Abstract
Bipolar disorder is a severe mental illness characterized by mood swings of elation and depression. Family, twin, and adoption studies suggest a complex genetic etiology that may involve multiple susceptibility genes and an environmental component. To identify chromosomal loci contributing to vulnerability, we have conducted a genome-wide scan on ≈396 individuals from 22 multiplex pedigrees by using 607 microsatellite markers. Multipoint nonparametric analysis detected the strongest evidence for linkage at 13q32 with a maximal logarithm of odds (lod) score of 3.5 (P = 0.000028) under a phenotype model that included bipolar I, bipolar II with major depression, schizoaffective disorder, and recurrent unipolar disorder. Suggestive linkage was found on 1q31-q32 (lod = 2.67; P = 0.00022) and 18p11.2 (lod = 2.32; P = 0.00054). Recent reports have linked schizophrenia to 13q32 and 18p11.2. Our genome scan identified other interesting regions, 7q31 (lod = 2.08; P = 0.00099) and 22q11-q13 (lod = 2.1; P = 0.00094), and also confirmed reported linkages on 4p16, 12q23-q24, and 21q22. By comprehensive screening of the entire genome, we detected unreported loci for bipolar disorder, found support for proposed linkages, and gained evidence for the overlap of susceptibility regions for bipolar disorder and schizophrenia.
Bipolar disorder is a complex neuropsychiatric disease of unknown genetic etiology. Theories concerning multigenic inheritance involving multiple genes that exert varying magnitudes of effect on overall susceptibility have been advanced, but these genes have yet to be identified. Early linkage studies on bipolar disorder have been marked with uncertainty (1), possibly because of difficulties related to the power of detecting weak- to moderate-effect genes (2–4), to other confounding factors, or to spurious reported findings. Recent advances, which include the availability of dense maps containing highly informative markers, the use of extended pedigrees or larger sample collections, and the application of newer analytical methods, have propelled reports showing evidence of linkage to chromosomes 4p16 (5), 12q23-q24 (6), 18pcen-qcen (7, 8), 18q22-q23 (9), and 21q22.3 (10).
Previous chromosomal screens on our 22 multiply affected pedigrees elicited evidence of linkage to chromosome 18 (7, 8) and support for the proposed linkage on 21q22.3 (11). In contrast, our earlier scans that used sparsely spaced and moderately informative markers have not uncovered areas of linkage (12, 13). In the present study, we performed a high-density genome scan by using highly polymorphic markers to identify chromosomal regions that contribute to the genetic risk of bipolar disorder. Here, we report evidence for bipolar-disorder susceptibility loci, support of prior linkages, and possible overlaps of regions implicated in bipolar disorder and schizophrenia.
METHODS
Pedigrees.
A panel of 22 multiplex pedigrees was used for the genome screening, and the diagnostic and ascertainment methods used for 21 of these have been described in detail elsewhere (14). These families included ≈365 informative persons (i.e., those persons whose genotype can be determined either directly or indirectly; ref. 14). The 22nd family was the “right extension” of the Old Order Amish pedigree 110 (7).
To address the issue of uncertainty regarding the spectrum of illness that is inherited, two hierarchical models of the affected phenotype were used in linkage analysis. For affection status model (ASM) I, the definition of the affected category was restricted to bipolar I, bipolar II with major depression, and schizoaffective disorder. For the second model, ASM II, the phenotype definition was broadened to include those individuals with two or more episodes of major depression. Each pedigree had a minimum of four affected individuals under ASM II. There were 396 individuals that were available for genotyping, including 117 with ASM I and 159 with ASM II diagnoses.
Although we excluded branches of a pedigree when a person marrying into the pedigree had any of the diagnoses included in ASM I and ASM II, bilineality could not be ruled out, particularly if a multigenic model with common susceptibility alleles is assumed. However, when nonparametric methods are used, this issue may not be relevant. Bilineal families may reduce power but would not be expected to increase the incidence of false-positive results.
The power of this pedigree series to detect linkage has been examined in a prior study; this study used assumed parameters, because the true genetic model for bipolar disorder is not known (14). Simulations of linkage using 21 of the 22 families included in the present genome scan were performed under a dominant transmission model and ASM II by using the simlink program (15). Assuming the presence of heterogeneity, there was >50% power when at least 25% of the families are linked, for a marker with a polymorphism information content of 0.7 at Θ = 0.01. In a multigenic model, power depends on the effect of the susceptibility gene and whether it is acting in a multiplicative or additive fashion with other predisposing genes.
Genotyping.
The microsatellite markers used for genotyping consisted of tandem repeated dinucleotides and tetranucleotides, as well as a few trinucleotides. Markers were chosen from the following databases: the Cooperative Human Linkage Center (CHLC; www.chlc.org); the CHLC/Weber screening set, versions 6 and 8 (Research Genetics, Huntsville, AL); Genethon (ftp://ftp.genethon.fr/pub/Gmap/Nature-1995/); the Center for Medical Research of the Marshfield Medical Research Foundation (www.marshmed.org/genetics/); and the Genetic Location Database (ref. 16; cedar.genetics.soton.ac.uk/public_html/). A few markers were taken from the map created by the National Institutes of Health/Centre d’Étude du Polymorphisme Humain (Paris) Collaborative Mapping Group (mapper.wustl.edu/basemaps/index.html). The Marshfield map included almost all of the markers, and, in other instances, intermarker distance and order were deduced from the pedigrees by using crimap [Green, P., Falls, K. & Crooks, S. (1990) crimap Documentation (Department of Genetics, School of Medicine, Washington University, St. Louis), Version 2.4; linkage.rockefeller.edu/soft/crimap/].
With few exceptions, PCR amplification was done by using one set of cycling conditions: 95°C for 5 min for the initial denaturation, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. A final extension of 72°C for 10 min was done. Markers that failed under these conditions were replaced by other markers mapping to the same region. The controls CEPH 1333-01 and 1333-02 were placed in specific lanes with each set of samples to obtain consistent assignment of allele sizes from gel to gel. Most of the genotypings were radioactivity-based, and each autoradiogram was read by two independent readers who were blind to diagnosis. As size standards, we used an M13 sequence ladder and a 123-bp ladder. Automated genotyping was done with an Applied Biosystems (ABI) 373 sequencer (Perkin–Elmer), and alleles were scored with the genescan and genotyper programs. Unlabeled and fluorescently labeled primers were purchased from Research Genetics, although a few were synthesized through BioServe Biotechnology (Gaithersburg, MD). Mendelian inconsistencies were resolved before linkage analysis, and marker allele frequencies were computed from individuals marrying into the pedigree.
Linkage Analysis.
Linkage was evaluated under the affected phenotype classifications of ASM I and ASM II. Pointwise affected-sib-pair (ASP) analysis was done by including all ASPs by using the sibpal program, version 3.0, in the sage package (17). Multipoint analyses were performed by using aspex [Hinds, D. & Risch, N. (1996) aspex, version 1.62, sib_phase package; ftp://lahmed.stanford.edu/pub/aspex] and genehunter-plus (ghp), version 1.1 (18, 19). Sex-averaged maps were used for multipoint analyses. For chromosome 18, sex-specific maps were used in additional calculations. For the ghp analysis, individuals that were uninformative or that were connected to the pedigree by more than two uninformative generations were deleted. The right extension of the Amish pedigree 110 was divided into two families. Pedigrees were analyzed using the all function of ghp, which examines all individuals simultaneously and assigns a higher score when more of them share the same allele by descent. The probability of the observed sharing is calculated by using a semiparametric logarithm of odds (lod) score as described (19).
ghp analysis also was done after splitting the pedigrees into nuclear families (ghpfam). It has been shown through simulation that in ghp analysis, more distant relatives provide less information when a susceptibility allele is common (20). The reverse is true when the allele is rare. ghpfam was used when there was a significant difference between ghp and aspex data. If the results from ghpfam are similar to ghp, then the differences likely were caused by the different algorithms. On the other hand, if the ghpfam results are similar to aspex, then the family structure being analyzed likely is affecting the results.
Parametric lod score analysis was conducted by using fastlink, version 3.0 P [Schaffer, A. A., Gupta, S. K., Shriram, K. & Cottingham, R. W., Jr. (1997); ftp://fastlink.nih.gov/pub/fastlink]. Pairwise lod score calculations were done under a dominant model with 85% and 50% penetrance and under a recessive model with 85% penetrance.
Because we have employed different methods of analysis under two phenotype definitions, the most significant signals in any specific region may be inflated because of multiple testing. Biases in each analysis caused by deviations from assumptions also could occur. We believe, however, that the consistency of results derived from different analytical methods is important to consider, particularly if two or more methods detect lod scores of >2 in the same region. Moreover, support from other studies on independently ascertained families is critical to strengthening the validity of a finding.
RESULTS AND DISCUSSION
Genome-Wide Screening Data Analyzed by Nonparametric Methods.
We have screened 607 polymorphic loci covering all 22 autosomes and the X chromosome, with an average marker spacing of ≈6 centimorgans (cM) and an average heterozygosity of ≈60%. Model-free calculations identified clusters of loci in several regions of the genome that displayed excess identical-by-descent (IBD) sharing.
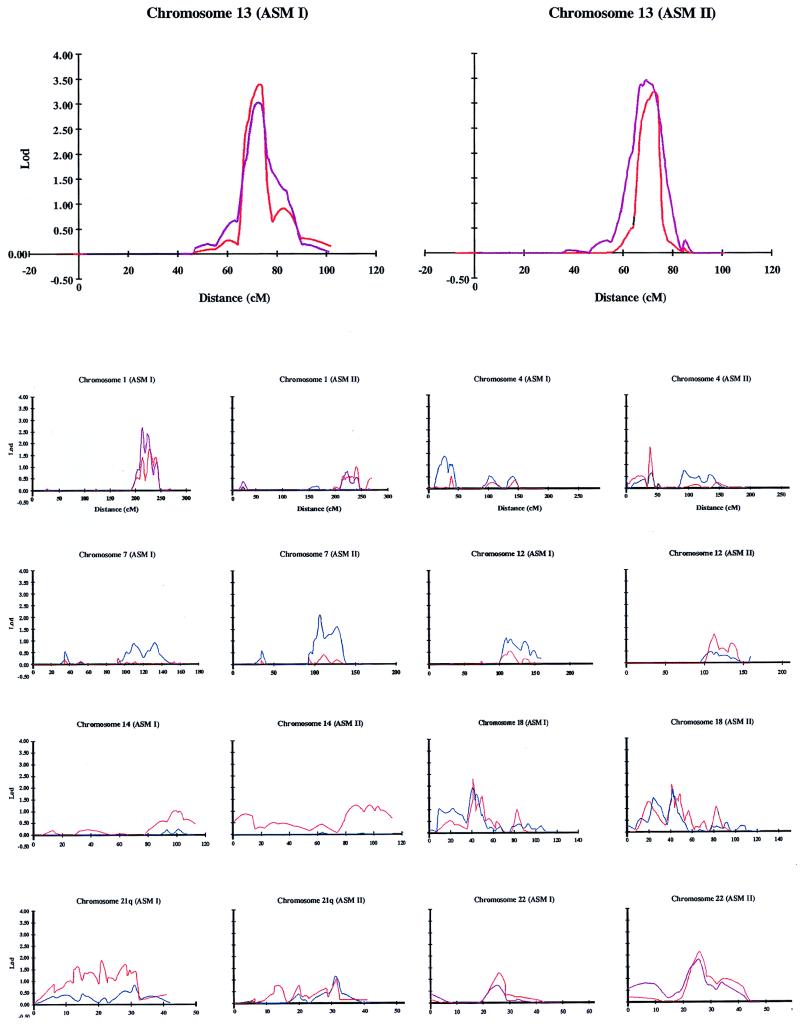
The strongest evidence for linkage derived from nonparametric analysis centered at chromosome 13q32, the only region that yielded a lod score of >3 (Table 1; Fig. 1). Pointwise analysis depicted excess IBD sharing in an ≈17-cM region maximizing at D13S1271 (64%; P = 0.0002) under ASM I. Analysis by ghpfam using nuclear families detected the highest multipoint lod at the D13S1252–D13S1271 interval (lod = 3.5; P = 0.000028) under ASM II. aspex localized the maximal peak in the D13S1271–D13S779 region for both ASM I (lod = 3.4; P = 0.000039) and ASM II (lod = 3.3; P = 0.000051). Broadening the classification of affected phenotypes to include recurrent major depression did not have a significant effect on the lod score. Based on statistical criteria proposed by Lander and Kruglyak (21), these values approach significant linkage (lod = 3.6; P = 0.000022) for a genome scan. Consistent with this finding was an excess sharing in this region in the 97 bipolar pedigrees of the National Institute of Mental Health Genetics Initiative (22). These data suggest a prominent role for a gene encoded by 13q32 in the genetic basis of affective disorder in these pedigrees. It is noteworthy that the susceptibility region on 13q32 overlapped the most significant region in a schizophrenia series (23). Examination of the diagnosis in the current pedigrees showed that there was at least one individual with schizoaffective disorder or schizophrenia in 15 of the families. Conceivably, the 13q32 gene is a predisposing factor for psychoses.
Table 1.
Multipoint linkage data derived through aspex showing lod of >1 (P ≤ 0.01)
| Chromosome | Region | ASM I (P) | ASM II (P) |
|---|---|---|---|
| 1q25-q32 | S471–S237 | 1.78 (0.0021)* | |
| 2pter | S2976 | 2.0 (0.0012) | |
| 2p25-p24 | S1400–S1360 | 1.11 (0.012) | |
| 4p16-p14 | S2408–S2632 | 1.77 (0.0022) | |
| 5q33-q35 | S498–S408 | 1.16 (0.01) | 1.7 (0.0026) |
| 11q13 | S1883–S913 | 1.89 (0.0016) | |
| 12q22-q24 | S1343–S2070 | 1.24 (0.0084) | |
| 13q32 | S1271–S779 | 3.4 (0.000039) | 3.3 (0.000051)† |
| 14q24 | S1434–S65 | 1.04 (0.014) | |
| 14q24-q32.2 | S617–S1434 | 1.23 (0.0087) | |
| 18p11.2 | S1150–S71 | 2.32 (0.00054) | 2.03 (0.0011) |
| 21q22.1 | S1254–S65 | 1.85 (0.0018) | |
| 22q11-q13 | S689–S685 | 1.26 (0.008) | 2.1 (0.00094) |
GHPfam yielded lod = 2.67 (P = 0.00022) at D1S1660–D1S1678.
GHPfam yielded lod = 3.5 (P = 0.000028) at D13S1252–D13S1271.
Figure 1.
Multipoint nonparametric analysis of a genome scan on chromosomes 13, 1, 4, 7, 12, 14, 18, 21, and 22. The affected phenotype models ASM I and ASM II are defined in the text. Plots were derived through aspex (red), ghpfam (purple), and ghp (blue).
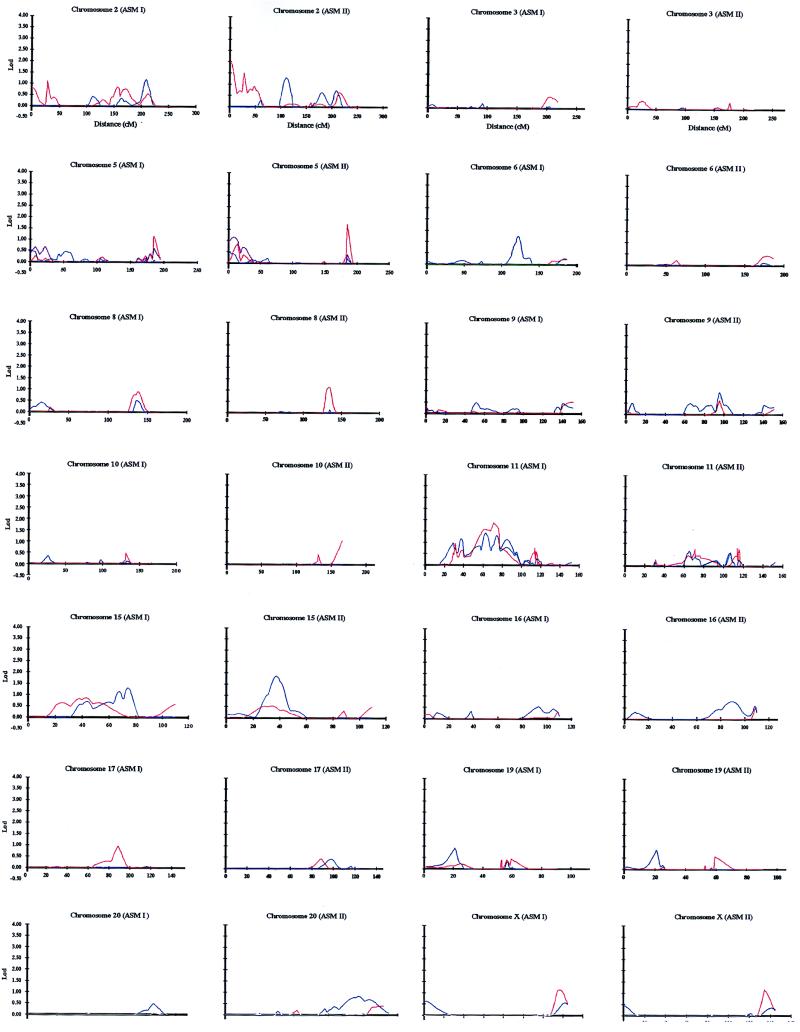
Regions on chromosomes 1, 7, 18, and 22 showed excessive allele sharing, yielding maximal multipoint lods of >2 but <3 (Table 1; Fig. 2), with values for 1q and 18p exceeding suggestive linkage (21). Pointwise analysis disclosed elevated IBD sharing, in an ≈30-cM region spanning 1q25-q42, which was more pronounced under ASM I. Addition of recurrent major depression to the stringent phenotypes, as in ASM II, may increase power, because it expands the number included in the affected category. On the contrary, this addition could have the opposite effect, because the genetic heterogeneity of the sample would be magnified. Multipoint analysis by ghpfam produced a peak of allele sharing at 1q25-q32 (ASM I lod = 2.67; P = 0.00022). Complementing this finding was a study that showed a frequent fragile site on 1q32 in bipolar patients (24). Together, these results raise the possibility that disruption of a gene on 1q32 contributes to overall susceptibility to bipolar disorder.
Figure 2.
Multipoint nonparametric analysis of a genome scan on chromosomes 2, 3, 5, 6, 8, 9, 10, 11, 15, 16, 17, 19, 20, and X. ASM I and ASM II are defined in the text. Analysis was done with aspex (red) and ghp (blue).
The candidate region on chromosome 18 encompassed a large area surrounding the centromere (7, 8). We increased marker coverage to an average spacing of 3.2 cM and localized the highest peak between D18S1150 and D18S71 (ASM I lod = 2.32; P = 0.00054) at 18p11.2 (25) by using aspex (Fig. 1). There was no significant discrepancy between the peak profiles for ASM I and ASM II. A telomeric, less prominent peak at ≈18p11.3 (ASM II ghp lod = 1.44; P = 0.005) and another small peak at ≈18q21 (lod = ≈1) also were noted. These weak signals may be relevant because an inversion involving 18p11.3 and 18q21.1 seemed to cosegregate with affective disorder in some families (26), and 18q21 included a proposed region for bipolar disorder (27, 28).
Additional multipoint aspex analysis was performed by using both male- and female-specific maps (data not shown). Both analyses generated peak height and location that were roughly similar to those found when the sex-averaged map was used (Fig. 1). A previous study postulated a parent-of-origin effect on bipolar disorder in this chromosome (27). We extended the analysis to calculate parental-specific lods by using sex-specific maps. Paternal allele sharing under ASM I was increased, consistent with a previous finding on these pedigrees (29). However, this increase seemed to be caused by a single large family in which the paternal haplotype was shared (data not shown).
A recent study implicated 18p11.2 in psychoses (30). Evidence for linkage and association in schizophrenia families increased when affective diagnoses were included in the affected category (30). The emergence of overlaps for bipolar disorder and schizophrenia loci on 13q32 (23) and 18p11.2 (30) is consistent with family studies that showed an increased risk among first degree relatives of schizophrenia probands for schizoaffective and recurrent unipolar disorders (31, 32).
Neither sibpal nor aspex detected a region of excess IBD sharing on chromosome 7. In contrast, ghp gave a lod of 2.08 (P = 0.00099) under ASM II between D7S1799 and D7S501 at 7q31. Coincident with this location was an area with elevated allele sharing in the bipolar series of the National Institute of Mental Health Genetics Initiative (33). The divergent results derived through aspex and ghp may be ascribed to random chance or to a rare susceptibility allele that is more likely to be detected when extended pedigrees are studied (20).
On chromosome 22, the maximal signal spanned D22S689 and D22S685 on 22q11-q13 (ASM II aspex lod = 2.1; P = 0.00094). In this region, microdeletions have been found in velo-cardio-facial syndrome, a dysmorphism associated with an enhanced prevalence of psychoses (34). Significant linkage to bipolar disorder (J. Kelsoe, personal communication) and support for a susceptibility locus in schizophrenia (23, 35) have been reported for this region of chromosome 22.
We have generated evidence supporting proposed linkages (21) on chromosomes 4p16 (see below), 12q23-q24, and 21q22 (Table 1; Fig. 1). The 12q23-q24 region has been linked to Darier’s disease (36), which was found to cosegregate with affective disorder (37). Screening of loci in the region identified significant linkage (21) to bipolar disorder in Danish kindreds (6) and in an extended French-Canadian pedigree (N. Barden, personal communication). Confirmatory data were detected in our families (ASM II aspex lod = 1.24; P = 0.0084). We have extended our earlier study on 21q22 (11) and found a broad region with excess sharing that included a proposed vulnerability locus (10), maximizing at D21S1254–D21S65 (ASM I aspex lod = 1.85; P = 0.0018).
Nonparametric analysis detected other loci with marginal signals (lod of >1 but <2; Table 1; Fig. 2). Excess IBD sharing (lod = 1.77) was found on 5q33-q35 (Table 1), and by two point analysis under a dominant transmission (85% penetrance), D5S462 gave a lod of 1.8 (Table 2). Lod scores of >1 under a dominant model have been reported for markers mapping to this region (38). Excess sharing was displayed by a marker mapping to 5q35 in the National Institute of Mental Health Genetics Initiative bipolar pedigrees (39). The increase in IBD sharing on chromosome 11q13 (lod = ≈1.9) seemed to be supported by parametric analysis, because D11S4076, located in this region, gave a lod of 1.55 under a dominant transmission (Table 2). A cautious interpretation of these signals is warranted, because several methods of analysis under two phenotype models were employed; therefore, signals could have arisen from random statistical fluctuations.
Table 2.
Pairwise parametric lod scores >1.5 (θ = 0.2) for the entire pedigree series
| Chromosome | Locus | ASM I | ASM II |
|---|---|---|---|
| 1q32 | GATA124F08 | 2.37 (dom, 85% pen) | |
| 5q35 | D5S462 | 1.80 (dom, 85% pen) | 1.54 (dom, 85% pen) |
| 7p15-q22 | D7S492 | 1.59 (dom, 50% pen) | |
| 10q25 | D10S187 | 1.69 (rec, 85% pen) | |
| 11p15-p14 | D11S915 | 1.84 (dom, 50% pen) | |
| 11p14-p13 | D11S904 | 1.62 (dom, 50% pen) | |
| 11q12.3-q13.2 | D11S4076 | 1.55 (dom, 50% pen) | |
| 13q32 | D13S1271 | 2.06 (rec, 85% pen) | 1.69 (rec, 85% pen) |
| 14q11-q13 | D14S1060 | 2.19 (rec, 85% pen) | |
| 14q32.1-q32.2 | D14S1434 | 1.79 (rec, 85% pen) | |
| 18p11.2 | D18S1353 | 1.78 (dom, 85% pen) | |
| 18p11.2 | D18S40 | 1.70 (dom, 50% pen) | |
| 21q22.1-p22.3 | D21S267 | 1.57 (rec, 85% pen) | |
| 21q22.3-qter | D21S212 | 1.79 (rec, 85% pen) |
Parametric analysis was done with fastlink (see Methods). dom, dominant; rec, recessive, pen, penetrance.
Genome-Wide Screening Data Analyzed by a Parametric Method.
Parametric calculations identified two loci giving lods of >3 in the two largest pedigrees. Pedigree 0048 had 39 members that were genotyped, with six affected under ASM I (14). On 4p16, a region that showed linkage to bipolar disorder in Scottish families (5), D4S2632 gave a lod of 3.24 (Θ = 0) for ASM I under a dominant, 50% penetrance model. Model-free analysis by aspex showed elevated allele sharing within the D4S2408–D4S2632 interval (ASM II lod = 1.77; P = 0.0022; Table 1; Fig. 2). Hence, these data support the reported linkage (5), and the gene on 4p16 may confer a major effect on susceptibility in pedigree 0048.
Pedigree 1482 had 30 members genotyped with 11 affected under ASM II (14). At D14S617 on 14q23-q32.1, this pedigree gave a lod of 3.19 (Θ = 0; dominant transmission; 85% penetrance) under ASM II. Approximately 8 cM distal to D14S617 on 14q32.1-q32.2, D14S1434 displayed a total lod score of 1.79 for the entire pedigree series under ASM I and a recessive, 85% penetrance model (Table 2). aspex analysis displayed increased sharing in the region (ASM II lod = 1.23; P = 0.0087; Table 1; Fig. 1). At a more proximal location, D14S1060, which maps to 14q11-q13, a region involved in a balanced translocation in a patient with bipolar disorder (40), displayed a lod of >2 under ASM II and a recessive transmission (Table 2). It is unclear whether these signals would be supported on further scrutiny of other pedigree series.
Markers that produced parametric lods of >1.5 for the entire panel tended to reinforce findings from nonparametric analysis on 1q32, 13q32, 18p11.2, and 21q22 (Table 2), even though parameters involved in the genetic inheritance of bipolar disorder are not known. GATA124F08 on 1q32 yielded a lod of >2, assuming a dominant mode of transmission, and D13S1271 on 13q32 yielded a lod of >2 under a recessive model. Similarly, the peak heterogeneity lod in a schizophrenia series for a marker on 13q32 was found by using a recessive model (23). The highest parametric lods and IBD sharing on chromosome 18 were found in loci mapping to 18p11.2 (Fig. 1). The distal portion of 21q also had markers with lods of >1.5 under a recessive model, which is consistent with findings from model-free analysis. Despite the concurrence of signals derived from different analytical methods and some apparent corroborating evidence from other studies, verification of linkage requires additional confirmation from other pedigree series and must await cloning of genes manifesting variants that cosegregate with the affected phenotype.
By implementing a high-density genome scan, we detected potential susceptibility loci for bipolar disorder on 13q32, 1q32, and 18p11.2 and found support for proposed predisposing regions. Our data, coupled with independent findings, suggest the existence of shared loci for bipolar disorder and schizophrenia and may provide evidence for the continuum model of psychosis (41).
Acknowledgments
We thank the patients and their family members for their participation in the study.
ABBREVIATIONS
- lod
logarithm of odds
- ASM
affection status model
- ASP
affected sib pair
- ghp
the genehunter-plus program
- ghpfam
ghp analysis with pedigrees split into nuclear families
- cM
centimorgan
- IBD
identical by descent
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Risch N, Botstein D. Nat Genet. 1996;12:351–353. doi: 10.1038/ng0496-351. [DOI] [PubMed] [Google Scholar]
- 2.Hauser E R, Boehnke M, Guo S-W, Risch N. Genet Epidemiol. 1996;13:117–137. doi: 10.1002/(SICI)1098-2272(1996)13:2<117::AID-GEPI1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Suarez B K, Hampe C L, Van Eerdewegh P. In: Genetic Approaches to Mental Disorders. Gershon E S, Cloninger C R, editors. Washington, DC: American Psychiatric Institute; 1994. pp. 23–46. [Google Scholar]
- 4.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 5.Blackwood D H R, He L, Morris S W, McLean A, Whitton C, Thomson M, Walker M T, Woodburn K, Sharp C M, Wright A F, et al. Nat Genet. 1996;12:427–430. doi: 10.1038/ng0496-427. [DOI] [PubMed] [Google Scholar]
- 6.Ewald H, Degn B, Mors O, Kruse T A. Psychiatr Genet. 1998;8:131–140. doi: 10.1097/00041444-199800830-00002. [DOI] [PubMed] [Google Scholar]
- 7.Berrettini W H, Ferraro T N, Goldin L R, Weeks D E, Detera-Wadleigh S D, Nurnberger J I, Jr, Gershon E S. Proc Natl Acad Sci USA. 1994;91:5918–5921. doi: 10.1073/pnas.91.13.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berrettini W H, Ferraro T N, Choi H, Goldin L R, Detera-Wadleigh S D, Muniec D, Hsieh W-T, Hoehe M, Guroff J J, Kazuba D, et al. Arch Gen Psychiatry. 1997;54:32–39. doi: 10.1001/archpsyc.1997.01830130031006. [DOI] [PubMed] [Google Scholar]
- 9.Freimer N B, Reus V I, Escamilla M E, McInnes L A, Spesny M, Leon P, Service S K, Smith L B, Silva S, Rojas E, et al. Nat Genet. 1996;12:436–441. doi: 10.1038/ng0496-436. [DOI] [PubMed] [Google Scholar]
- 10.Straub R E, Lehner T, Luo Y, Loth J E, Shao W, Sharpe L, Alexander J R, Das K, Simon R, Fieve R R, et al. Nat Genet. 1994;8:291–296. doi: 10.1038/ng1194-291. [DOI] [PubMed] [Google Scholar]
- 11.Detera-Wadleigh S D, Badner J A, Goldin L R, Berrettini W H, Sanders A R, Rollins D Y, Turner G, Moses T, Haerian H, Muniec D, et al. Am J Hum Genet. 1998;58:1279–1285. [PMC free article] [PubMed] [Google Scholar]
- 12.Berrettini W H, Detera-Wadleigh S D, Goldin L R, Martinez M, Hsieh W-T, Hoehe M, Choi H, Muniec D, Ferraro T N, Guroff J J, et al. Psychiatr Genet. 1991;2:191–208. [Google Scholar]
- 13.Detera-Wadleigh S D, Hsieh W-T, Berrettini W H, Goldin L R, Rollins D Y, Muniec D, Grewal R, Guroff J J, Turner G, Coffman D, et al. Am J Med Genet. 1994;54:206–218. doi: 10.1002/ajmg.1320540307. [DOI] [PubMed] [Google Scholar]
- 14.Berrettini W H, Goldin L R, Martinez M M, Maxwell E, Smith A L, Guroff J J, Kazuba D M, Nurnberger J I, Jr, Hamovit J, Simmons-Alling S, et al. Psychiatr Genet. 1991;2:125–160. [Google Scholar]
- 15.Boehnke M. Am J Hum Genet. 1986;39:513–527. [PMC free article] [PubMed] [Google Scholar]
- 16.Collins A, Frezal J, Teague J, Morton N E. Proc Natl Acad Sci USA. 1996;93:14771–14775. doi: 10.1073/pnas.93.25.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Department of Epidemiology and Biostatistics. sage, Statistical Analysis for Genetic Epidemiology. Cleveland: Case Western Reserve Univ.; 1997. , Release 3.0. [Google Scholar]
- 18.Kruglyak L, Daly M J, Reeve-Daly M P, Lander E S. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 19.Kong A, Cox N J. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badner J A, Gershon E S, Goldin L R. Am J Hum Genet. 1998;63:880–888. doi: 10.1086/302007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander E S, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 22.Stine O C, McMahon F J, Chen L, Xu J, Meyers D A, MacKinnon D F, Simpson S, McInnis M G, Rice J P, Goate A, et al. Am J Med Genet. 1997;74:263–269. [PubMed] [Google Scholar]
- 23.Blouin J-L, Dombroski B A, Nath S K, Lasseter V K, Wolyniec P S, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, et al. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 24.Turecki G, Smith M, Mari J J. Am J Med Genet. 1995;60:179–182. doi: 10.1002/ajmg.1320600302. [DOI] [PubMed] [Google Scholar]
- 25.Esterling L E, Matise T C, Sanders A R, Yoshikawa T, Overhauser J, Gershon E S, Moskowitz M, Detera-Wadleigh S D. Mol Psychiatry. 1997;2:501–504. doi: 10.1038/sj.mp.4000317. [DOI] [PubMed] [Google Scholar]
- 26.Mors O, Ewald H, Blackwood D, Muir W. Br J Psychiatry. 1997;170:278–280. doi: 10.1192/bjp.170.3.278. [DOI] [PubMed] [Google Scholar]
- 27.Stine O C, Xu J, Koskela R, McMahon F, Gschwend M, Friddle C, Clark C D, McInnis M G, Simpson S G, Breschel T S, et al. Am J Hum Genet. 1995;57:1384–1394. [PMC free article] [PubMed] [Google Scholar]
- 28.McMahon F J, Hopkins P J, Xu J, McInnis M G, Shaw S, Cardon L, Simpson S G, MacKinnon D F, Stine O C, Sherrington R, et al. Am J Hum Genet. 1997;61:1397–1404. doi: 10.1086/301630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershon E S, Badner J A, Detera-Wadleigh S D, Ferraro T N, Berrettini W H. Am J Med Genet. 1996;67:202–207. doi: 10.1002/(SICI)1096-8628(19960409)67:2<202::AID-AJMG11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 30.Schwab S G, Hallmayer J, Lerer B, Albus M, Borrmann M, Hönig S, Strauss M, Segman R, Lichtermann D, Knapp M, et al. Am J Hum Genet. 1998;63:1139–1152. doi: 10.1086/302046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gershon E S, DeLisi L E, Hamovit J, Nurnberger J I, Jr, Maxwell M E, Schreiber J, Dauphinais D, Dingman C W, II, Guroff J J. Arch Gen Psychiatry. 1988;45:328–336. doi: 10.1001/archpsyc.1988.01800280038006. [DOI] [PubMed] [Google Scholar]
- 32.Maier W, Lichtermann D, Minges J, Hallmayer J, Heun R, Benkert O, Levinson D. Arch Gen Psychiatry. 1993;50:871–883. doi: 10.1001/archpsyc.1993.01820230041004. [DOI] [PubMed] [Google Scholar]
- 33.Detera-Wadleigh S D, Badner J A, Yoshikawa T, Sanders A R, Goldin L R, Turner G, Rollins D Y, Moses T, Guroff J J, Kazuba D, et al. Am J Med Genet. 1997;74:254–262. doi: 10.1002/(sici)1096-8628(19970531)74:3<254::aid-ajmg4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 34.Pulver A E, Nestadt G, Goldberg R, Shprintzen R J, Lamacz M, Wolyniec P S, Morrow B, Karayiorgou M, Antonarakis S E, Housman D. J Nerv Ment Dis. 1994;182:476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Schizophrenia Collaborative Linkage Group (Chromosome 22) Am J Med Genet. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Craddock N, Dawson E, Burge S, Parfitt L, Mant B, Roberts Q, Daniels J, Gill M, McGuffin P, Powell J, et al. Hum Mol Genet. 1993;2:1941–1943. doi: 10.1093/hmg/2.11.1941. [DOI] [PubMed] [Google Scholar]
- 37.Craddock N, Owen M, Burge S, Kurina B, Thomas P, McGuffin P. Br J Psychiatry. 1994;164:355–358. doi: 10.1192/bjp.164.3.355. [DOI] [PubMed] [Google Scholar]
- 38.Coon H, Jensen S, Hoff M, Holik J, Plaetke R, Reimherr F, Wender P, Leppert M, Byerley W. Am J Hum Genet. 1993;52:1234–1249. [PMC free article] [PubMed] [Google Scholar]
- 39.Edenberg H J, Foroud T, Conneally P M, Sorbel J J, Carr K, Crose C, Willig C, Zhao J, Miller M, Bowman E, et al. Am J Med Genet. 1997;74:238–246. [PubMed] [Google Scholar]
- 40.Overhauser J, Berrettini W H, Rojas K. Psychiatr Genet. 1998;8:53–56. doi: 10.1097/00041444-199800820-00004. [DOI] [PubMed] [Google Scholar]
- 41.Crow T J. Br J Psychiatry. 1986;149:419–429. doi: 10.1192/bjp.149.4.419. [DOI] [PubMed] [Google Scholar]