Abstract
Poly(ADP-ribose) polymerases (PARPs) catalyze the post-translational modification of proteins with poly(ADP-ribose). Two PARP isoforms, PARP-1 and PARP-2, display catalytic activity by contact with DNA-strand breaks and are involved in DNA base-excision repair and other repair pathways. A body of correlative data suggests a link between DNA damage-induced poly(ADP-ribosyl)ation and mammalian longevity. Recent research on PARPs and poly(ADP-ribose) yielded several candidate mechanisms through which poly(ADP-ribosyl)ation might act as a factor that limits the rate of ageing.
INTRODUCTION
Poly(ADP-ribose) (PAR), the reaction product of the large family of poly(ADP-ribose) polymerases (PARPs), has been discovered in the 1960s (1). Since then, this molecule and its producers have made an astounding career from an obscure side reaction of cellular DNA damage to the ‘Swiss army knife’ for the maintenance of genomic stability. The areas of involvement of PARPs range from the modulation of DNA repair and the regulation of chromatin structure (2,3) to transcriptional regulation (4–8), mitotic spindle organization (9–12), telomere maintenance (13–16), regulation of trafficking (17–19), involvement in multi-drug resistance (20), to cell death activation (21,22). Understanding the underlying mechanisms of PARP-mediated regulation is a prerequisite for interpretation of the results linking PARP activity to mammalian ageing. In this review, we are focussing on the aspects related with ageing processes, including cancer.
Ageing and the connection to PARP
Ageing is a multi-factorial process and has been defined as time-dependent general decline in physiological function of an organism, associated with a progressively increasing risk of morbidity and mortality. It is apparent that during ageing different organs are losing their functional reserve and plasticity and become less able to fulfil their physiological function, especially under conditions of stress. Molecular hallmarks are (i) changes in extracellular components (i.e. collagen-matrix, deposits in the vascular system), (ii) changes in cellular metabolism (i.e. DNA repair, protein surveillance) as well as (iii) cellular functionality (senescence and cell death). To maintain their ability to act and respond in an appropriate way, cells have to protect their genomic information from the constant attack of internal and external damaging agents. DNA repair and DNA damage signalling pathways are major factors determining the cellular fate. PARPs have been shown to be a central hinge in maintaining genomic stability. PARP activity is important for repair of DNA single- and double-strand breaks, facilitates mitosis by interaction of PAR with the spindle apparatus, is involved in the decision of life and death after genotoxic insults, helps regulating the integrity of the chromosomal ends, and is a crucial mediator in inflammatory responses, thus covering all three points mentioned above. Disturbances in any of these pathways may lead to senescence or cell death, thus decreasing the functionality of the harbouring organ. Another detrimental outcome may be cancer initiation. How are PARPs able to integrate so many tasks? The following sections will discuss this point by point.
PARP and cell death
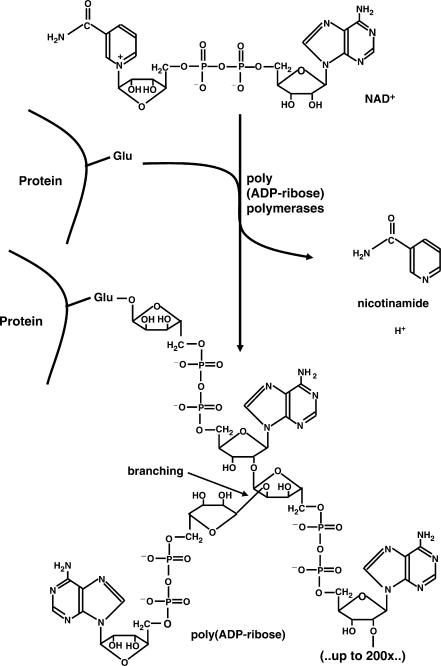
PARPs use NAD+ as a substrate from which they cleave off nicotinamide and form, via repeated reaction cycles, a polymer of ADP-ribose units, which can be branched (23,24) (Figure 1). Interestingly the main target proteins (‘acceptors’) are PARPs themselves, which is referred to as PARP automodification. Cellular PAR formation is dramatically stimulated after exposure to DNA-damaging agents that induce single- or double-strand breaks in DNA (25). This kind of induced PAR formation depends on the activation of two abundantly expressed members of the PARP family, i.e. PARP-1 and PARP-2, with PARP-1 activity accounting for ∼90% of the polymer produced under these conditions (26). Under conditions of genotoxic exposure poly(ADP-ribose) undergoes a rapid turnover, since, in parallel with the synthesis of polymer, its enzymatic catabolism is highly active. This is catalyzed by poly(ADP-ribose) glycohydrolase (PARG) and leads to the formation of free monomeric ADP-ribose. The latter compound can be recycled for NAD+ synthesis, but this requires large amounts of ATP, and as a result cellular NAD+ levels drop significantly in this process.
Figure 1.
Synthesis and structure of poly(ADP-ribose). β-NAD+ is used as a substrate by poly(ADP-ribose) polymerases. Nicotinamide is released and the residual ADP-ribose moieties are joined in an α-glycosidic manner, covalently attached to a glutamate or aspartate of the acceptor protein. Chain lengths can be up to 200 residues with the possibility of branching every 50 units.
For a long time, the response to genotoxic insults was thought to be the only primary action of PARP(s), and indeed a large body of data has accumulated showing that, perhaps surprisingly, both PARP overactivation (21,27) and the inhibition of the moderate, ‘physiological’ PARP activity induced by low numbers of DNA strand breaks increase cell death. Interestingly, PARP inhibition potentiates the incidence of genomic instability induced by genotoxic exposure (28–30) while PARP-1 overexpression has a suppressive effect (31,32). An obvious explanation could be that inhibited PARP-1 and PARP-2 cannot contribute to repair pathways anymore or have even negative functions. Therefore, unrepaired lesions either are substrates for other repair processes like homologous recombination (HR) or lead to cell death. In summary, inactive PARP leads to massive cell death due to unrepaired DNA damages and subsequent apoptosis, with escaping cells being genetically altered. On the other hand, hyperactive PARP restricts genomic instability, but kills the cell by energy failure; survivors are not or only moderately altered genetically.
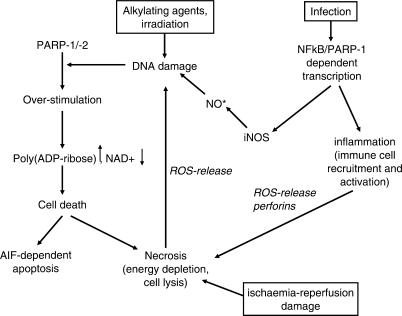
Cell death by active PARP is mediated by at least two separate mechanisms. First, massive activation of PARP by excess DNA damage leads to a drop in cellular NAD+ levels. In order to regenerate this very important cellular energy carrier, ATP has to be consumed in large amounts and cells die from necrosis. On the other hand, PARP activity can induce caspase-independent apoptosis (Figure 2). Recent data suggest that PAR releases apoptosis-inducing factor (AIF) from the inter-membrane space of mitochondria (22,33,34). AIF then translocates into the nucleus and induces high-molecular weight DNA fragmentation (35). Which mechanism of cell death is used probably depends on the cellular system and the stimulus applied. During classical apoptosis there is an initial burst of PAR production (36,37) followed by cleavage of PARP-1 and PARP-2, probably in order to preserve energy for the apoptosis execution programme by stopping PARP activation and thus NAD+ consumption. Regardless of the precise mechanism by which cell death occurs, this phenomenon has an important impact in pathophysiological settings (Figure 2). In models of ischaemia-reperfusion damage, PARP inhibitors have been used to minimize the affected area (38,39). The tissue damage accompanied by ischaemia leads to massive cell death and release of reactive oxygen species (ROS), which damage surrounding cells, activate PARP-1/-2 and induce further cell death, thus spreading the necrotic area. Interfering with this vicious cycle by PARP inhibition obviously has beneficial effects. Additionally, PARP-1 is not only involved in the cellular reaction to ROS, but also in ROS production (see next section).
Figure 2.
Cell death induced by the activity of poly(ADP-ribose) polymerases. DNA damage induced by various stimuli triggers the formation of PAR by PARP-1/-2. Depending on the cell type and the energy capacity, PAR can either directly induce cell death by AIF release or by energy depletion. In this process, necrotic cells release themselves ROS, accelerating the effect. Ischaemia-reperfusion damage acts on the same line. PARP-1 also interacts with NFκB in inflammation processes, which induce cellular NO formation by inducible NO-synthase (iNOS) and accumulation of reactive products. Also, cytokine production and subsequent immune-cell stimulation is under control of NFκB, leading to target cell lysis with release of pro-inflammatory cytokines and death of surrounding cells.
PARP-1 and transcription
In 1983, Roeder and colleagues identified PARP-1 as a transcription factor (TFII-C) relevant for regulation of transcriptional initiation (4). Subsequently, PARP-1 has been shown to interact with and modify several classical transcription factors (5,40,41). Most strikingly, Parp1 (formerly called ADPRT) knockout mice are protected from septic shock (42,43) and some pathological effects in various inflammatory diseases (see above). This depends partially on the requirement of PARP-1 for NFκB-coupled transactivation. NFκB is the major transcriptional regulator in the immune system. Failure to activate it dampens the immune response, which may be advantageous in settings of deleterious over-stimulation of the immune system. For example, the gene encoding inducible nitric oxide synthase is a prominent target of NFκB transactivation. The product of this enzyme, nitric oxide, and downstream reaction products such as peroxynitrite are potent DNA-damaging agents, which lead to PARP-1/-2 stimulation. PARPs consume NAD+ during this process with potentially lethal outcome (see above). PARP inhibition has been shown to rescue cells and tissues in different experimentally induced pathological conditions such as colitis (44,45), MPTP-induced Parkinson syndrome (46), ischaemia-reperfusion damage (47–50) and type-I diabetes (51–53). These disease-associated features could be referred to as the ‘ageing-promoting dark backyard’ of PARPs (Figure 2). On the other hand, fully functional NFκB is indispensable for normal activation of the immune response. Ageing of the immune system leads to a less pronounced defensive response to infectious agents and therefore to increased mortality. This important branch of involvement of PARPs in ageing needs to be analysed more closely than it has been in the past.
PARP and DNA repair
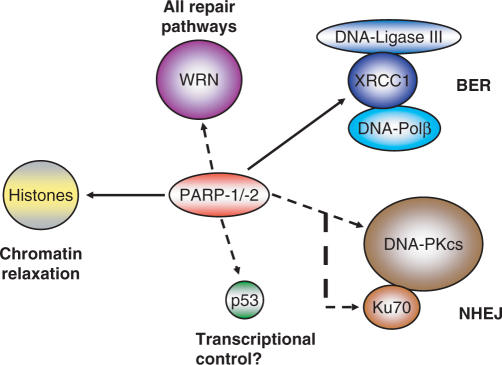
The best understood function of PARP-1 and PARP-2 is regulation of the base excision repair pathway (BER), especially the decision between short-patch and long-patch repair. DNA-base excision repair occurs in a ‘passing the baton’ fashion (54,55). After recognition and excision of the damaged base by type II or type I base glycosylases, the sugar-phosphate backbone of DNA is cleaved on the 5′ side either by the same or an additional enzyme, respectively. The 5′dRPase activity of DNA polymerase β cuts out the residual sugar moiety and fills in the gap in a one-nucleotide synthesis step, followed by sealing the nicked DNA by DNA-ligase III (short-patch repair). Blocked terminal ribose residues can lead to a switch to long-patch repair either by stimulating the strand-replacement synthesis activity of polymerase β or by alternative usage of the processive DNA polymerase δ. The resulting DNA overhang is cleaved by flap endonuclease 1(FEN-1) and the nicked DNA is ligated. In-vitro data point to a necessity of PARP-1 for the activation of the long patch pathway (56–58). If direct repair by DNA polymerase β is blocked, PARP-1 helps out by switching to the alternative route by poly(ADP-ribosyl)ation and attracting auxiliary proteins. Furthermore, PARP-1 stimulates the strand-displacement synthesis activity of DNA polymerase β with the help of FEN-1 (59). In vivo, the chromatin relaxing activity of PAR may aid in this process and is therefore likely to be involved in short-patch BER also. Indeed, even slightly poly(ADP-ribosyl)ated PARP-1 is able to recruit the scaffold protein XRCC1 (60–62), which interacts with DNA polymerase β as well as with DNA ligase III. Interfering with poly(ADP-ribosyl)ation by pharmacological inhibition, dominant negative competition or genetic knockout leads to genomic instability, as mentioned above. If single-strand breaks arising during the repair process are not ligated, the next round of replication may produce a double-strand break, which is potentially lethal (63). Therefore, cells activate the double-strand break repair machinery, i.e. either homologous recombination (HR, during S/G2 phase of the cell cycle), or non-homologous end-joining (NHEJ). Surprisingly, PARP-1 and PAR also interact with DNA-dependent protein kinase (DNA-PK), a heterotrimeric enzyme complex belonging to the family of phosphatidylinositol-3-kinase-like kinases (PIKK) involved in DNA damage signalling. The catalytic subunit DNA-PKcs and also the Ku70 autoantigen are in vitro targets for covalent ADP-ribosylation (and also for non-covalent PAR binding), and vice versa. PARP-1 is a substrate for phosphorylation by DNA-PK (64–66) (Figure 3). The activity of DNA-PKcs is stimulated by poly(ADP-ribosyl)ation. The rationale for the interaction between these two enzymes is not clear. Perhaps PARP-1 facilitates, by polymer formation, chromatin opening and therefore better access of DNA-PK to the break, or weak poly(ADP-ribosyl)ation might induce a conformational change in the 3D structure of the kinase, thus increasing its activity. It was shown recently that the activity of another PIKK-member, ATM (ataxia telangiectasia mutated), is positively modulated by PAR (67). Thus, it becomes more and more evident that PARP-1 and PARP-2 are general DNA damage sensing and repair regulating factors.
Figure 3.
Direct interactions of PARP-1/-2 with other proteins important in repair. PARP-1/-2 and their product PAR interact with several other proteins involved in different repair pathways. Histones are a prominent target of poly(ADP-ribosyl)ation, releasing them from DNA. A full-line arrow marks interactions shown for both PARPs, whereas dashed arrows indicate established interactions between PARP-1 and other proteins.
The impact of PARP-1 on genomic stability is exploited in cancer therapy. It was reported that simultaneous treatment of tumours with DNA-damaging agents like temozolomide and PARP inhibitors increases the efficacy of the chemotherapeutic drug (68–70). Inactivated PARP blocks the repair of induced DNA lesions and therefore triggers cell death more efficiently (71). Recent experimental data take these findings one step further. Tumour cells with non-functional BRCA-1/-2 proteins can be sensitized to cell death by simple PARP inhibition even in the absence of any additional treatment (72–74). The single-strand breaks that are accumulating in this setting are converted into double-strand breaks by DNA replication. As the BRCA proteins are integral components of the signalling and repair machinery responsive to this kind of lesions, repair by HR is blocked, so the cells will die as a result of the persistent damage. These findings, however, have been challenged recently (75), and it was suggested that the observed results may have been due to unspecific effects of PARP inhibition.
PARP localizing to specific intracellular compartments
Seemingly PARPs are not only regulating DNA repair pathways, as they have also been found at chromosome-organizing regions like the centromere, the telomere or the centrosome. PARP-1 has been shown to localize to the centromere, and centromeric proteins have been shown to be a substrate for covalent poly(ADP-ribosyl)ation (3) but the impact of this modification has been elusive so far.
PARP-2 seems to be specifically involved in X-chromosomal stability, as Parp2 knockout mice showed a decreased frequency of female pups born (76). Cytogenetic analyses revealed a selectively increased embryonic lethality in females due to X-chromosomal instability.
PARP-1 seems to be involved in maintaining euploidy of mammalian cells as Parp1 knockout cells in culture show increased aneuploidy; the same is the case for wild-type cells under chronic treatment with PARP inhibitors (77–79). This effect depends on a deregulation of centrosome duplication, leading to increased numbers of spindle-pole bodies and, as a consequence, aberrant mitosis. One hallmark of cancer is aneuploidy. Likewise, fibroblasts from Parp1 knockout mice show increased polyploidy, raising the question if PARP-1 is also involved in mitotic checkpoints (77). Also, the product of PARPs, poly(ADP-ribose), has been shown to be an integral component of mitotic spindles, responsible for their stability (9,10).
At chromosomal ends, a protein complex designated as ‘shelterin’ or ‘telosome’ forms together with roughly 20 kb of the repetitive hexanucleotide DNA (T2AG3) a protective cap, the telomere (80,81). Telomeric DNA ends in a single-stranded 3′ extension, which is able to fold back and invade the double-stranded region of the same sequence. This plasmid-like structure is called t-loop and has been detected in vitro as well as in vivo (82,83). Therefore, the normally highly recombinogenic combination of a DNA double-strand break with a single-stranded extension is masked and shielded from the repair machinery to facilitate genomic stability. Two specific proteins bind the telomeric double strand, i.e. telomeric repeat binding factor (TRF) 1 and 2. Whereas TRF-1 is responsible for telomere length regulation by a protein counting mechanism (84,85), TRF-2 stabilizes the t-loop and is therefore indispensable for the functionality of the telomere (85–87). Expression of a dominant-negative version of TRF-2 immediately uncaps telomeres, followed by initiation of repair and subsequently senescence or chromosomal end-to-end fusions and massive cell death. Another feature of TRF-2 is its direct inhibition of ATM (ataxia telangiectasia mutated), the key protein kinase in activating DNA double-strand repair (88). On the other hand, the t-loop inhibits progression of the replication fork in vitro (89). Therefore, this secondary structure has to be resolved in concert with cell cycle progression in order to enable complete replication through telomeric DNA. In analogy to TRF-1, where the tankyrases 1 and 2 (PARP-5 subfamily) have been shown to dislodge TRF-1 from DNA by poly(ADP-ribosyl)ation (13,90,91), TRF-2 interacts with PARP-1 (15,16) and in a special case with PARP-2 (14). Displacing TRF-2 from the DNA would result in an ‘open-state’ telomere without t-loop, not hindering progression of the replication machinery anymore. Thus, timed on and off shuttling of TRF-2 is important for suppressing unscheduled repair (recombination) as well as telomere replication, keeping telomeric functions intact and avoiding induction of a damage signal as discussed above. The WRN protein, a helicase, may aid in the process of resolving the t-loop after TRF-2 release (discussed below).
PARP-1, WRN and p53: three proteins, one mission—genomic stability
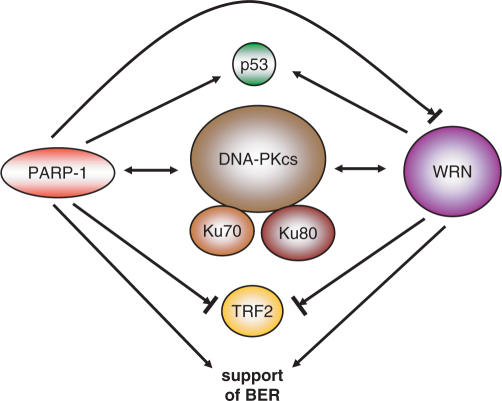
Interaction of PARP and PAR with proteins already known to affect ageing adds an additional layer to the involvement of PARP in this process. The most prominent example is the modulation of the Werner syndrome protein (WRN) activity by PARP-1 but not PARP-2 (92). In mice, double knockout of Parp1 and Wrn leads to increased sister-chromatid exchange and an earlier onset of tumorigenesis than in each single knockout (93). Together with BLM (Blooms syndrome), RTD/RecQ4 (Rothmund–Thompson disorder) and other proteins, WRN belongs to the family of RecQ helicases (94). As an exception within this family, WRN also contains an exonuclease activity. Patients with the rare recessive disorder Werner syndrome show accelerated ageing after the onset of puberty with symptoms including cataracts, greying and loss of hair, atherosclerosis, osteoporosis, diabetes mellitus and a higher incidence of sarcomas, melanomas and meningiomas. On the molecular level, Werner syndrome cells show increased numbers of chromosomal translocations and deletions, an extended S-phase due to persistent DNA damage, a higher sensitivity to DNA-damaging agents, accelerated telomere loss and shortened life span in culture. WRN associates with many proteins involved in DNA repair, replication and recombination like proliferating cell nuclear antigen (PCNA), replication protein A (RPA), all three DNA-PK subunits (Ku70, Ku 80 and DNA-PKcs), p53, DNA polymerases β and δ (95), flap-endonuclease 1 (FEN-1) (96,97), telomeric repeat-binding factor 2 (TRF-2) (98,99) and PARP-1 (92). WRN interacts with most of these proteins via its RecQ homology domain. WRN acts in vitro on secondary structures that arise during replication block, DNA repair and at the telomeric t-loop (100). In BER, WRN seems to activate the long patch pathway by direct stimulation of DNA polymerase δ and the strand displacement synthesis activity of DNA polymerase β (95). It also stimulates the activity of FEN-1 leading to more efficient cleavage of the overhanging displaced DNA strand, the ‘flap’, which is a by-product of the synthesis. The strong interdependence of WRN and PARP-1 is underlined by the fact that cells from WRN patients show a modified pattern of poly(ADP-ribosyl)ated proteins, i.e. less heteromodification of other proteins yet unchanged automodification of PARP-1 (101). Interaction with Ku70/Ku80 stimulates WRN exonuclease activity (102), which may be required to resolve secondary structures or trim blocked substrates in combination with the helicase to facilitate NHEJ. On telomeres, WRN directly binds to TRF-2 and is necessary in telomere regulation, i.e. opening up the t-loop (103). This is supported by the observation that cells from WRN patients display chromosomal fusions and faster telomere shortening, features similar to expression of a dominant-negative version of TRF-2. Besides the direct interaction between PARP-1 and WRN, the two proteins share many binding partners, such as p53, DNA-PK and TRF-2. Both proteins are especially involved in the long-patch pathway of BER. Both have a positive impact on telomere length. In contrast, whereas Ku70/80 stimulates WRN's exonuclease activity, PARP-1 inhibits both WRN functions (Figure 4). This is probably not due to direct modification of WRN protein, although one publication reported WRN as a substrate for poly(ADP-ribosyl)ation. All three functional units (PARP-1, WRN, DNA-PK with Ku70/80 and DNA-PKcs) can form a complex, in which polymer-modified Ku70/80 shows a decreased stimulation of WRN (102). Upon automodification of PARP-1, the inhibition of WRN exonuclease and helicase activity is released. Therefore, PARP-1 may regulate the timing of WRN activity towards its substrates, i.e. giving other repair proteins a trial before WRN steps in.
Figure 4.
Interaction map between PARP-1 and the Werner syndrome protein WRN. The two proteins share many overlapping interaction pathways. There is a reciprocal interaction with DNA-PK (double-headed arrow), interaction with p53, stimulation and/or modulation of base-excision repair (BER, one-headed arrow), and inhibition of TRF-2 DNA binding (blocked arrow). PARP-1 also inhibits WRN functions if in an unmodified state.
Interaction of p53, the ‘guardian of the genome’, with PARP-1 and PAR is complex and not fully understood (104–112). Whereas poly(ADP-ribosyl)ation seems to stabilize p53, it also inhibits the important transcriptional activity of p53. Moreover, p53 can bind PAR non-covalently at three sites, two of which are located in the DNA-binding domain and one in the C-terminal oligomerization domain. The function of PAR bound to p53 has to be investigated more deeply.
PARP and ageing
Fifteen years ago, Grube and Bürkle showed that DNA strand break-dependent PARP activity in permeabilized peripheral blood mononuclear cells (PBMC) correlates positively with the maximal life span in mammalian species (113). Human PBMC displayed a 5-fold increase in PAR production compared to rat PBMC, yet expressed the same amount of PARP-1 protein. Subsequent experiments with recombinant PARP-1 proteins from man and rat revealed a 2-fold higher automodification level in human PARP-1 although classical enzymatic parameters like Vmax and Km were not different (114). The interplay with other proteins either relaying the PAR signal or being directly influenced by it probably enhances evolutionary divergences. Interestingly, both in humans and rats, PARP activity declined with donor age. On the other hand, a study using immortalized lymphocytes proved that PARP activity was higher in cells from centenarians than in appropriate controls (115). Significance was even further increased if activity was normalized not to total protein but to PARP-1 content alone. Thus, a high (but probably tightly controlled) PARP activity may be beneficial for a prolonged life by suppressing genomic instability and tumorigenesis. Therefore, it was obvious to screen for polymorphisms within the human PARP-1 gene locus (Parp1). Although several polymorphisms in Parp1 have been reported, only a minority of them change the amino acids sequence and none of them could be correlated with ‘successful’ ageing as exemplified by centenarians in a first experimental setting (116). Intriguingly, recent papers report that the V762A polymorphism, originally reported by Cottet and colleagues (116), is indeed associated with diminished PARP-1 activity (117) and increased risk of some but not all types of cancer (118,119). Moreover, a duplication within a PARP1 pseudogene at 13q33 seems to be a pre-disposition marker for increased cancer risk (120). Also, a polymorphic tract of CA repeats in the promoter region seems to pre-dispose to rheumatoid arthritis in the Spanish population (121), although it is not clear if this variance influences transcription and/or activity levels. But still, as valid data are missing that integrate successful ageing and molecular mechanisms responsible for higher PARP activity, the factors determining the positive association remain elusive.
SUMMARY
PARP activity may positively influence ‘healthy ageing’ through several pathways. The classical view is that this is mediated by facilitating and regulating DNA strand break repair. Also, the role of PARP activity in triggering cell death either by energy depletion or by induction of AIF may protect the individual from the uncontrolled outgrowth of potentially mutated cells after genotoxic insult. PARP-1 and PARP-2, however, not only stabilize the genome by their repair activity, but have also been found at other locations. PARP-1 and PARP-2 were detected at the centromere, and centromeric proteins have been shown to be a substrate for poly(ADP-ribosyl)ation. Additionally, PARP-1 is a part of the centrosome, i.e. the spindle organizer, and pharmacological inhibition of PARP-1 or Parp1 knockout leads to increased rates of aneuploidy in cultured cells. Also, it has been shown that PAR is a stabilizing component of the spindle, although its precise function and its origin has not been clarified yet. Last but not least, PARP-1 interacts with the telomere DNA double-strand binding protein TRF-2. TRF-2 is responsible for telomeric stability and suppression of unscheduled activity of the double-strand break repair machinery by maintaining the t-loop; it is a key component of the proteinaceous complex called shelterin/telosome, which integrates telomere length regulation as well as protective activities. On the other hand, the t-loop inhibits passing of the replication fork, and therefore dislodging the proteins holding the t-loop in place is necessary for complete duplication of the genome. As a candidate mechanism, PARP-1 could modify TRF-2 and target it for destruction, counteracting accelerated telomere shortening and accompanied senescence or cell death. In mice, PARP-2 is involved especially in female X-chromosome stability. Independent of its genome-stabilizing functions, PARP-1 also impacts on transcription. The interaction of PARP-1 with the key regulator of the immune system, NFκB, and the dependence of the latter on the presence of PARP-1 for its activity (at least in the setting of some NFκB-regulated promoters) impacts strongly on the functionality of this important line of defence against deleterious invasion of pathogenic micro-organisms. The importance of an intact PAR-system including PARP-1 and PARP-2 is underlined by the fact that double-knockout mice display embryonic lethality (76). Therefore, discrimination between the partially overlapping functions of PARP-1 and PARP-2 is difficult, and redundancy may mask aspects that are normally accounted for by one of the two proteins in an in vivo setting.
In conclusion, there are two major branches of how PARP activity can influence the organismal ageing process: First, there is modulation of the immune system via interaction with NFκB, which could lead to a better fitness in the constant battle with pathogens our bodies have to fight. Second, there is maintenance of genomic stability, subdivided in several pathways. (i) Regulation of BER (via interaction with XRCC1); (ii) modulating DNA double-strand break repair (DNA-PK, ATM); (iii) telomere stability (TRF-2/shelterin); (iv) spindle organization and stability (centrosome, PAR at mitotic microtubules, X-chromosome) and (v) death of cells with heavily damaged DNA (AIF, energy depletion). All of these factors merge into the suppression of mutations potentially leading to tumorigenesis. But—as the other side of the coin—it also can disturb tissue renewal, leading to reduced functional plasticity of organs. As PARP activity declines with the age of the individual, the regulative interplay may not be effective enough to maintain cellular fitness. The loss of functional cells from tissues either by senescence or cell death will compromise the respective organ functions (Figure 5). Along the same lines, successful ageing of centenarians may be supported by high PARP activity leaving regulatory circuits intact and preventing cancer formation.
Figure 5.
Poly(ADP-ribose) as a non-proteinaceous tumour-suppressor. Poly(ADP-ribosyl)ation not only regulates repair-pathways as BER and DNA double-strand break repair (DNA-PK, ATM), but also contributes to other aspects of maintaining genomic stability by PAR interaction with the centrosome and (heterochromatic) chromosome regions like the telomere and the centromere. Blocked lines describe factors inhibited by PARP activity and arrows stand for positive correlation. The effects in the middle column lead to either cancer initiation or accelerated tissue ageing. Taken together, the cellular poly(ADP-ribose) system is an important factor in suppressing tumorigenesis or tissue ageing and is needed for maintaining cellular functionality.
ACKNOWLEDGEMENT
Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (International Research Training Group 1331 Konstanz/Zürich, “Cell-based Characterization of Disease Mechanisms in Tissue Destruction and Repair”).
Conflict of interest statement. None declared.
REFERENCES
- 1.Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963;11:39–43. doi: 10.1016/0006-291x(63)90024-x. [DOI] [PubMed] [Google Scholar]
- 2.Earle E, Saxena A, MacDonald A, Hudson DF, Shaffer LG, Saffery R, Cancilla MR, Cutts SM, Howman E, et al. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet. 2000;9:187–194. doi: 10.1093/hmg/9.2.187. [DOI] [PubMed] [Google Scholar]
- 3.Saxena A, Saffery R, Wong LH, Kalitsis P, Choo KH. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly (ADP-ribosyl)ated. J. Biol. Chem. 2002;277:26921–26926. doi: 10.1074/jbc.M200620200. [DOI] [PubMed] [Google Scholar]
- 4.Slattery E, Dignam JD, Matsui T, Roeder RG. Purification and analysis of a factor which suppresses nick-induced transcription by RNA polymerase II and its identity with poly(ADP-ribose) polymerase. J. Biol. Chem. 1983;258:5955–5959. [PubMed] [Google Scholar]
- 5.Oei SL, Griesenbeck J, Schweiger M, Ziegler M. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 1998;273:31644–31647. doi: 10.1074/jbc.273.48.31644. [DOI] [PubMed] [Google Scholar]
- 6.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol. Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 7.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 8.Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 10.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 11.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 2005;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 13.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 14.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, et al. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell. Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 16.Gomez M, Wu J, Schreiber V, Dunlap J, Dantzer F, Wang Y, Liu Y. PARP1 Is a TRF2-associated poly(ADP-ribose)polymerase and protects eroded telomeres. Mol. Biol. Cell. 2006;17:1686–1696. doi: 10.1091/mbc.E05-07-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 18.Yeh TY, Sbodio JI, Tsun ZY, Luo B, Chi NW. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem. J. 2006 doi: 10.1042/BJ20060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh TY, Meyer TN, Schwesinger C, Tsun ZY, Lee RM, Chi NW. Tankyrase recruitment to the lateral membrane in polarized epithelial cells: regulation by cell-cell contact and protein poly(ADP-ribosyl)ation. Biochem. J. 2006;399:415–425. doi: 10.1042/BJ20060713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger NA, Sims JL, Catino DM, Berger SJ. Poly(ADP-ribose) polymerase mediates the suicide response to massive DNA damage: studies in normal and DNA-repair defective cells. Princess Takamatsu Symp. 1983;13:219–226. [PubMed] [Google Scholar]
- 22.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 23.Bürkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 24.Bürkle A. DNA repair and PARP in aging. Free Radic. Res. 2006;40:1295–1302. doi: 10.1080/10715760600915288. [DOI] [PubMed] [Google Scholar]
- 25.Bürkle A. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 2001;163:1–5. doi: 10.1016/s0304-3835(00)00694-7. [DOI] [PubMed] [Google Scholar]
- 26.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 27.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl Acad. Sci. USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Küpper JH, Müller M, Jacobson MK, Tatsumi-Miyajima J, Coyle DL, Jacobson EL, Bürkle A. trans-dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against gamma-irradiation and N-methyl-N'-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol. Cell. Biol. 1995;15:3154–3163. doi: 10.1128/mcb.15.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald D, De Murcia G, De Murcia JM. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc. Natl Acad. Sci. USA. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah GM, Poirier D, Desnoyers S, Saint-Martin S, Hoflack JC, Rong P, ApSimon M, Kirkland JB, Poirier GG. Complete inhibition of poly(ADP-ribose) polymerase activity prevents the recovery of C3H10T1/2 cells from oxidative stress. Biochim. Biophys. Acta. 1996;1312:1–7. doi: 10.1016/0167-4889(96)00004-3. [DOI] [PubMed] [Google Scholar]
- 31.Van Gool L, Meyer R, Tobiasch E, Cziepluch C, Jauniaux JC, Mincheva A, Lichter P, Poirier GG, Bürkle A, et al. Overexpression of human poly(ADP-ribose) polymerase in transfected hamster cells leads to increased poly(ADP-ribosyl)ation and cellular sensitization to gamma irradiation. Eur. J. Biochem. 1997;244:15–20. doi: 10.1111/j.1432-1033.1997.00015.x. [DOI] [PubMed] [Google Scholar]
- 32.Meyer R, Müller M, Beneke S, Küpper JH, Bürkle A. Negative regulation of alkylation-induced sister-chromatid exchange by poly(ADP-ribose) polymerase-1 activity. Int. J. Cancer. 2000;88:351–355. doi: 10.1002/1097-0215(20001101)88:3<351::aid-ijc5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 33.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl Acad. Sci. USA. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- 36.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 37.Negri C, Donzelli M, Bernardi R, Rossi L, Bürkle A, Scovassi AI. Multiparametric staining to identify apoptotic human cells. Exp. Cell Res. 1997;234:174–177. doi: 10.1006/excr.1997.3591. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Zingarelli B, Szabo C. Effect of genetic disruption of poly (ADP-ribose) synthetase on delayed production of inflammatory mediators and delayed necrosis during myocardial ischemia-reperfusion injury. Shock. 2000;13:60–66. doi: 10.1097/00024382-200013010-00011. [DOI] [PubMed] [Google Scholar]
- 39.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 40.Oei SL, Griesenbeck J, Schweiger M, Babich V, Kropotov A, Tomilin N. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biochem. Biophys. Res. Commun. 1997;240:108–111. doi: 10.1006/bbrc.1997.7621. [DOI] [PubMed] [Google Scholar]
- 41.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell. Mol. Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasko G, Mabley JG, Nemeth ZH, Pacher P, Deitch EA, Szabo C. Poly(ADP-ribose) polymerase is a regulator of chemokine production: relevance for the pathogenesis of shock and inflammation. Mol. Med. 2002;8:283–289. [PMC free article] [PubMed] [Google Scholar]
- 44.Liaudet L, Szabo A, Soriano FG, Zingarelli B, Szabo C, Salzman AL. Poly (ADP-ribose) synthetase mediates intestinal mucosal barrier dysfunction after mesenteric ischemia. Shock. 2000;14:134–141. doi: 10.1097/00024382-200014020-00010. [DOI] [PubMed] [Google Scholar]
- 45.Jijon HB, Churchill T, Malfair D, Wessler A, Jewell LD, Parsons HG, Madsen KL. Inhibition of poly(ADP-ribose) polymerase attenuates inflammation in a model of chronic colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2000;279:G641–G651. doi: 10.1152/ajpgi.2000.279.3.G641. [DOI] [PubMed] [Google Scholar]
- 46.Mandir AS, Przedborski S, Jackson-Lewis V, Wang ZQ, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, et al. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc. Natl Acad. Sci. USA. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, et al. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 48.Pieper AA, Walles T, Wei G, Clements EE, Verma A, Snyder SH, Zweier JL. Myocardial postischemic injury is reduced by polyADPripose polymerase-1 gene disruption. Mol. Med. 2000;6:271–282. [PMC free article] [PubMed] [Google Scholar]
- 49.Mota-Filipe H, Sepodes B, McDonald MC, Cuzzocrea S, Pinto R, Thiemermann C. The novel PARP inhibitor 5-aminoisoquinolinone reduces the liver injury caused by ischemia and reperfusion in the rat. Med. Sci. Monit. 2002;8:BR444–BR453. [PubMed] [Google Scholar]
- 50.Fiorillo C, Ponziani V, Giannini L, Cecchi C, Celli A, Nediani C, Perna AM, Liguori P, Nassi N, et al. Beneficial effects of poly (ADP-ribose) polymerase inhibition against the reperfusion injury in heart transplantation. Free Radic. Res. 2003;37:331–339. doi: 10.1080/1071576021000055262. [DOI] [PubMed] [Google Scholar]
- 51.Szabo C. PARP as a drug target for the therapy of diabetic cardiovascular dysfunction. Drug News Perspect. 2002;15:197–205. doi: 10.1358/dnp.2002.15.4.840052. [DOI] [PubMed] [Google Scholar]
- 52.Szabo C, Zanchi A, Komjati K, Pacher P, Krolewski AS, Quist WC, LoGerfo FW, Horton ES, Veves A. Poly(ADP-Ribose) polymerase is activated in subjects at risk of developing type 2 diabetes and is associated with impaired vascular reactivity. Circulation. 2002;106:2680–2686. doi: 10.1161/01.cir.0000038365.78031.9c. [DOI] [PubMed] [Google Scholar]
- 53.Beneke S, Burkle A. Poly(ADP-ribosyl)ation, PARP, and aging. Sci. Aging Knowledge Environ. 2004;2004:re9. doi: 10.1126/sageke.2004.49.re9. [DOI] [PubMed] [Google Scholar]
- 54.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 55.Mitra S, Boldogh I, Izumi T, Hazra TK. Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ. Mol. Mutagen. 2001;38:180–190. doi: 10.1002/em.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, Oliver J, Rolli V, Menissier-de Murcia J, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 57.Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39:7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 58.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 59.Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001;276:32411–32414. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- 60.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem. Pharmacol. 2003;66:1547–1554. doi: 10.1016/s0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 64.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 65.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 66.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 67.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 68.Boulton S, Pemberton LC, Porteous JK, Curtin NJ, Griffin RJ, Golding BT, Durkacz BW. Potentiation of temozolomide-induced cytotoxicity: a comparative study of the biological effects of poly(ADP-ribose) polymerase inhibitors. Br. J. Cancer. 1995;72:849–856. doi: 10.1038/bjc.1995.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tentori L, Leonetti C, Scarsella M, d'Amati G, Portarena I, Zupi G, Bonmassar E, Graziani G. Combined treatment with temozolomide and poly(ADP-ribose) polymerase inhibitor enhances survival of mice bearing hematologic malignancy at the central nervous system site. Blood. 2002;99:2241–2244. doi: 10.1182/blood.v99.6.2241. [DOI] [PubMed] [Google Scholar]
- 70.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 71.Beneke S, Diefenbach J, Burkle A. Poly(ADP-ribosyl)ation inhibitors: promising drug candidates for a wide variety of pathophysiologic conditions. Int. J. Cancer. 2004;111:813–818. doi: 10.1002/ijc.20342. [DOI] [PubMed] [Google Scholar]
- 72.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 73.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 74.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to Poly(ADP-Ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 75.De Soto JA, Wang X, Tominaga Y, Wang RH, Cao L, Qiao W, Li C, Xu X, Skoumbourdis AP, et al. The inhibition and treatment of breast cancer with poly (ADP-ribose) polymerase (PARP-1) inhibitors. Int. J. Biol. Sci. 2006;2:179–185. doi: 10.7150/ijbs.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simbulan-Rosenthal CM, Haddad BR, Rosenthal DS, Weaver Z, Coleman A, Luo R, Young HM, Wang ZQ, Ried T, et al. Chromosomal aberrations in PARP(-/-) mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc. Natl Acad. Sci. USA. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanai M, Uchida M, Hanai S, Uematsu N, Uchida K, Miwa M. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem. Biophys. Res. Commun. 2000;278:385–389. doi: 10.1006/bbrc.2000.3801. [DOI] [PubMed] [Google Scholar]
- 79.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-Ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 81.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 82.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 83.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 87.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 88.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cook BD, Dynek JN, Chang W, Shostak G, Smith S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 2002;22:332–342. doi: 10.1128/MCB.22.1.332-342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, Dawut L, Bohr VA. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harrigan JA, Opresko PL, von Kobbe C, Kedar PS, Prasad R, Wilson SH, Bohr VA. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 96.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 98.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 99.Machwe A, Xiao L, Orren DK. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 100.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 103.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 104.Kumari SR, Mendoza-Alvarez H, Alvarez-Gonzalez R. Functional interactions of p53 with poly(ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: covalent poly(ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the M(r) 85,000 proteolytic fragment. Cancer Res. 1998;58:5075–5078. [PubMed] [Google Scholar]
- 105.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 106.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Smulson ME. Poly(ADP-ribosyl)ation of p53 during apoptosis in human osteosarcoma cells. Cancer Res. 1999;59:2190–2194. [PubMed] [Google Scholar]
- 107.Mendoza-Alvarez H, Alvarez-Gonzalez R. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- 108.Simbulan-Rosenthal CM, Rosenthal DS, Luo RB, Samara R, Jung M, Dritschilo A, Spoonde A, Smulson ME. Poly(ADP-ribosyl)ation of p53 in vitro and in vivo modulates binding to its DNA consensus sequence. Neoplasia. 2001;3:179–188. doi: 10.1038/sj.neo.7900155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wesierska-Gadek J, Schmid G. Poly(ADP-ribose) polymerase-1 regulates the stability of the wild-type p53 protein. Cell. Mol. Biol. Lett. 2001;6:117–140. [PubMed] [Google Scholar]
- 110.Wesierska-Gadek J, Wojciechowski J, Schmid G. Central and carboxy-terminal regions of human p53 protein are essential for interaction and complex formation with PARP-1. J. Cell. Biochem. 2003;89:220–232. doi: 10.1002/jcb.10521. [DOI] [PubMed] [Google Scholar]
- 111.Wesierska-Gadek J, Wojciechowski J, Schmid G. Phosphorylation regulates the interaction and complex formation between wt p53 protein and PARP-1. J. Cell. Biochem. 2003;89:1260–1284. doi: 10.1002/jcb.10569. [DOI] [PubMed] [Google Scholar]
- 112.Wieler S, Gagne JP, Vaziri H, Poirier GG, Benchimol S. Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J. Biol. Chem. 2003;278:18914–18921. doi: 10.1074/jbc.M211641200. [DOI] [PubMed] [Google Scholar]
- 113.Grube K, Bürkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc. Natl Acad. Sci. USA. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beneke S, Alvarez-Gonzalez R, Bürkle A. Comparative characterisation of poly(ADP-ribose) polymerase-1 from two mammalian species with different life span. Exp. Gerontol. 2000;35:989–1002. doi: 10.1016/s0531-5565(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 115.Muiras ML, Müller M, Schächter F, Bürkle A. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J. Mol. Med. 1998;76:346–354. doi: 10.1007/s001090050226. [DOI] [PubMed] [Google Scholar]
- 116.Cottet F, Blanche H, Verasdonck P, Le Gall I, Schachter F, Bürkle A, Muiras ML. New polymorphisms in the human poly(ADP-ribose) polymerase-1 coding sequence: lack of association with longevity or with increased cellular poly(ADP-ribosyl)ation capacity. J. Mol. Med. 2000;78:431–440. doi: 10.1007/s001090000132. [DOI] [PubMed] [Google Scholar]
- 117.Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem. Biophys. Res. Commun. 2007;354:122–126. doi: 10.1016/j.bbrc.2006.12.162. [DOI] [PubMed] [Google Scholar]
- 118.Zhang X, Miao X, Liang G, Hao B, Wang Y, Tan W, Li Y, Guo Y, He F, et al. Polymorphisms in DNA base excision repair genes ADPRT and XRCC1 and risk of lung cancer. Cancer Res. 2005;65:722–726. [PubMed] [Google Scholar]
- 119.Zhai X, Liu J, Hu Z, Wang S, Qing J, Wang X, Jin G, Gao J, Shen H. Polymorphisms of ADPRT Val762Ala and XRCC1 Arg399Glu and risk of breast cancer in Chinese women: a case control analysis. Oncol. Rep. 2006;15:247–252. [PubMed] [Google Scholar]
- 120.Lyn D, Cherney BW, Lalande M, Berenson JR, Lichtenstein A, Lupold S, Bhatia KG, Smulson M. A duplicated region is responsible for the poly(ADP-ribose) polymerase polymorphism, on chromosome 13, associated with a predisposition to cancer. Am. J. Hum. Genet. 1993;52:124–134. [PMC free article] [PubMed] [Google Scholar]
- 121.Pascual M, Lopez-Nevot MA, Caliz R, Ferrer MA, Balsa A, Pascual-Salcedo D, Martin J. A poly(ADP-ribose) polymerase haplotype spanning the promoter region confers susceptibility to rheumatoid arthritis. Arthritis Rheum. 2003;48:638–641. doi: 10.1002/art.10864. [DOI] [PubMed] [Google Scholar]