Abstract
Clostridium thermocellum polynucleotide kinase-phosphatase (CthPnkp) catalyzes 5′ and 3′ end-healing reactions that prepare broken RNA termini for sealing by RNA ligase. The central phosphatase domain of CthPnkp belongs to the dinuclear metallophosphoesterase superfamily exemplified by bacteriophage λ phosphatase (λ-Pase). CthPnkp is a Ni2+/Mn2+-dependent phosphodiesterase-monoesterase, active on nucleotide and non-nucleotide substrates, that can be transformed toward narrower metal and substrate specificities via mutations of the active site. Here we characterize the Mn2+-dependent 2′,3′ cyclic nucleotide phosphodiesterase activity of CthPnkp, the reaction most relevant to RNA repair pathways. We find that CthPnkp prefers a 2′,3′ cyclic phosphate to a 3′,5′ cyclic phosphate. A single H189D mutation imposes strict specificity for a 2′,3′ cyclic phosphate, which is cleaved to form a single 2′-NMP product. Analysis of the cyclic phosphodiesterase activities of mutated CthPnkp enzymes illuminates the active site and the structural features that affect substrate affinity and kcat. We also characterize a previously unrecognized phosphodiesterase activity of λ-Pase, which catalyzes hydrolysis of bis-p-nitrophenyl phosphate. λ-Pase also has cyclic phosphodiesterase activity with nucleoside 2′,3′ cyclic phosphates, which it hydrolyzes to yield a mixture of 2′-NMP and 3′-NMP products. We discuss our results in light of available structural and functional data for other phosphodiesterase members of the binuclear metallophosphoesterase family and draw inferences about how differences in active site composition influence catalytic repertoire.
INTRODUCTION
Incision of the phosphodiester backbone of DNA or RNA can generate either 3′-OH/5′PO4 or 3′-PO4(or 2′,3′ cyclic phosphate)/5′-OH breaks. Whereas 3′-OH/5′-PO4 termini can be sealed directly by polynucleotide ligases, 3′-PO4(or 2′,3′ cyclic phosphate)/5′-OH ends must be ‘healed’ before they can be sealed. Healing entails two steps: (i) phosphorylation of the 5′-OH terminus to form a 5′-PO4 and (ii) removal of the 3′-PO4 to form a 3′-OH terminus. End-healing is carried out by polynucleotide kinase-phosphatase (Pnkp) enzymes, of which bacteriophage T4 Pnkp is the prototype (1–7). Additional Pnkps have been characterized from diverse viral and cellular sources (8–12). Clostridium thermocellum Pnkp (CthPnkp) is the exemplary bacterial end-healing enzyme. CthPnkp is a trifunctional protein composed of N-terminal kinase, central phosphoesterase and C-terminal adenylyltransferase domains (12–14). Trifunctional and bifunctional homologs (containing only the kinase and phosphoesterase domains) of CthPnkp are found in several other bacterial genera. The CthPnkp kinase module catalyzes phosphoryl transfer from ATP to the 5′-OH terminus of DNA or RNA polynucleotides. The phosphoesterase domain releases Pi from 2′-PO4, or 3′-PO4 or 2′,3′ cyclic phosphate mononucleotides. Packaging of the kinase and phosphatase activities within a single polypeptide suggested an RNA repair function for CthPnkp, analogous to that of T4 Pnkp (15,16). Indeed, we showed recently that CthPnkp is able to heal a broken tRNA with 2′,3′ cyclic phosphate and 5′-OH termini, yielding 3′-OH and 5′-PO4 ends that are substrates for sealing by T4 RNA ligase 1 (17).
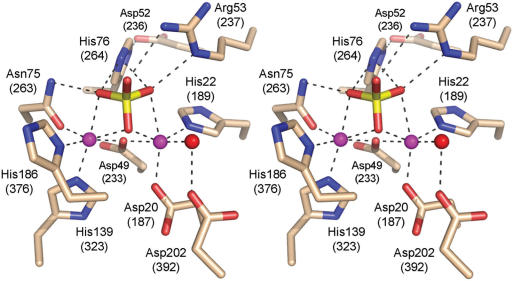
CthPnkp and its bacterial homologs differ from their viral and eukaryal analogs with respect to the structure and mechanism of the phosphatase domain. Viral and eukaryal Pnkp phosphatases belong to the DXDXT superfamily of enzymes that catalyze phosphoester hydrolysis via formation of a covalent aspartyl-phosphate intermediate (7,18,19). The CthPnkp phosphatase belongs to the calcineurin-like metallophosphoesterase superfamily, whose members characteristically utilize a dinuclear metal center to catalyze phosphoester hydrolysis (20,21). Bacteriophage λ phosphatase (λ-Pase) is one of the best-studied members of this superfamily and uses either Ni2+ or Mn2+ to catalyze phosphoester hydrolysis in various phosphopeptides, phosphoproteins and nonspecific organic phosphomonoester substrates such as p-nitrophenyl phosphate and naphthyl phosphate (22–24). Although λ-Pase is commonly dubbed a phosphoprotein phosphatase, its physiological substrates are not known. The crystal structure of λ-Pase bound to a sulfate ion (that likely mimics the phosphate in the product complex) shows two manganese ions coordinated by histidines, aspartates, an asparagine, a water molecule and three sulfate oxygens (Figure 1) (25). The phosphatase domain of CthPnkp is homologous to λ-Pase (13) and contains counterparts of all of the residues that bind to the metal ions and coordinate the sulfate/phosphate in the active site (Figure 1).
Figure 1.
The active site of λ phosphatase. Stereo view of the active site of λ-Pase (protomer C) from the crystal structure of Voegtli et al. (25) (PDB accession code 1G5B). The amino acid side chains coordinating the binuclear metal cluster and the sulfate ion are shown. The corresponding amino acids of CthPnkp are indicated in parentheses. The manganese ions are colored magenta. Water is colored red.
Previous studies highlighted CthPnkp as a Ni2+/Mn2+-dependent phosphodiesterase-monoesterase that can be transformed toward narrower metal and substrate specificities via mutations of the active site (12–14). Indeed, analyses of an extensive collection of mutants showed that the phosphodiesterase and monoesterase reactions rely on overlapping but different ensembles of active site functional groups and that the requirements for the phosphodiesterase and monoesterase activities can vary according to the divalent cation cofactor (13,14). For example, whereas seven active site side chains (Asp187, His189, Asp233, Asn263, His323, His376 and Asp392) are required for Ni2+-dependent hydrolysis of p-nitrophenyl phosphate, the Mn2+-dependent phosphomonoesterase requires two additional residues: Arg237 and His264. The CthPnkp phosphodiesterase converts bis-p-nitrophenyl phosphate to p-nitrophenol and p-nitrophenyl phosphate. The Ni2+-dependent phosphodiesterase activity of CthPnkp requires the same seven side chains as the Ni2+-dependent phosphomonoesterase. However, the Mn2+-dependent phosphodiesterase activity does not require His189, Arg237 or His264, each of which is critical for the Mn2+-dependent phosphomonoesterase (14).
The metal and substrate specificities of CthPnkp can be altered dramatically by mutations of the active site (14). For example, mutations H189A, H189D and D392N transform CthPnkp into a Mn2+-dependent phosphodiesterase devoid of monoesterase activity. The H189E change results in a Mn2+/Ni2+-dependent phosphodiesterase. Mutations H376N, H376D and D392E convert the enzyme into a Mn2+-dependent phosphodiesterase-monoesterase. Such effects attest to the plasticity of the catalytic repertoire, and perhaps the catalytic mechanism.
Here we extend our studies of CthPnkp substrate specificity in two directions. We show that CthPnkp readily hydrolyzes p-nitrophenyl phenylphosphonate utilizing the same ensemble of active site residues required for cleaving the phosphodiester bis-p-nitrophenyl phosphate. This result signifies that a phosphorus center with only three oxygen atoms suffices for phosphoester hydrolysis by CthPnkp. We analyze in depth the 2′,3′ cyclic phosphodiesterase of CthPnkp, in light of the fact that 2′,3′ cyclic phosphate termini are the predominant products of the known purposeful RNA damage pathways (15,26–34). We find that wild-type CthPnkp prefers a 2′,3′ cyclic phosphate to a 3′,5′ cyclic phosphate. Moreover, the H189D mutation enforces absolute specificity for a 2′,3′ cyclic phosphate, which it hydrolyzes to yield a 2′-NMP as the sole product.
We also characterize a phosphodiesterase activity of λ-Pase, the first time, to our knowledge, that such an activity has been reported for this enzyme. λ-Pase catalyzes the release of p-nitrophenol from bis-p-nitrophenyl phosphate and p-nitrophenyl phenylphosphonate. λ-Pase also has cyclic phosphodiesterase activity with nucleoside 2′,3′ cyclic phosphates, which it hydrolyzes to form a mixture of 2′-NMP and 3′-NMP products.
MATERIALS AND METHODS
Materials
p-nitrophenyl phosphate, p-nitrophenyl phenylphosphonate, bis-p-nitrophenyl phosphate, thymidine 5′-monophosphate p-nitrophenyl ester, p-nitrophenyl phosphorylcholine, dimethyl p-nitrophenyl phosphate and p-nitrophenol were purchased from Sigma. cAMP, cGMP and cUMP were purchased from Sigma. Concentrations of nucleotide stock solutions were determined by UV absorbance. Malachite green reagent (35) was purchased from BIOMOL Research Laboratories, Plymouth Meeting, PA.
Recombinant CthPnkp
Wild-type CthPnkp and mutated versions with single amino acid substitutions were produced in Escherichia coli as His10-tagged fusions and purified from soluble bacterial extracts by Ni-agarose chromatography as described previously (13,14).
Recombinant λ phosphatase
The open reading frame encoding λ-Pase was amplified from bacteriophage genomic DNA with primers that introduced an NdeI site at the start codon and a BamHI site 3′ of the stop codon. The PCR product was digested with NdeI and BamHI and inserted into pET16b to generate an expression plasmid encoding the λ-Pase polypeptide fused to an N-terminal His10 tag. The insert was sequenced to exclude the acquisition of unwanted coding changes during amplification or cloning. The pET-λ-Pase plasmid was transformed into E. coli BL21(DE3). A 200 ml culture was grown at 37°C in Luria-Bertani medium containing 0.1 mg/ml ampicillin until the A600 reached ∼0.6. The culture was adjusted to 0.3 mM isopropyl-β-d-thiogalatopyranoside and incubated at 37°C for 3 h with continuous shaking. Cells were harvested by centrifugation, and the pellet was stored at –80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 20 ml of buffer A (50 mM Tris–HCl, pH 7.5, 0.25 M NaCl, 10% sucrose). Lysozyme, PMSF and Triton X-100 were added to final concentrations of 1 mg/ml, 0.5 mM and 0.1%, respectively. The lysate was sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extract was applied to a 1 ml column of Ni-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with buffer A. The column was washed with 10 ml of the same buffer and then eluted stepwise with 4 ml aliquots of 50, 100 and 200 mM imidazole in buffer B (50 mM Tris–HCl, pH 8.0, 0.25 M NaCl, 10% glycerol, 0.05% Triton X-100). The polypeptide compositions of the column fractions were monitored by SDS–PAGE. The His10-λ-Pase polypeptide was recovered predominantly in the 200 mM imidazole fraction. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The λ-Pase preparation was stored at –80°C.
RESULTS
CthPnkp catalyzes hydrolysis of p-nitrophenyl phenylphosphonate
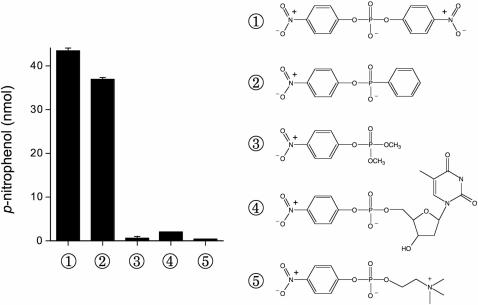
CthPnkp is adept at hydrolyzing both nucleotide and non-nucleotide phosphomonoester/phosphodiester substrates. To further query the non-nucleotide substrate specificity of CthPnkp, we assayed the enzyme for activity with the phosphoester substrates bis-p-nitrophenyl phosphate, p-nitrophenyl phenylphosphonate, dimethyl-p-nitrophenyl phosphate, thymidine 5′-monophosphate-p-nitrophenyl ester, and p-nitrophenyl phosphorylcholine (Figure 2). CthPnkp hydrolyzed p-nitrophenyl phenylphosphonate to a similar extent as bis-p-nitrophenyl phosphate, thereby signifying that three oxygen atoms linked to the phosphorus center sufficed for phosphoester hydrolysis (Figure 2). CthPnkp did not release p-nitrophenol from any of the other phosphodiesters or triester substrates tested.
Figure 2.
Substrate specificity of CthPnkp. Reaction mixtures (25 µl) containing 50 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 1.5 µg of CthPnkp and 10 mM substrate as specified were incubated for 30 min at 45°C. The reactions were quenched by adding 20 mM EDTA and then 0.9 ml of 1 M Na2CO3. Release of p-nitrophenol was determined by measuring A410 and interpolating the value to a p-nitrophenol standard curve. The extents of formation of p-nitrophenol are plotted at left. The chemical structures of the substrates bis-p-nitrophenyl phosphate ①, p-nitrophenyl phenylphosphonate ②, dimethyl-p-nitrophenyl phosphate ③, thymidine 5′-monophosphate-p-nitrophenyl ester ④ and p-nitrophenyl phosphorylcholine ⑤ are shown at right.
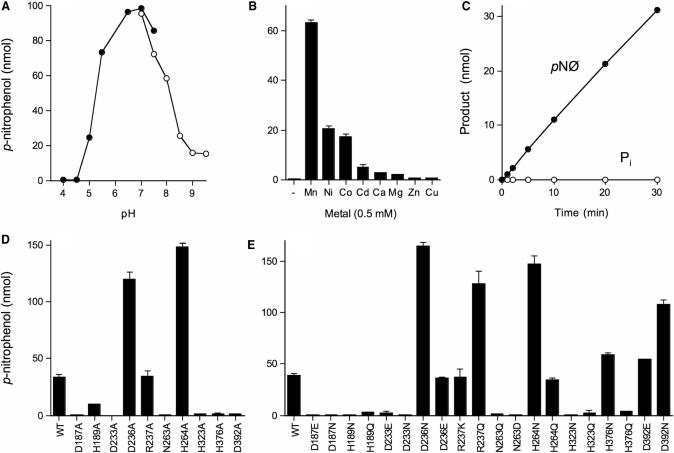
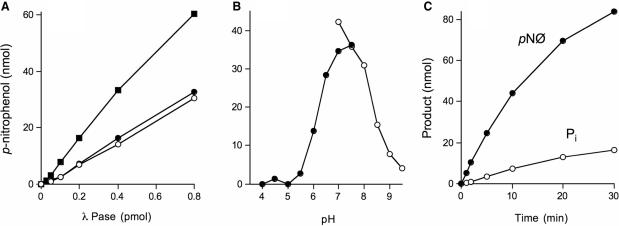
It was surprising to us that CthPnkp had such vigorous activity with the phosphonate substrate, given the extensive contacts of the homologous λ-Pase with the four sulfate oxygens in the crystal structure (Figure 1). Consequently, we examined in greater detail the requirements for the reaction of CthPnkp with p-nitrophenyl phenylphosphonate. Activity was optimal at pH 7.0 in the presence of Mn2+ and declined sharply at pH < 5.5 or >8.0 (Figure 3A). A divalent metal ion was required. Mn2+ (0.5 mM) supported optimal activity while Ni2+ and Co2+ (0.5 mM) supported one-third the activity of Mn2+. Magnesium, calcium, cobalt, cadmium, copper and zinc (at 0.5 mM concentration) were either inactive or much less active than manganese (Figure 3B). From a double-reciprocal plot of the dependence of p-nitrophenol production as a function of p-nitrophenyl phenylphosphonate substrate concentration in Tris–HCl buffer at pH 7.0 and 0.5 mM MnCl2, we calculated a Km of 67 mM p-nitrophenyl phenylphosphonate and a kcat of 1670 min−1 (data not shown). A comparison to the kinetic parameters reported previously (14) for Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate by CthPnkp (Km 88 mM; kcat 1390 min−1) indicates that the absence of the fourth oxygen at the phosphorus center had little impact on substrate binding or catalysis.
Figure 3.
Requirements for hydrolysis of p-nitrophenyl phenylphosphonate. (A) Reaction mixtures (25 µl) containing 50 mM Tris–acetate (filled circle) or Tris–HCl (open circle) at the pH specified, 0.5 mM MnCl2, 10 mM p-nitrophenyl phenylphosphonate and 1 µg wild-type CthPnkp were incubated for 30 min at 45°C. (B) Reaction mixtures (25 µl) containing 50 mM Tris–HCl (pH 7.0), 10 mM p-nitrophenyl phenylphosphonate, 1 µg of CthPnkp and either no divalent cation (lane –) or 0.5 mM MnCl2, NiCl2, CoCl2, CdCl2, CaCl2, MgCl2, ZnCl2 or CuCl2 were incubated for 30 min at 45°C. Each datum in the bar graph is the average of two separate experiments. S.E. bars are shown. (C) Reaction mixtures (350 µl) containing 50 mM Tris–HCl (pH 7.0), 10 mM p-nitrophenyl phenylphosphonate, 0.5 mM MnCl2 and 7 µg CthPnkp were incubated at 45°C. Two aliquots (25 µl) were withdrawn at the times specified, quenched immediately with EDTA, and assayed for p-nitrophenol (filled circle) and inorganic phosphate (open circle). (D and E) Reaction mixtures (25 µl) containing 50 mM Tris–HCl (pH 7.0), 0.5 mM MnCl2, 10 mM p-nitrophenyl phenylphosphonate and 0.5 µg wild-type or mutant CthPnkp as specified were incubated for 30 min at 45°C. Each datum in the bar graph is the average of two separate experiments. S.E. bars are shown.
There are two potential routes to the generation of p-nitrophenol from p-nitrophenyl phenylphosphonate. Cleavage of the P–O bond would directly yield benzyl phosphate and p-nitrophenol. Cleavage of the P–C bond would generate benzene and p-nitrophenyl phosphate, the latter species being readily hydrolyzed by CthPnkp to p-nitrophenol and Pi (13). To discriminate the two pathways, we carried out a kinetic analysis measuring the formation of p-nitrophenol and phosphate. We observed only the release of p-nitrophenol over the 30 min reaction (Figure 3C). We surmise that CthPnkp hydrolyzes p-nitrophenyl phenylphosphonate to form benzyl phosphate and p-nitrophenol.
Active site requirements for hydrolysis of p-nitrophenyl phenylphosphonate
To gauge which active site constituents are needed for hydrolysis of the phosphonate substrate, we tested a collection of 10 alanine mutants of CthPnkp (13). Six of the alanine mutations reduced activity to ≤5% of the wild-type enzyme (Figure 3D). The residues defined thereby as essential for Mn2+-dependent hydrolysis of p-nitrophenyl phenylphosphonate included 6/7 of the predicted direct or water-mediated ligands of the dinuclear metal cluster: Asp187, Asp233, Asn263, His323, His376 and Asp392. Loss of the seventh metal ligand, His189, reduced activity to 30% of wild-type. Removal of Arg237 had no impact on hydrolysis of p-nitrophenyl phenylphosphonate. Deletion of the Asp236 and His264 side chains beyond the β carbon resulted in a several-fold increase in activity (Figure 3D). These alanine effects are similar to what we observed previously for Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate (14).
Structure-function relationships at the active site were inferred from the effects of 20 conservative mutations (Figure 3E). Asp187, Asp233, Asn263 and His323 were strictly essential, insofar as the conservative substitutions did not restore activity over the level seen for the respective alanine mutants. Replacing His376 with asparagine fully revived activity, but glutamine was of no benefit. The Asn376 Nδ amide nitrogen is a putative mimetic of the His376 Nδ atom that coordinates a manganese ion in the active site (Figure 1). The essential Asp392 side chain, which coordinates a metal-bound water, can be replaced functionally by either glutamate or glutamine, either of which has the potential to coordinate a water (Figure 3E). It is notable that whereas the H189A change elicited a modest decrement in activity, the conservative substitutions with asparagine and glutamine were more deleterious.
H264N mimicked the gain of function seen with H264A, while H264Q had wild-type activity (Figure 3E). Reference to the active site structure suggests that the loss of the hydrogen bond of the His264 Nε to the phosphate oxygens is probably responsible for the increased activity (Figure 1). The Nε contacts would be mimicked by glutamine but not by asparagine. D236N elicited a gain of function over wild-type CthPnkp, similar to D236A, while D236E had wild-type activity (Figure 3E), indicating that the loss of the negatively charged carboxylate is the likely factor in the activity increase. Finally, the R237K mutant resembled R237A in maintaining wild-type activity, yet the R237Q change stimulated activity 3-fold (Figure 3E). We previously noted similar effects of conservative changes on the Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate (14). We surmise that CthPnkp utilizes the same constellation of active site residues for the hydrolysis of the phosphoester linkage in bis-p-nitrophenyl phosphate and p-nitrophenyl phenylphosphonate.
Cyclic phosphodiesterase activity of CthPnkp
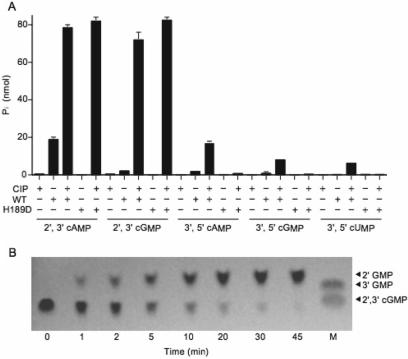
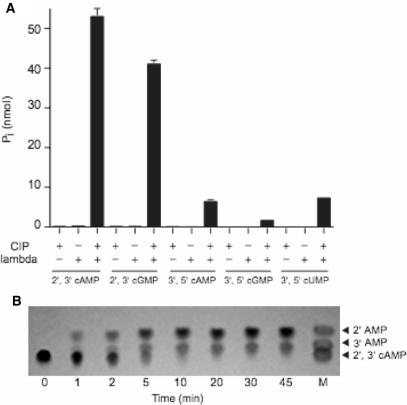
The two sequential healing reactions of CthPnkp at a 2′,3′ cyclic phosphate end can be assayed by comparing the Mn2+-dependent release of Pi from 10 mM 2′,3′ cAMP by CthPnkp alone versus a duplicate sample in which the CthPnkp reaction products are treated with calf intestinal phosphatase (CIP) (17). In the experiment shown in Figure 4A, CthPnkp converted 20% of the input 2′,3′ cAMP to Pi. Treatment of the CthPnkp reaction product with CIP increased the yield of Pi to 78% of the input 2′,3′ cAMP substrate. (Treatment with CIP alone released <2% of the Pi from 2′,3′ cAMP.) These results confirm that a nucleoside 2′,3′ cyclic phosphate is converted by CthPnkp to a phosphomonoester prior to the release of Pi. To query whether CthPnkp displays specificity towards a particular nucleotide and whether the enzyme discriminates between a five-membered 2′,3′ cyclic phosphate and a six-membered 3′,5′ cyclic phosphate, we reacted CthPnkp with 2′,3′ cGMP, 3′,5′ cAMP, 3′,5′ cGMP and 3′,5′ cUMP. CthPnkp converted 76% of the input 2′,3′ cGMP to a CIP-sensitive phosphomonoester, but only 3% of the input nucleotide was hydrolyzed completely to release Pi (Figure 4A). CthPnkp displayed much weaker activity as a 3′,5′ cyclic phosphodiesterase, converting only 16%, 8% and 6% of the input 3′,5′ cAMP, 3′,5′ cGMP and 3′,5′ cUMP substrates to CIP-sensitive phosphomonoesters, respectively (Figure 4A). There was virtually no CthPnkp-catalyzed release of Pi from any of the 3′,5′ cyclic nucleotide substrates (Figure 4A).
Figure 4.
Hydrolysis of cyclic phosphodiester substrates by CthPnkp. (A) Reaction mixtures (10 µl) containing 50 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 10 mM substrate as specified and either 4 µg wild-type CthPnkp, 1.4 µg CthPnkp-H189D, or calf intestine phosphatase (CIP; 1 U) where indicated by + were incubated for 30 min at 45°C. The reactions were quenched by adding EDTA (20 mM final concentration) and then 1 ml of malachite green reagent. Release of phosphate was determined by measuring A620 and interpolating the value to a phosphate standard curve. Each datum in the bar graph is the average of two separate experiments. S.E. bars are shown. (B) Kinetics. Reaction mixtures (per 10 µl) containing 50 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 10 mM 2′,3′ cGMP and 1.4 µg of CthPnkp-H189D were incubated at 45°C. Samples (10 µl) were withdrawn at the times specified and quenched immediately with EDTA. Aliquots (1 µl) of each sample were applied to a cellulose-F TLC plate (EMD chemicals). Markers 2′,3′ cGMP and 3′GMP (5 nmol each) were spotted in lane M. The TLC plate was developed with buffer containing saturated ammonium sulfate/3 M sodium acetate/isopropanol (80/6/2). The nucleotides were visualized by photography under UV light.
We reported previously that certain mutations in the CthPnkp phosphoesterase active site alter the reaction outcome. For example, an H189D change abolishes the phosphomonoesterase activity without affecting the phosphodiesterase (14,17). The experiment in Figure 4A verifies that CthPnkp-H189D failed to release inorganic phosphate from 2′,3′ cAMP, while converting 80% of the input 2′,3′ cAMP to a monoester that was hydrolyzed by CIP. CthPnkp-H189D was equally active in hydrolyzing 2′,3′ cGMP to a CIP-sensitive monoester, but it was virtually inert as a 3′,5′ cyclic phosphodiesterase with 3′,5′ cAMP, 3′,5′ cGMP and 3′,5′ cUMP substrates. Thus, the H189D change strongly enhances the selectivity of CthPnkp cyclic phosphodiesterase for 2′,3′ linkages.
We showed recently that CthPnkp-H189D cleaves the phosphodiester linkage of 2′,3′ cAMP to yield 2′-AMP as the sole product (17). To probe the outcome of the reaction with 2′,3′ cGMP, we performed cellulose TLC analysis of the reaction mixture as a function of reaction time in the presence of CthPnkp-H189D only (no CIP). The TLC plate was developed with buffer containing saturated ammonium sulfate/3 M sodium acetate/isopropanol (80/6/2), in which the order of migration away from the origin (Rf) is 2′,3′ cGMP <3′-GMP < 2′-GMP (36,37). This experiment revealed a single-step conversion of 2′,3′ cGMP to a faster-moving product that migrated ahead of 3′-GMP, at a position corresponding to 2′-GMP (Figure 4B). Thus, the selectivity of CthPnkp cyclic phosphodiesterase for cleavage of the P–O3′ bond was not affected by the identity of the nucleoside base.
Active site requirements for hydrolysis of 2′,3′ cAMP by CthPnkp
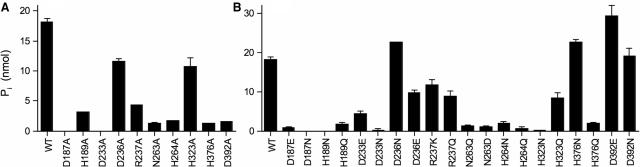
Wild-type CthPnkp and the 10 alanine and 20 conservative mutants were assayed for hydrolysis of 10 mM 2′,3′ cAMP in the presence of CIP. The D187A and D233A changes abolished the cyclic phosphodiesterase activity; the N263A, H264A, H376A and D392A proteins were 7–10% as active as wild-type CthPnkp and H189A was 17% as active (Figure 5A). These mutational effects roughly parallel the trends seen for CthPnkp activity with p-nitrophenyl phenylphosphonate (Figure 3D). However, the modest effect of the H323A change on cyclic phosphate hydrolysis (60% of wild-type activity; Figure 5A) contrasted with its ablation of p-nitrophenyl phenylphosphonate hydrolysis (Figure 3D) and bis-p-nitrophenyl phosphate hydrolysis (14). On the other hand, the D236A, R237A and H264A changes that either stimulated or had no effect on activity with p-nitrophenyl phenylphosphonate were modestly to severely inhibitory to the cyclic phosphodiesterase: D236A, R237A and H264A were 64%, 24% and 10% as active as wild-type CthPnkp in cleaving 2′,3′ cAMP (Figure 5A). These results suggest that CthPnkp relies on a different ensemble of functional groups for hydrolysis of 2′,3′ cyclic nucleotide and acyclic non-nucleotide diester/phosphonate substrates. In particular, cyclic phosphodiesterase function appears to lean more heavily on residues Arg237 and His264 that contact the sulfate oxygens in the λ-Pase active site (Figure 1).
Figure 5.
Mutational effects on 2′,3′ cyclic phosphodiesterase activity. Reaction mixtures (25 µl) containing 50 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 5 mM 2′,3′ cAMP, 1 U CIP and 0.5 µg CthPnkp as specified were incubated for 30 min at 45°C. (A) Effects of alanine mutations. (B) Effects of conservative substitutions. Each datum in the bar graph is the average of two separate experiments. S.E. bars are shown.
Conservative changes at Asp187, Asp233, Asn263 and His264 had little or no salutary effects compared to the respective alanine mutants (Figure 5B). Whereas replacing His189 with glutamine or asparagine reduced activity compared to H189A (Figure 5), activity was restored to greater than wild-type levels when His189 was replaced by aspartate or glutamate (see below). Similar mutational effects at His189 were observed for Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate (14). The D392E and D392N changes restored cyclic phosphodiesterase activity to 160% and 100% of the wild-type level, respectively (Figure 5B), again highlighting the hydrogen bonding interaction with the metal-bound water (Figure 1). Whereas the H376N substitution fully revived the phosphodiesterase activity, H376Q mimicked the alanine mutant (Figure 5B). Again, the results underscore the essentiality of the Nδ atomic contacts of this side chain.
CthPnkp His323 is the counterpart of His139 in λ-Pase, which coordinates manganese via Nε (Figure 1). Although H323A and H323Q had 60% and 47% of the wild-type cyclic phosphodiesterase activity, respectively, the conservative asparagine substitution abolished activity. It is conceivable that, in the absence of the histidine, a water molecule can act as a ligand to the metal. Glutamine can support activity by mimicking the Nε contact to metal. However, the shorter asparagine side chain might be unable to reach the metal coordination sphere, while also hampering entry of a water molecule.
Kinetic parameters for CthPnkp 2′,3′ cyclic phosphodiesterase
The Mn2+-dependent hydrolysis of 2′,3′ cAMP displayed a hyperbolic dependence on nucleotide concentration (data not shown). From a double reciprocal plot, we calculated a Km of 18 mM 2′,3′ cAMP and a kcat of 536 min−1 (Table 1). Kinetic parameters were also determined for proteins mutated at residues His189, Asp233, Asp236, Arg237, His323, His376 and Asp392 (Table 1). The H189A change increased Km slightly (29 mM) while reducing kcat (87 min−1) by a factor of 6 compared to wild-type CthPnkp. It was striking that the H189D and H189E mutations increased kcat 4-fold (2060 min−1) and 2-fold (1030 min−1) respectively without significantly affecting Km. These His189 mutations had similar suppressive or enhancing effects on kcat for Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate (14). We surmise that CthPnkp prefers an oxygen metal ligand to a nitrogen at position 189 for catalysis of phosphodiester hydrolysis of both nucleotide and non-nucleotide substrates.
Table 1.
Kinetic Parameters for Hydrolysis of 2′,3′ cAMP by CthPnkp
| Protein | Km (mM) | kcat (min−1) |
|---|---|---|
| Wild-type | 18 ± 1.4 | 536 ± 24 |
| H189A | 29 ± 1 | 87 ± 6 |
| H189D | 20 ± 1 | 2060 ± 480 |
| H189E | 28 ± 2 | 1030 ± 79 |
| D233E | 62 ± 6.5 | 195 ± 43 |
| D236A | 7.8 ± 1.5 | 272 ± 12 |
| D236N | 6.6 ± 0.2 | 346 ± 14 |
| D236E | 18 ± 1.1 | 362 ± 20 |
| R237A | 77 ± 10 | 513 ± 1 |
| R237K | 100 ± 7.5 | 1570 ± 94 |
| R237Q | 60 ± 6.5 | 1210 ± 10 |
| H323A | 32 ± 4.3 | 546 ± 34 |
| H323Q | 24 ± 2.6 | 390 ± 87 |
| H376N | 4.7 ± 0.1 | 308 ± 18 |
| H376D | 3.9 ± 0.7 | 192 ± 46 |
| D392A | 100 ± 2 | 106 ± 4.5 |
| D392E | 18 ± 0.8 | 958 ± 100 |
| D392N | 31 ± 9.1 | 1120 ± 240 |
Reaction mixtures (25 µl) containing 50 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 0.1 unit of CIP, increasing concentrations (0.625, 1.25, 2.5, 5, 10, or 20 mM) of 2′,3′ cAMP, and a fixed amount of one of the indicated CthPnkp proteins were incubated for 15 min at 45°C. Pi production was plotted as a function of substrate concentration, and then Km and kcat values were calculated from double-reciprocal plots of the data. The values in the table are averages from two independent substrate titration experiments.
Changing the Asp392 to alanine reduced both substrate affinity (Km 100 mM) and kcat (106 min−1), such that the catalytic efficiency of D392A (kcat/Km) was 28-fold lower than wild-type CthPnkp. The conservative D392E and D392N changes increased kcat by ∼2-fold compared to wild-type, with little impact on Km (Table 1). We infer that a carboxylate oxygen (of Glu or Asp) or an amide oxygen (of Asn) at position 392 can productively coordinate the metal-bound water in the active site during phosphodiester hydrolysis. Asp233 is predicted to coordinate both manganese ions of the metal cluster (Figure 1). Whereas the activity of the D233A and D233N mutants was too low for kinetic parameters to be determined, we found that the D233E change increased Km to 62 mM and decreased kcat to 195 min−1, thereby lowering catalytic efficiency by a factor of 9. We presume this reflects steric hindrance by the longer glutamate side chain. Replacing the metal-binding His323 with alanine (Km 32 mM; kcat 546 min−1) or glutamine (Km 24 mM; kcat 390 min−1) had only a modest impact on the kinetic parameters for 2′,3′ cAMP hydrolysis. As discussed in the preceding section, we suspect that water can occupy the octahedral metal complex in lieu of the Nε atoms of histidine or glutamine.
Replacing the metal-binding His376 with asparagine enhanced substrate affinity (Km 4.7 mM) while only modestly diminishing kcat (308 min−1), the net result being that catalytic efficiency of H376N was 2-fold higher than wild-type CthPnkp. We also determined kinetic parameters for mutant protein H376D, which retains activity in Mn2+-dependent hydrolysis of p-nitrophenyl phosphate and bis-p-nitrophenyl phosphate, but is no longer able to use Ni2+ as a cofactor (14). The H376D change increased substrate affinity (Km 3.9 mM), but this was offset by a decrease in kcat (192 min−1), so that the catalytic efficiency of H376D was ∼60% higher than the wild-type enzyme. These results imply that the role of the manganese-coordinating Nδ atom of His376 can be performed reasonably well by either Nδ of asparagine or Oδ of aspartate. The observed higher affinity of H376N and H376D for the phosphodiester substrate might also elicit a decrease in the off-rate of the Pi product, thereby accounting for the observed decrease in steady-state kcat of these mutants.
Arg237 makes a predicted bidentate interaction with the phosphate oxygens (Figure 1). The R237A, R237K and R237Q changes increased Km by 4-fold (77 mM), 6-fold (100 mM) and 3-fold (60 mM), respectively, thereby highlighting the importance of the phosphate contacts for substrate binding affinity. Yet, Arg237 makes remarkably little contribution to the maximal rate of hydrolysis of the cyclic phosphodiester, insofar as the alanine mutation had no impact on kcat (513 min−1), while the R237K and R237Q changes actually increased kcat by 3-fold (1570 min−1) and 2-fold (1210 min−1), respectively. The observed lower affinity of R237K and R237Q for the phosphodiester substrate might also increase the Pi product off-rate, which could explain the observed increases in kcat of these two mutants.
Replacing the Asp236 with alanine (Km 7.8 mM; kcat 272 min−1) or asparagine (Km 6.6 mM; kcat 346 min−1) increased enzyme affinity for 2′,3′ cAMP, which was countered by modest decrements in kcat, such that net catalytic efficiency was equal to (D236A) or slightly better than (D236N) the wild-type enzyme. The parameters for D236E (Km 18 mM; kcat 362 min−1) were similar to wild-type CthPnkp. The small impact of Asp236 mutations on catalysis of the cyclic phosphodiesterase reaction is consistent with the benign effects of the same changes on Mn2+-dependent hydrolysis of bis-p-nitrophenyl phosphate (14) and the inference from structure of λ-Pase that Asp236 makes no direct contact to the metals or the phospho-substrate (Figure 1). Rather, Asp236 contacts His264, which in turn coordinates two of the sulfate/phosphate oxygens.
Phosphodiesterase activity of λ-Pase
The structure of λ-Pase is known (25) and its hydrolysis of the generic monoesterase substrate p-nitrophenyl phosphate has been studied in depth (38–40). λ-Pase has phosphoprotein and phosphopeptide phosphatase activity in vitro, but it is not clear that phosphoproteins are the physiological substrates for this enzyme. λ-Pase is reportedly unable to hydrolyze phosphoamino acids P-Ser, P-Thr or P-Tyr (23,24). We can find little in the literature regarding the capacity of λ-Pase to hydrolyze nucleotide or nucleic acid phosphomonoesters, beyond a citation that it does not hydrolyze 5′-AMP (23). There is no report, to our knowledge, of the ability of λ-Pase to hydrolyze phosphodiesters.
Prompted by the primary structure similarity between λ-Pase and the phosphatase domain of CthPnkp, we queried if recombinant His-tagged λ-Pase could hydrolyze a non-nucleotide phosphodiester linkage. The enzyme readily converted bis-p-nitrophenyl phosphate (10 mM) and p-nitrophenyl phenylphosphonate (10 mM) to p-nitrophenol in the presence of 0.2 mM MnCl2 (Figure 6A). The extent of product formation was proportional to the amount of input enzyme. We calculated specific activities of 44 and 47 s−1 for bis-p-nitrophenyl phosphate and p-nitrophenyl phenylphosphonate respectively, which compared favorably with the specific activity of 100 s−1 observed for hydrolysis of p-nitrophenyl phosphate (Figure 6A). Hydrolysis of 10 mM bis-p-nitrophenyl phosphate by λ-Pase was optimal at pH 7.0–7.5 in Tris buffer and declined sharply at pH <6.0 and >8.5 (Figure 6B). Hydrolysis of p-nitrophenyl phosphate was optimal at pH 8.0 in Tris–HCl buffer (data not shown). Substrate titration experiments revealed a 2-fold higher affinity of λ-Pase for p-nitrophenyl phosphate (Km = 5.0 ± 0.1 mM; data not shown) than bis-p-nitrophenyl phosphate (Km = 10 ±1.6 mM; data not shown).
Figure 6.
Phosphodiesterase activity of λ-Pase. (A) Reaction mixtures (25 µl) containing 0.2 mM MnCl2, 10 mM DTT and either 100 mM Tris–HCl pH 8.0 and 10 mM p-nitrophenyl phosphate (filled square) or 100 mM Tris–HCl pH 7.0 and 10 mM bis-p-nitrophenyl phosphate (open circle) or p-nitrophenyl phenylphosphonate (filled circle), and λ-Pase as specified were incubated for 15 min at 37°C. (B) Reaction mixtures (25 µl) containing 100 mM Tris–acetate (filled circle) or Tris–HCl (open circle) at the pH specified, 0.5 mM MnCl2, 10 mM DTT, 10 mM bis-p-nitrophenyl phosphate and 2 pmol λ-Pase were incubated for 10 min at 37°C. (C) Kinetics. Reaction mixtures (per 25 µl) containing 100 mM Tris–HCl (pH 7.0), 0.2 mM MnCl2, 10 mM DTT, 10 mM bis-p-nitrophenyl phosphate and 2 pmol λ-Pase were incubated at 37°C. Two aliquots (25 µl) were withdrawn at the times specified, quenched immediately with EDTA and assayed for p-nitrophenol (filled circle) and inorganic phosphate (open circle).
Cleavage of the phosphodiester linkage of bis-p-nitrophenyl phosphate results in the formation of p-nitrophenyl phosphate and the release of p-nitrophenol. The reaction product p-nitrophenyl phosphate is a substrate for the phosphomonoesterase activity of λ-Pase. In the event that λ-Pase performs serial phosphodiesterase and monoesterase reactions with bis-p-nitrophenyl phosphate via a processive mechanism, we would expect to see the simultaneous release of both p-nitrophenol and phosphate in a molar ratio of 2 : 1. However, if the p-nitrophenyl phosphate product is released from the active site after the initial phosphodiesterase reaction, then it must compete with the excess bis-p-nitrophenyl phosphate to rebind to the enzyme. In the latter scenario, we expect that the rate of p-nitrophenol production will exceed that of Pi release by a greater than 2:1 ratio. A kinetic analysis revealed simultaneous accumulation of both products over a period of 30 min, with an 8:1 ratio of p-nitrophenol to Pi (Figure 6C). We surmise that the phosphodiesterase and monoesterase reactions of λ-Pase with bis-p-nitrophenyl phosphate are not processive.
Cyclic phosphodiesterase activity of λ-Pase
The finding that λ-Pase has both phosphomonoesterase and diesterase activities underscores the similarity to CthPnkp and raises the issue of whether λ-Pase might also act on nucleotide monoester and diester substrates. We found that λ-Pase was unable to hydrolyze a nucleoside phosphomonoester, insofar as we detected no release of Pi from 10 mM 2′-AMP, 3′-AMP and 5′-AMP by λ-Pase in the presence of 0.5 mM manganese (data not shown). We then tested λ-Pase for cyclic phosphodiesterase activity with 10 mM 2′,3′ cAMP, 2′,3′ cGMP, 3′,5′ cAMP, 3′,5′ cGMP or 3′,5′ cUMP substrates in the presence of 0.5 mM manganese (Figure 7). λ-Pase had vigorous 2′,3′ cyclic phosphodiesterase activity, converting 53% of the input 2′,3′ cAMP and 42% of the input 2′,3′ cGMP to a monoester that was sensitive to CIP (Figure 7A). λ-Pase was less effective as a 3′,5′ cyclic phosphodiesterase, converting only 6%, 2% and 7% of the input 3′,5′ cAMP, 3′,5′ cGMP or 3′,5′ cUMP to CIP-sensitive monoesters. No phosphate was released from any of the cyclic phosphate nucleotides by λ-Pase in the absence of CIP (Figure 7A), as expected given our finding that the enzyme was inactive with nucleoside phosphomonoesters.
Figure 7.
Cyclic phosphodiesterase activity of λ-Pase. (A) Reaction mixtures (10 µl) containing 100 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 10 mM DTT, 10 mM substrate as specified and either 32 pmol λ-Pase or 1 U CIP (where indicated by +) were incubated for 30 min at 37°C. Each datum in the bar graph is the average of two separate experiments. S.E. bars are shown. (B) Reaction mixtures (per 10 µl) containing 100 mM Tris–HCl (pH 7.5), 0.5 mM MnCl2, 10 mM DTT, 10 mM 2′,3′ cAMP and 64 pmol λ-Pase were incubated at 37°C. Aliquots (10 µl) were withdrawn at the times specified and quenched immediately with EDTA. One micro liter of each sample was applied to a cellulose-F TLC plate. Markers 2′-AMP, 3′-AMP and 2′,3′ cAMP (5 nmol each) were spotted in lane M. The TLC plate was developed with buffer containing saturated ammonium sulfate/3 M sodium acetate/isopropanol (80/6/2). The nucleotides were visualized by photography under UV light.
The outcome of the cyclic phosphodiesterase reaction with 2′,3′ cAMP was probed by TLC analysis of the reaction mixture as a function of reaction time in the presence of λ-Pase only (no CIP). This experiment revealed simultaneous conversion of 2′,3′ cAMP to two faster-moving products comigrating with 3′-AMP and 2′-AMP (Figure 7B). Thus, the λ-Pase does not display the selectivity of CthPnkp-H189D for cleavage of the P–O3′ bond of 2′,3′ cyclic nucleotides. Rather λ-Pase can hydrolyze either the P–O3′ or P–O2′ linkage to yield a mixture of 3′-NMP and 2′-NMP end products.
DISCUSSION
Our studies of the substrate specificity and active site of CthPnkp phosphoesterase, and the present extension of this analysis to λ-Pase, provide new insights to the catalytic repertoire of the calcineurin-type binuclear metallophosphoesterase family. The prevailing view that members of this family are dedicated to the hydrolysis of either phosphomonoesters or phosphodiesters is vitiated by the clear evidence that CthPnkp and λ-Pase are adept at cleaving both monoester and diester substrates. Moreover, the activity of both enzymes with a phosphoester phosphonate attests to a requirement for only three oxygens around the phosphorus center that is the target of hydrolytic attack.
The ensemble of CthPnkp active site functional groups needed for Mn2+-dependent hydrolysis of the phosphoester phosphonate (Figure 3) is the same as for cleavage of bis-p-nitrophenyl phosphate, but is distinct from that required for Mn2+-dependent hydrolysis of the p-nitrophenyl phosphate monoester (14). The monoesterase reaction relies on phosphate-binding residues Arg237 and His264, which are not essential for the cleavage of bis-p-nitrophenyl phosphate or p-nitrophenyl phenylphosphonate. The two classes of substrates differ in the charge on the ground state (dianionic for the monoester, monoanionic for the diester and phosphonate), and presumably in the transition-state, so that is possible that charge neutralization by the Arg and His side chains assumes greater importance for the monoesterase reaction than for the diesterase or phosphonate cleavage reactions. This inference is consistent with the fact that Mre11, a family member that catalyzes a Mn2+-dependent DNA phosphodiesterase reaction, has no equivalent of Arg237 in its active site (41).
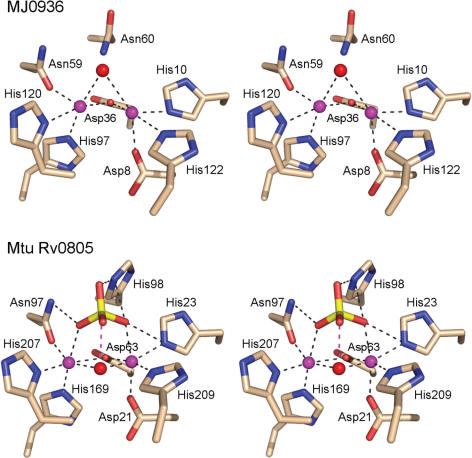
CthPnkp is not adept at hydrolyzing p-nitrophenyl diesters of thymidine or choline. Because there is no clear chemical distinction at the phosphorus center or leaving group between these two poor substrates and the good diester substrate bis-p-nitrophenyl phosphate, it is likely that the thymidine and choline groups interfere with productive binding of the substrate in the active site, either because of steric clashes or the positive charge on the choline quaternary amine. It is important to stress that the particular selectivity of CthPnkp for certain p-nitrophenyl diesters does not apply to all other calcineurin-type binuclear metallophosphoesterases. For example, the Methancoccus jannaschii MJ0936 protein is a phosphodiesterase with vigorous activity in hydrolyzing bis-p-nitrophenyl phosphate and p-nitrophenyl phosphorylcholine, and weaker activity in cleaving thymidine 5′-monophosphate p-nitrophenyl ester (42). Though possessed of a phosphodiesterase activity, MJ0936 reportedly has no detectable cyclic phosphodiesterase activity with the 2′,3′ or 3′,5′ forms of cAMP or cGMP (42). Thus, the specificity of the MJ0936 phosphodiesterase contrasts sharply with that of CthPnkp and λ-Pase, both of which display diesterase activity with 2′,3′ cyclic nucleotides. It is also noteworthy that MJ0936, which is structurally homologous to λ-Pase, is cited as being unable to cleave the monoester substrate p-nitrophenyl phosphate (42), thereby further distinguishing it from λ-Pase and CthPnkp.
Inspection of the crystal structure of manganese-bound MJ0936 (14, PDB 1S3N) reveals similarity of its active site to that of λ-Pase, with the critical difference being that MJ0936 has no equivalent of the arginine corresponding to CthPnkp Arg237 or λ-Pase Arg53 (this residue is a leucine in MJ0936) (Figure 8). As noted above, CthPnkp Arg237 is essential for phosphomonoesterase activity, but dispensable for the phosphodiesterase function. Thus we can posit a simple route to the evolution of the diesterase-only activity of MJ0936 (and Mre11) from a Mn2+-dependent diesterase-monoesterase ancestor (like CthPnkp) by mutation of the arginine and consequent disabling of the monoesterase. The MJ0936 active site is also remarkable for having an asparagine in lieu of the phosphate-coordinating histidine residue: His264 in CthPnkp and His76 in λ-Pase (Figure 8). The selective inhibitory effect of the CthPnkp H264N mutation on cyclic phosphodiesterase activity (while stimulating CthPnkp phosphodiesterase activity with bis-p-nitrophenyl phosphate) provides a plausible explanation for why MJ0936 is defective in cleaving cAMP and cGMP.
Figure 8.
Active sites of binuclear metallophosphodiesterases MJ0936 and Rv0805. Stereo views of the active site of Methanoccus jannaschii MJ0936 (top panel; PDB 1S3N) and Mycobacterium tuberculosis Rv0805 (bottom panel; PDB 2HY1) oriented similarly to the λ-Pase active site depicted in Figure 1. The amino acid side chains coordinating the binuclear metal cluster (and the phosphate ion in Rv0805) are shown. The metal ions are colored magenta. Water is colored red. Unlike CthPnkp and λ-Pase, the MJ0936 and Rv0805 phosphodiesterases have no arginine side chain in their active sites. Whereas the cyclic nucleotide phosphodiesterase Rv0805 has a phosphate-coordinating histidine corresponding to CthPnkp His264, the analogous residue is asparagine in MJ0936, which is unable to hydrolyze cyclic nucleotides. The water-mediated metal contact of Asp202 in λ-Pase is replaced by a direct interaction of the metal with a histidine in both MJ0936 and Rv0805.
It is instructive to compare CthPnkp to the Mycobacterium tuberculosis Rv0805 protein, which is a cyclic nucleotide phosphodiesterase member of the binuclear metallophosphoesterase family (43,44). Rv0805 catalyzes Mn2+-dependent cleavage of bis-p-nitrophenyl phosphate (kcat 250 min−1), but is 400-fold less active with p-nitrophenyl phosphate. Rv0805 can hydrolyze 3′,5′ cAMP to 5′-AMP and is also active with 3′,5′ cGMP (43). However, its ability to hydrolyze 2′,3′ cyclic nucleotides (the preferred substrates for the CthPnkp and λ-Pase cyclic nucleotide phosphodiesterase activities) has not been reported. The crystal structure of Rv0805 with metals and a phosphate anion in the active site (44) is notable for a bridging water coordinated by both metals and situated 3 Å from the phosphorus atom (Figure 8). The almost perfectly apical orientation of this water to the putative ‘leaving’ oxygen atom (169°) implicates the metal-bridged water as the nucleophile in the hydrolysis reaction. The absence of an arginine in Rv0805 at the position corresponding to Arg237(Arg53) of CthPnkp(λ-Pase) could account for its lack of phosphomonoesterase activity. The retention of a histidine equivalent to His264 of CthPnkp that contacts two phosphate oxygens in the Rv0805 crystal structure (Figure 8), in a manner analogous to the sulfate contacts of His76 in λ-Pase (Figure 1), underscores the likely role of this histidine side chain as a determinant of cyclic nucleotide phosphodiesterase activity.
The E. coli YfcE protein is a recently characterized Mn2+-dependent phosphodiesterase that cleaves bis-p-nitrophenyl phosphate, thymidine 5′-monophosphate-p-nitrophenyl ester, and p-nitrophenyl phosphorylcholine, but is unable to hydrolyze 2′,3 ′ or 3′,5′ cyclic nucleic phosphodiesters or any of 50 different phosphomonoesters, including p-nitrophenyl phosphate (45). The crystal structure of YfcE with metals and sulfate in the active site reveals that it lacks an exact counterpart of Arg237 of CthPnkp, although a different arginine does donate a single hydrogen bond to the bound sulfate in only one of the four YfcE protomers in the asymmetric unit (45). The position equivalent to His264 of CthPnkp is occupied by a cysteine in YfcE.
The studies presented here documenting the vigorous activity of CthPnkp at 2′,3′ cyclic phosphate ends, the prior demonstration that CthPnkp can heal 2′,3′ cyclic phosphate termini in a broken tRNA (17), and the bundling of the 2′,3′ cyclic phosphodiesterase with a 5′ end-healing function (5′-OH polynucleotide kinase) in a single polypeptide (12) all weight in favor of a role for CthPnkp in RNA repair, although the identity of the putative RNA repair substrate(s) remains uncharted. Similarly, the physiological substrates of λ-Pase are unknown and, in light of the findings herein, it cannot be taken for granted that λ-Pase acts exclusively a monoesterase. Indeed, given its cyclic phosphodiesterase activity and preference for 2′,3′ cyclic phosphate nucleotides over 3′,5′ derivatives, the prospect that λ-Pase acts on nucleic acids cannot be dismissed.
ACKNOWLEDGEMENTS
This work was supported by grant GM42498 from the U.S. National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by NIG grant GM42498.
Conflict of interest statement. None declared.
REFERENCES
- 1.Richardson CC. Phosphorylation of nucleic acid by enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl Acad. Sci. USA. 1965;54:158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novogrodsky A, Hurwitz J. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: phosphorylation at 5′-hydroxyl termini. J. Biol. Chem. 1966;241:2923–2932. [PubMed] [Google Scholar]
- 3.Novogrodsky A, Tal M, Traub A, Hurwitz J. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: further properties of the 5′-hydroxyl polynucleotide kinase. J. Biol. Chem. 1966;241:2933–2943. [PubMed] [Google Scholar]
- 4.Cameron V, Uhlenbeck OC. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977;16:5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- 5.Wang LK, Shuman S. Mutational analysis defines the 5′-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LK, Lima CD, Shuman S. Structure and mechanism of T4 polynucleotide kinase-an RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Yin S, Shuman S. Characterization of polynucleotide kinase/phosphatase enzymes from mycobacteriophages Omega and Cjw1 and vibriophage KVP40. J. Biol. Chem. 2004;279:26358–26396. doi: 10.1074/jbc.M403200200. [DOI] [PubMed] [Google Scholar]
- 9.Blondal T, Hjorleifsdottir S, Aevarsson A, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermansdottir AG, Hreggvidsson GO, Smith AV, et al. Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J. Biol. Chem. 2005;280:5188–5194. doi: 10.1074/jbc.M409211200. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Martins A, Shuman S. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′-kinase and polynucleotide 3′-phosphatase activities. J. Biol. Chem. 2004;279:18220–18231. doi: 10.1074/jbc.M313386200. [DOI] [PubMed] [Google Scholar]
- 12.Martins A, Shuman S. An end-healing enzyme from Clostridium thermocellum with 5′-kinase, 2′, 3′ phosphatase, and adenylyltransferase activities. RNA. 2005;11:1271–1280. doi: 10.1261/rna.2690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keppetipola N, Shuman S. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA. 2006;12:73–82. doi: 10.1261/rna.2196406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keppetipola N, Shuman S. Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase/phosphatase. J. Biol. Chem. 2006;281:19251–19259. doi: 10.1074/jbc.M602549200. [DOI] [PubMed] [Google Scholar]
- 15.Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc. Natl Acad. Sci. USA. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keppetipola N, Nandakumar J, Shuman S. Reprogramming the tRNA –splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007;35:3624–3630. doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J. Biol. Chem. 1998;273:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- 19.Zhu H, Smith P, Wang LK, Shuman S. Structure-function analysis of the 3′ phosphatase component of T4 polynucleotide kinase/phosphatase. Virology. 2007;366:126–136. doi: 10.1016/j.virol.2007.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg J, Huang H, Kwon Y, Greengard P, Nairn AC, Kuriyan J. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 21.Rusnak F, Mertz P. Calcineurin: Form and function. Physiol. Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 22.Cohen PTW, Cohen P. Discovery of a protein phosphatase activity encoded in the genome of bacteriophage λ. Biochem. J. 1989;260:931–934. doi: 10.1042/bj2600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barik S. Expression and biochemical properties of a protein serine/threonine phosphatase encoded by bacteriophage λ. Proc. Natl Acad. Sci. USA. 1993;90:10633–10637. doi: 10.1073/pnas.90.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuo S, Clemens JC, Hakes DJ, Barford D, Dixon JE. Expression, purification, crystallization, and biochemical characterization of a recombinant protein phosphatase. J. Biol. Chem. 1993;268:17754–17761. [PubMed] [Google Scholar]
- 25.Voegtli WC, White DJ, Reiter NJ, Rusnak F, Rosenzweig AC. Structure of the bacteriophage λ Ser/Thr protein phosphatase with sulfate ion bound in two coordination modes. Biochemistry. 2000;39:15365–15374. doi: 10.1021/bi0021030. [DOI] [PubMed] [Google Scholar]
- 26.Blanga-Kanfi S, Amitsur M, Azem A, Kaufmann G. PrrC-anticodon nuclease: functional organization of a prototypal bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez TN, Sidrauski C, Dörfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 29.Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl Acad. Sci. USA. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graille M, Mora L, Buckingham RH, van Tilbeurgh H, de Zamaroczy M. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 2004;23:1474–1482. doi: 10.1038/sj.emboj.7600162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin YL, Elias Y, Huang RH. Structural and mutational studies of the catalytic domain of colicin E5: a tRNA-specific ribonuclease. Biochemistry. 2005;44:10494–10500. doi: 10.1021/bi050749s. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 33.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Huang B, Esberg A, Johanson M, Byström AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 36.Genshik P, Billym E, Saniewicz M, Filipowicz W. The human RNA 3’-terminal phosphate is a member of a new family of proteins conserved in eukarya, bacteria, and archaea. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2. J. Biol. Chem. 2004;279:36819–36827. doi: 10.1074/jbc.M405120200. [DOI] [PubMed] [Google Scholar]
- 38.Zhuo S, Clemens JC, Stine RL, Dixon JE. Mutational analysis of a Ser/Thr phosphatase: identification of residues important in phosphoesterase substrate binding and catalysis. J. Biol. Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]
- 39.Mertz P, Yu L, Sikkink R, Rusnak F. Kinetic and spectroscopic analysis of mutants of a conserved histidine in the metallophosphatases calcineurin and λ protein phosphatase. J. Biol. Chem. 1997;272:21296–21302. doi: 10.1074/jbc.272.34.21296. [DOI] [PubMed] [Google Scholar]
- 40.Hoff RH, Mertz P, Risnak F, Hengge AC. The transition state of the phosphoryl-transfer reaction catalyzed by the lambda Ser/Thr protein phosphatase. J. Am. Chem. Soc. 1999;121:6382–6390. [Google Scholar]
- 41.Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA. Structural biochemistry and interaction architechture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Yakunin AF, Kuznetsova E, Busso D, Pufan R, Proudfoot M, Kim R, Kim SH. Structural and functional characterization of a novel phosphodiesterase from Methanococcus janaschii. J. Biol. Chem. 2004;279:31854–31826. doi: 10.1074/jbc.M401059200. [DOI] [PubMed] [Google Scholar]
- 43.Shenoy AR, Sreenath N, Podobnik M, Kovacevic M, Visweswariah SS. The Rv0805 gene from Mycobacterium tuberculosis encodes a 3′-5′-cyclic nucleotide phosphodiesterase: biochemical and mutational analysis. Biochemistry. 2005;44:15695–15704. doi: 10.1021/bi0512391. [DOI] [PubMed] [Google Scholar]
- 44.Shenoy AR, Capuder M, Draskovic P, Lamba D, Visweswariah SS, Podobnik M. Structural and biochemical analysis of the Rv0805 cyclic nucleotide phoshodiesterase from Mycobacterium tuberculosis. J. Mol. Biol. 2007;365:211–225. doi: 10.1016/j.jmb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Miller DJ, Shuvalova L, Evdokimova E, Savchenko A, Yakunin AF, Anderson WF. Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene. Protein Science. 2007;16:1338–1348. doi: 10.1110/ps.072764907. [DOI] [PMC free article] [PubMed] [Google Scholar]