AJ, Collins T. Inducible PDGF A-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am. J. Physiol. 1997;273:H1415–H1426. doi: 10.1152/ajpheart.1997.273.3.H1415. [DOI] [PubMed] [Google Scholar]
- 19.Gashler AL, Bonthron DT, Madden SL, Rauscher F.J., III, Collins T, Sukhatme VP. Human platelet-derived growth factor A chain is transcriptionally repressed by the Wilms tumor suppressor WT1. Proc. Natl Acad. Sci. USA. 1992;89:10984–10988. doi: 10.1073/pnas.89.22.10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang ZY, Madden SL, Deuel TF, Rauscher F.J., III The Wilms’ tumor gene product, WT1, represses transcription of the platelet-derived growth factor A-chain gene. J. Biol. Chem. 1992;267:21999–22002. [PubMed] [Google Scholar]
- 21.Khachigian LM, Santiago FS, Rafty LA, Chan O.L.-W, Delbridge GJ, Bobik A, Collins T, Johnson AC. GC factor 2 represses platelet-derived growth factor A-chain gene transcription and is itself induced by arterial injury. Circ. Res. 1999;84:1258–1267. doi: 10.1161/01.res.84.11.1258. [DOI] [PubMed] [Google Scholar]
- 22.Rafty LA, Santiago FS, Khachigian LM. NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter. EMBO J. 2002;21:334–343. doi: 10.1093/emboj/21.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma D, Xing Z, Liu B, Pedigo NG, Zimmer SG, Bai Z, Postel EH, Kaetzel DM. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J. Biol. Chem. 2002;277:1560–1567. doi: 10.1074/jbc.M108359200. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZY, Lin XH, Nobuyoshi M, Qiu QQ, Deuel TF. Binding of single-stranded oligonucleotides to a non-B-form DNA structure results in loss of promoter activity of the platelet-derived growth factor-A chain gene. J. Biol. Chem. 1992;267:13669–13674. [PubMed] [Google Scholar]
- 25.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides, Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- 31.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 32.Sun XG, Cao EH, He YJ, Qin JF. Spectroscopic comparison of different DNA structures formed by oligonucleotides. J. Biomol. Struct. Dyn. 1999;16:863–872. doi: 10.1080/07391102.1999.10508298. [DOI] [PubMed] [Google Scholar]
- 33.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurizot JC. Circular dichroism of nucleic acids: nonclassical conformations and modified oligonucleotides. In: In: Berova N, Nakanishi K, Woody RW, editors. Circular Dichroism: Principles and Applications. 2nd ed. Wiley-VCH, New York: 2000. pp. 719–736. [Google Scholar]
- 35.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petraccone L, Erra E, Esposito V, Randazzo A, Mayol L, Nasti L, Barone G, Giancola C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 37.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 38.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 39.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 43.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 44.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 47.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henderson E, Hardin CC, Walk SK, Tinoco I.Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine–guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 49.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 50.Vorlickova M, Kypr J. Conformational variability of poly(dA-dT)·poly(dA-dT) and some other deoxyribonucleic acids includes a novel type of double helix. J. Biomol. Struct. Dyn. 1985;3:67–83. doi: 10.1080/07391102.1985.10508399. [DOI] [PubMed] [Google Scholar]
- 51.Kypr J, Fialova M, Chladkova J, Tumova M, Vorlickova M. Conserved guanine–guanine stacking in tetraplex and duplex DNA. Eur. Biophys. J. 2001;30:555–558. doi: 10.1007/s002490100174. [DOI] [PubMed] [Google Scholar]
- 52.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc. 1999;121:3561–3570. [Google Scholar]
- 53.Pietras K, Sjoblom T, Rubin K, Heldin CH, Ostman A. PDGF receptors as cancer drug targets. Cancer Cell. 2003;3:439–443. doi: 10.1016/s1535-6108(03)00089-8. [DOI] [PubMed] [Google Scholar]
- 54.George D. Platelet-derived growth factor receptors: a therapeutic target in solid tumors. Semin. Oncol. 2001;28:27–33. [PubMed] [Google Scholar]
- 55.Madhusudan S, Ganesan TS. Tyrosine kinase inhibitors in cancer therapy. Clin. Biochem. 2004;37:618–635. doi: 10.1016/j.clinbiochem.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 56.Hofer MD, Fecko A, Shen R, Setlur SR, Pienta KG, Tomlins SA, Chinnaiyan AM, Rubin MA. Expression of the platelet-derived growth factor receptor in prostate cancer and treatment implications with tyrosine kinase inhibitors. Neoplasia. 2004;6:503–512. doi: 10.1593/neo.04157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 58.Kumar N, Maiti S. Quadruplex to Watson-Crick duplex transition of the thrombin binding aptamer: a fluorescence resonance energy transfer study. Biochem. Biophys. Res. Commun. 2004;319:759–767. doi: 10.1016/j.bbrc.2004.05.052. [DOI] [PubMed] [Google Scholar]