Abstract
Ionizing radiation induces clustered DNA damage, which presents a challenge to the cellular repair machinery. The repair efficiency of a single-strand break (SSB) is ∼4× less than that for repair of an abasic (AP) site when in a bistranded cluster containing 8-oxoG. To explore whether this difference in repair efficiency involves XRCC1 or other BER proteins, synthetic oligonucleotides containing either an AP site or HAP1-induced SSB (HAP1-SSB) 1 or 5 bp 5′ or 3′ to 8-oxoG on the opposite strand were synthesized and the repair investigated using either nuclear extracts from hamster cells proficient (AA8) or deficient (EM7) in XRCC1 or purified BER proteins. XRCC1 is important for efficient processing of an AP site in clustered damage containing 8-oxoG but does not affect the already low repair efficiency of a SSB. Ligase I partly compensates for the absence of the XRCC1/ligaseIII during short-patch BER of an AP site when in a cluster but only weakly if at all for a HAP1-SSB. The major difference between the repair of an AP site and a HAP1-SSB when in a 8-oxoG containing cluster is the greater efficiency of short-patch BER with the AP site compared with that for a HAP1-SSB.
INTRODUCTION
DNA base lesions can be generated in cells by reactive oxygen species, ionizing radiation or radiomimetic agents, such as bleomycin, a drug commonly used in chemotherapy. In particular, it has been proposed that radiation produces a significant proportion of DNA damage in the form of clustered DNA damage (1,2). Clustered DNA damage exists when a combination of two or more damaged bases, apurinic/apyrimidinic (AP) sites or single-strand breaks (SSB) are produced within about one or two helical turns of the DNA by a single radiation track. Many of these lesions are chemically indistinguishable from lesions produced endogenously by reactive oxygen species formed during oxygen metabolism. In mammalian cells, the yield of non-DSB clusters is about four to eight times that of prompt DSB with γ-irradiation (3–6) and only a small sub-class of these non-DSB clustered damaged sites are converted at early times into DSB post-irradiation (7).
We and others have shown that the mutagenic potential of bistranded clustered damage sites containing a mixture of AP sites, 8-oxoG or dihydrothymine (DHT) lesions in Escherichia coli is higher in comparison with that of the isolated lesions (8–13). These studies emphasize the importance of the type of lesions, inter-lesion distance and relative orientation of lesions within a cluster for the effective processing of clustered damage sites in the cell. More recently the efficiency/abundance of the base glycosylases, such as endonuclease III in E. coli, has been identified to play a decisive role in the initial stages of processing DHT/8-oxoG clusters, removing DHT to give an intermediate with an abasic site or SSB opposing 8-oxoG (11). However, some types of clustered damage sites may lead to a lethal DSB formed during attempted repair of the site (8,11,12).
One of the major pathways to repair base lesions, AP sites and SSB is the base excision repair (BER) pathway (14,15), which minimizes the biological consequences of single lesions in cells. The BER pathway has evolved to repair single DNA lesions, such as those produced by endogenous reactive oxygen species, and is largely responsible for the removal of many lesions induced by ionizing radiation. It has been hypothesized that radiation-induced clustered damage sites are less repairable than the isolated base lesions and are particularly harmful to cells (2). A number of recent studies using model systems of clustered DNA damage sites comprised of bistranded lesions have been verified experimentally that the BER pathway of prokaryotes and eukaryotes is challenged by clustered DNA damage (16–18). Using cell extracts or purified proteins, it has been shown that a complex interplay exists between different repair activities in the processing of specific forms of base lesion within a clustered damage site, and this interplay determines the outcome of attempted repair (19–25). For example with a bistranded cluster containing two lesions, the repair of either an AP site or a SSB by xrs5 nuclear extracts is reduced when 8-oxoG is up to 5 bp upstream or downstream on the opposing DNA strand. As a consequence, the lifetime of the SSB is extended up to 8-fold (22). This impairment of the repair of a SSB is at least four times that is seen for repair of the AP site in a bistranded cluster containing 8-oxoG at the same inter-lesion separation. Moreover, it was proposed that the rate of repair of an AP site and SSB within clustered damage sites is reduced in part due to retardation of the DNA XRCC1/ligase III complex which results in a shift in the mechanism to long-patch pathway thereby contributing to retarded repair of the AP site/SSB (22,26). The difference in the efficiencies and the pathways between repair of an AP site and a SSB in a clustered damage site containing 8-oxoG are as yet not known.
Two potential candidate proteins involved in the BER and which may influence the difference in efficiency of repair of an AP site compared with that of a SSB in a clustered damage site are PARP and XRCC1. For instance, during short-patch BER of single lesions, XRCC1 has been reported to interact with apurinic/apyrimidic endonuclease (HAP1) (27), polymerase β (pol β) (28,29) and DNA ligase IIIα (30), whereas in single-strand break repair (SSBR), XRCC1 has been reported to interact with PARP-1 (28–32). These multiple interactions of XRCC1 with DNA repair proteins led to the proposal that XRCC1 acts as a scaffold protein that coordinates the enzymatic steps in both BER and SSBR (27,31,33–35). Additionally, ligase I may act as a backup to XRCC1/ligase III in sealing SSB intermediates during short-patch repair pathway of single lesions (36).
Building on our previous findings on the repairability of lesions in clustered damage sites (22,26), the aim of this study is to explore whether XRCC1, PARP or other BER proteins influence the difference in efficiency of repair of an AP site compared with that of a SSB in a bistranded clustered damage site containing 8-oxoG. The repair pathways of either an AP site or SSB in bistranded clusters with 8-oxoG were investigated using either nuclear extracts from hamster cell lines proficient (AA8) or deficient (EM7) in XRCC1 activity or purified proteins involved in BER. We show that XRCC1 but not PARP is important for efficient processing of an AP site in clustered damage sites containing 8-oxoG but does not affect the repair efficiency of SSBs which are already inefficiently repaired when in a clustered damage site. Moreover ligase I partly compensates for the absence of the XRCC1/ligaseIII complex in overall ligation by short-patch BER in the processing of an AP site when in a cluster.
MATERIALS AND METHODS
Substrate oligonucleotides
The oligonucleotides were purchased PAGE purified from Eurogentec. The sequences of the double-stranded oligonucleotides are presented in Table 1. Strand 1 contains either an AP site, HAP1-SSB (Table 1) or the corresponding undamaged base, as control, at the variable position Y. Strand 2, contains 8-oxoG at the fixed position X. The nomenclature of the relative position of the two lesions in the clustered DNA damage site (−5, −1, +1, +5) was developed by David-Cordonnier et al. (21) with a positive or negative number being assigned to each residue in the double-stranded oligonucleotide in Table 1. This number refers to the separation, in base pairs, of one lesion on strand 1 located 5′ (positive number) or 3′ (negative number) opposite to the lesion on strand 2.
Table 1.
Sequence of oligonucleotides used to generate the DNA clustered damage sites
| Position | Sequence | Strand | Oligo |
|---|---|---|---|
| −5 | 5′-ctcttagtcaggaaYatgtctctatgctgggagcaaaggc | 1 | A |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | 2 | ||
| −1 | 5′-ctcttagtcaggaatatgYctctatgctgggagcaaaggc | 1 | B |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | 2 | ||
| +1 | 5′-ctcttagtcaggaatatgtcYctatgctgggagcaaaggc | 1 | C |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | 2 | ||
| +5 | 5′-ctcttagtcaggaatatgtctctaYgctgggagcaaaggc | 1 | D |
| 3′-gagaatcagtccttatacaXagatacgaccctcgtttccg | 2 | ||
| Control | 5′-ctcttagtcaggaatatgtctctatgctgggagcaaaggc | 1 | Z |
| 3′-gagaatcagtccttatacaYagatacgaccctcgtttccg | 2 |
X represents 8-oxoG. Y represents an AP site or HAP1-SSB (following conversion of uracil to an AP site or to HAP1-SSB as described in Materials and Methods Section). −5 and −1 indicate the positions on the complementary strand of the X base 3′ from Y base. +1 and +5 are the positions on the complementary strand of the X base 5′ from the Y base. Control is control oligonucleotide containing AP site or HAP1-SSB as a single lesion.
Oligonucleotide labelling and annealing
Oligonucleotides (0.2 μg) were 5′-32P-end-labelled using 10 U of T4 polynucleotide kinase (Invitrogen, Paisley, UK), 15 μCi [γ-32P]ATP (6000 Ci/mmol, 10 mCi/ml, Perkin Elmer), incubated in 20 μl of buffer (70 mM Tris–HCl pH 7.6, 10 mM MgCl2, 100 mM KCl, 1 mM β-mercaptoethanol) for 30 min at 37°C. Oligonucleotides (0.2 μg) were 3′-end-labelled using 15 U of terminal deoxynucleotidyl transferase (TdT) (Invitrogen, Paisley, UK), 30 μCi [α-32P]dATP (5000 Ci/mmol, 10 mCi/ml, Perkin Elmer), incubated in 40 μl of TdT buffer [100 mM potassium cacodylate pH 7.2, 2 mM CoCl2, 200 μM 1, 4-dithiothreitol (DTT)] for 30 min at 37°C. The reaction was stopped by a subsequent incubation for 10 min at 70°C. The solution was subsequently centrifuged through a Quick Spin G-25 Sephadex column to remove unincorporated labelled nucleotides. Labelled oligonucleotide was hybridized with a 2-fold excess of the purified non-radiolabelled complementary strand in 1× annealing buffer (10 mM Tris–HCl pH 8, 1 mM EDTA). Annealing mixtures were heated to 95°C and slowly cooled to room temperature. Efficient annealing to give the double-stranded oligonucleotide was verified on a 12% native polyacrylamide gel.
Preparation of an AP site
The purified double-stranded oligonucleotides containing a uracil residue were treated with 1 U of uracil DNA glycosylase (UDG) (Invitrogen) in 100 μl of buffer [10 mM Tris–HCl (pH 7.5), 50 mM NaCl and 1 mM EDTA] for 30 min at 37°C to produce an AP site (26). The AP site containing oligonucleotides were used immediately in the repair assays.
Preparation of a HAP1-SSB
The AP-containing oligonucleotides were treated with 25 ng of HAP1 in 50 μl buffer (20 mM HEPES pH 7.9, 100 mM KCl, 1 mM MgCl2, 0.2 mM EDTA, 20% glycerol) for 30 min at 37°C to produce a SSB with 3′-OH and 5′-dRP termini, as described previously (37). Following treatment with HAP1, the SSB containing oligonucleotide was ethanol precipitated to remove excess HAP1 prior to undertaking the repair assays. These HAP1-SSB containing oligonucleotides were stored at 4°C and used within 2 weeks of preparation.
Preparation of nuclear extracts
An XRCC1 mutant cell line (EM7) and its parental Chinese hamster ovary cells (AA8) were kindly provided by Prof. John Thacker. Cultures were grown in α-MEM (Invitrogen) supplemented with 10% fetal bovine serum (Mycoplex, PAA Laboratories, Teddington, UK) and 100 U/ml penicillin (Gibco BRL, Maryland, USA) (38). The cells were harvested and the pelleted cells suspended in an equal volume of buffer (10 mM HEPES pH 7.9, 100 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) and incubated on ice for 15 min. The cytoplasmic membranes were broken by drawing the cell suspension into a 0.5 μm diameter needle 10 times. Following a brief centrifugation at 12 000g at 4°C, the supernatant was removed and the nuclear pellet resuspended in 2/3 volume high salt buffer (20 mM HEPES pH 7.9, 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) and incubated for 30 min with agitation on ice. Following centrifugation for 10 min at 12 000g at 4°C, the supernatant was dialysed twice over a total period of 16 h against 1 l of buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF). The protein concentration was determined using Bradford colorimetric technique and was found to be between 4.5 and 9 mg/ml for AA8 and between 7 and 16 mg/ml for EM7 cell extract. Aliquots of nuclear extracts were stored at −80°C.
Repair assays
The double-stranded oligonucleotides (10 000 c.p.m., 0.75 fmol) were incubated with 1 μg of either AA8 or EM7 nuclear extracts in 5 μl of repair buffer (70 mM Tris–HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 4 mM ATP, 40 mM phosphocreatine, 1.6 μg/ml phosphocreatine kinase), and dATP, dCTP, dGTP, dTTP (0.1 mM each) at 37°C for 0, 1, 5, 15, 30 and 60 min. The concentrations of extract were optimized from titration studies (data not shown). To stop the reactions, 5 μl of denaturating stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue and 0.025% xylene cyanol) was added. The samples were then subjected to electrophoresis on a 12% denaturating polyacrylamide gel containing 8 M urea in 1× TBE [89 mM Tris–HCl, 89 mM boric acid and 2 mM EDTA (pH 8.3)] for 90 min at a constant power of 90 W. The dried gel was exposed to a Bio-Rad PhosphorImager screen for visualization of repair products using phosphorimaging technology (Bio-Rad, Molecular Imager FX) and quantified with Quantity One software (Bio-Rad, Hercules, CA). When the time dependence of the repair of the AP site or HAP1-SSB is followed, the intensity of the bands representing either single-stranded DNA, single-stranded DNA with one, two or five bases added (before ligation; see the Results section), or rejoined DNA [ligation of the AP site or HAP1-SSB following addition of the missing base(s)] is expressed as a percentage of the total intensities for all bands. The efficiencies of repair of an AP site or of HAP1-SSB within clustered damage sites were compared with that for the repair of the respective single lesions in the control oligonucleotides. The errors represent standard deviations of the mean from at least three experiments.
Immunodepletion of DNA ligase III/XRCC1 complex from AA8 cells extracts
Immunodepletion was performed as previously described (36). Briefly 100 μl of AA8 nuclear extract (NE) (5 mg/ml in 25 mM HEPES at pH 7.9, 0.1 M KCl, 12 mM MgCl2, 17% glycerol, 1 mM EDTA and 2 mM DTT) was mixed with XRCC1 antibody (0.1 μg) (Abcam, ab1838) and incubated at 4°C for 2 h. A total of 50 μl of a 50% slurry of protein A/G agarose (Santa Cruz) beads in the same buffer was added, and incubation was continued for a further 2 h. The beads were then removed by centrifugation and the protein concentration of the immunodepleted nuclear extract was determined by the Bradford Protein assay (Bio-rad). The level of XRCC1 following immunodepletion was verified by Western blot.
Western blots
Western blots were performed by standard procedure as recommended by the vendor (Novex, San Diego, CA, USA). Blots were visualized using the ECL plus system (Amersham, Little Chalfont, UK). All experiments were repeated at least 3 times, and representative gels are shown.
Reconstitution of short-patch BER with purified proteins
The 32P-5′-end-labelled double-stranded oligonucleotides (10 000 c.p.m., 0.75 fmol) were incubated for 30 min at 37°C in the absence or presence of purified BER enzymes, e.g. 25 ng HAP1 or 25 ng HAP1 + 2 ng pol β or 25 ng HAP1 + 2 ng pol β + 3 ng ligase I in a 10 μl reaction solution containing 80 mM HEPES pH 7.9, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 200 μM EDTA, 4 mM ATP, 800 μg/ml bovine serum albumin, 40 μM each of dATP, dTTP, dGTP and dCTP. The reactions were stopped by the addition of 10 μl denaturating stop solution (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). The samples were subsequently incubated at 90°C for 3 min and then subjected to electrophoresis on a 20% denaturating polyacrylamide gel containing 8 M urea in 1× TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA pH 8.3). The bands in the dried gel were quantified as described above. The intensity of the bands representing the single-stranded DNA with base(s) added (before ligation) and ligated DNA (ligation of the HAP1-SSB following addition of the missing base) are expressed as a percentage of the total intensities for all bands.
Enzymatic assay for dRP lyase activity of pol β
dRP lyase activity of DNA pol β was determined under the same conditions as described above for reconstituted DNA repair. The indicated amounts of pol β were added to the reaction mixtures containing 0.75 fmol of 3′-end-labelled substrate duplex with an AP site pre-incised with HAP1. Following incubation for 30 min at 37°C all samples were treated with sodium borohydride (2 μl of 0.5 M NaBH4) for 10 min on ice, to stabilize the dRP sites and prevent their self-degradation during electrophoresis. Reactions were processed as described above, and the products were analysed by electrophoresis on 20% denaturing polyacrylamide gel.
Reconstitution of long-patch BER with purified proteins
Forty-mer oligonucleotides containing uracil residue at different positions (Table 1) were 3′-end-labelled as described above. Prior to the assembly of the excision reaction, the oligonucleotide substrates were pre-treated with uracil-DNA glycosylase (1U) in 100 μl of buffer [10 mM Tris–HCl (pH 7.5), 50 mM NaCl and 1 mM EDTA] for 30 min at 37°C. The resulting AP site was reduced by the addition of sodium borohydride (0.1 M) and after incubation on ice for 10 min, the reaction buffer was exchanged to TE by filtration through a Sephadex G25 spin column. The reactions were reconstituted in a reaction mixture (10 μl) that contained 80 mM HEPES pH 7.9, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 200 μM EDTA, 4 mM ATP, 800 μg/ml bovine serum albumin, 40 μM each of dATP, dTTP, dGTP and dCTP and the 32P-labelled oligonucleotide substrate (0.75 fmol), together with HAP1, pol β, FEN1 or Lig I at the amounts indicated in the figure legends. The reactions were initiated by adding substrate oligonucleotide either to the reaction mixture as above or to the mixture containing only HAP1 and the other proteins added sequentially as described in the results. After incubation for 30 min at 37°C, the reactions were stopped by addition of 10 μl of gel-loading buffer (98% formamide, 2 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). Following incubation at 90°C for 2 min, the reaction products were separated by electrophoresis in a 20% denaturating polyacrylamide gel. All experiments were repeated at least 3 times, and representative phosphorimages of the gels are shown.
RESULTS
Efficiency of repair of either an AP site or HAP1-SSB in bistranded clusters with 8-oxoG using AA8 nuclear extracts
Previous findings using xrs5 nuclear extracts showed that an AP site is repaired more efficiently than a SSB when in a bistranded cluster with 8-oxoG (22,26). To verify if similar differences in the repair efficiency between these two types of lesions in the corresponding clusters are seen with AA8 nuclear extracts, the efficiency of repair of an AP site and HAP1-SSB, when present within a clustered DNA damage site containing 8-oxoG (Table 1), was investigated as previously described (22,26). Figure S1 shows a representative profile from a phosphorimaging scan allowing each stage of BER to be monitored over 60 min, i.e. incision of an AP site within 1 min to a SSB, followed by addition of bases by polymerases and ultimately restoration of a repaired intact 40-mer. The time course of repair of the AP site following its incision to a SSB is similar to that of the control for each of the clusters (Figure 1A), whereas repair of the pre-formed HAP1-SSB when in the cluster is dramatically reduced using AA8 nuclear extracts (Figure 1B, representative gel is shown Figure S2), consistent with observations using xrs5 nuclear extracts (22,26). Furthermore, the repair of HAP1-SSB is likewise retarded by ≥4-fold (over the first 15 min) when 8-oxoG is positioned at +1, −1 and −5 compared with that of the HAP1-SSB control. The initial rate of rejoining of a HAP1-SSB is independent of the presence of 8-oxoG when at position +5.
Figure 1.
Effect of 8-oxoG on the repair of either an AP site or HAP1-SSB when incubated with AA8 nuclear extracts for various times. (A) Timescale for the repair of an AP site control (filled triangle, oligo Z) and when in a cluster with 8-oxoG at positions −1 (filled circle, oligo B), +1 (open circle, oligo C), −5 (filled square, oligo A), +5 (open square, oligo D). (B) Timescale for the repair of HAP1-SSB control (filled triangle, oligo Z) and when in a cluster with 8-oxoG at positions, −1 (filled circle, oligo B), +1 (open circle, oligo C), −5 (filled square, oligo A), +5 (open square, oligo D). To assist the reader, lines have been drawn between the points only to guide the eye and do not represent fitted curves. The error bars represent the SD of the mean from three experiments.
Efficiency of repair of either an AP site or HAP1-SSB in bistranded clusters with 8-oxoG using EM7 nuclear extracts deficient in XRCC1
To investigate whether the efficiency of repair of an AP site present in bistranded clusters with 8-oxoG depends upon the presence of XRCC1, repair assays were performed with EM7 nuclear extracts deficient in XRCC1. Compared with the corresponding findings with AA8 extracts (Figure 1A), the efficiency of repair of the AP site at positions +1 or −1 in the 8-oxoG containing cluster is reduced in the absence of XRCC1. When 8-oxoG is at position +1, the extent of repair of the AP site is reduced 3-fold at 15 min but to a lesser extent with the −1 cluster (Figure 2A and B). However, an important consideration is whether the composition of proteins and relative concentrations of BER proteins in AA8 and EM7 nuclear extracts are different such that the level of repair could reflect any differences seen. To address this, XRCC1-DNA ligase III was immunodepleted from AA8 nuclear extract using XRCC1 antibodies. As can be seen in Figure 3A, the level of XRCC1 is reduced to levels below that of detection in immunodepleted AA8 extracts. These immunodepleted AA8 extracts, AA8 ID (–XRCC1), were used to explore whether the efficiency of repair of an AP site in a cluster containing 8-oxoG at positions +1 or −1 is reduced in the absence of XRCC1 and comparable with the findings with EM7 nuclear extracts (Figure 2). As pol β interacts with and is partially co-precipitated with XRCC1, immunodepleted extracts contained ∼20% lower levels of pol β (data not shown), and therefore the rate of repair of the AP control with AA8 ID (–XRCC1) extracts is reduced by ∼15% relative to that with AA8 nuclear extracts (Figure 3B). The relative rate of repair of an AP-site control compared with that of an AP site in the presence of 8-oxoG at position −1 is similar in AA8 and AA8 ID (–XRCC1) nuclear extracts (Figure 3C). In contrast, the level of repair of the AP site in the presence of 8-oxoG in position +1 is reduced 2- to 3-fold at 60 min with AA8 ID (–XRCC1) relative to that with AA8 extracts (Figure 3C) but comparable with the corresponding level of repair with EM7 extracts (Figure 2B). Moreover, with AA8 ID (–XRCC1) nuclear extracts or AA8 WT extracts, accumulation of the SSB+1 base intermediate during repair of the AP site in the cluster with 8-oxoG in position +1 but not with the other clusters or AP control is consistent with retarded ligation (Figure 3D).
Figure 2.
(A) Representative denaturating polyacrylamide gel showing the rejoining of an AP site in the various oligonucleotides and (B) time-dependent repair of an AP site as a single lesion and when positioned 1 bp 3′ or 5′ to a fixed 8-oxoG by XRCC1 deficient cell line (EM7).
Figure 3.
Comparison of the rate of repair of an AP site as a single lesion and when positioned 1 bp 3′ or 5′ to a fixed 8-oxoG by wild type (AA8) and XRCC1-DNA lig III-immunodepleted AA8 nuclear extracts (AA8-ID). (A) Western blot analysis of wild-type nuclear extract (AA8), mock immunodepleted (Mock) and XRCC1 immunodepleted AA8 (ID) nuclear extracts with XRCC1 antibody. (B) Time dependence for repair (from three independent experiments) of a single AP site by mock immunodepleted (AA8 Mock) and XRCC1 immunodepleted AA8 (AA8 ID) nuclear extracts. (C) Time course for the rejoining of the AP site by mock immunodepleted (AA8 Mock) and XRCC1 immunodepleted AA8 (AA8 ID) nuclear extracts. AP-site control (filled triangle, oligo Z) and when in cluster with 8-oxoG at positions −1 (filled circle, oligo B) and +1 (open circle, oligo C). (D) Accumulation of intermediate bands during repair of an AP site in the presence of 8-oxoG following addition of one nucleotide. AP-site control (-filled triangle-), and when in a cluster with 8-oxoG at position −1 (-filled circle-) and +1 (-open circle-).
In contrast to the differences seen with an AP site, the efficiency of repair of a HAP1-SSB as a single lesion or when 8-oxoG is at positions −5, +5, +1 or −1 is not markedly affected by using XRCC1 deficient extracts EM7 or AA8 ID (–XRCC1) (data not shown). Therefore the presence or absence of XRCC1 does not influence the rate of repair of HAP1-SSBs although it should be noted that the repair is already very inefficient in XRCC1 containing extracts.
Repair of either an AP site or HAP1-SSB in a bistranded cluster with 8-oxoG occurs by both long- and short-patch BER
Oligonucleotides containing either an AP site or HAP1-SSB as a single lesion are repaired mainly via short-patch BER using AA8 extracts, as previously shown using xrs5 nuclear extract (22). During repair of an HAP1-SSB, and to a lesser extent an AP site within a cluster containing 8-oxoG, the sequential addition of more than one base occurs, representing a mix of short- and long-patch BER (Figures S1 and S2). To verify that addition of >1 base probably occurs during long-patch BER, the contribution of short- to long-patch repair was estimated by substituting a dideoxy version of the dNTP which if incorporated would be two bases downstream from the repair gap (ddATP for control cluster, ddATP for −5 cluster; ddCTP for −1; ddCTP for +1 and ddGTP for +5, respectively). In each case, the extent of rejoining of an AP-control or an AP site in a cluster with 8-oxoG at all positions is only reduced by ∼15–25% compared with that seen with the corresponding dNTP, confirming only a small contribution from long-patch repair (Figure S3A). A 30–50% reduction occurs in the extent of repair of HAP1-SSB as control at 60 min or when in a cluster with 8-oxoG at position +5. In contrast the low level of rejoining of HAP1-SSB in a cluster with 8-oxoG at position −5, −1 and +1 is slightly reduced (<15%) in the presence or absence of ddNTP when present as the second incoming base (Figure S4A). This independence on ddNTP for these clusters together with the reduced efficiency of repair of the HAP1-SSB control suggests that both short-patch and long-patch rejoining of the HAP1 SSB are inefficient even though repair intermediates representing addition of one or more bases were seen.
DNA ligase I as a backup during short-patch repair of an AP site in bistranded clusters with XRCC1 deficient extracts
Dianov et al. (36) showed that DNA ligase I is able to substitute for the XRCC1–DNA ligase III complex during short-patch BER of a single AP site. Therefore, we wanted to establish whether DNA ligase I could also act as a backup ligase during short-patch repair of an AP site in the bistranded clusters containing 8-oxoG or if repair switches in favour of long-patch BER for clustered DNA damage due to polymerases being able to compete more effectively with the short-patch ligation step. Using XRCC1-deficient extracts, the rate of rejoining of an AP site as control or when 8-oxoG is at position −5, +5 or −1 (15% lower) occurs at a similar rate in the presence of ddNTP, to minimize long-patch repair, or dNTP. Therefore ligation of an AP site occurs efficiently during short-patch BER of clustered DNA damage sites containing 8-oxoG in the absence of the XRCC1-DNA ligaseIII complex (Figure S3B). In contrast, the rejoining of an AP site with 8-oxoG at the +1 position in the presence or absence of XRCC1 occurs via both short and long patch (sequential addition of 2 bases) (Figure S5A). The use of XRCC1-deficient EM7 nuclear extracts and the appropriate ddNTP, to stall competition between the two pathways, has enabled us to verify that the short-patch rejoining (ligation) step is inhibited (Figure S3B) even though accumulation of bases added by the polymerases was observed on the gel (Figure 5S B–C). These data suggest the inability of DNA ligase I to compensate for XRCC1-DNA ligase III when 8-oxoG is at position +1 to the AP site.
Figure 5.
Effect of 8-oxoG on the efficiency of removal of the dRP residue by pol β. A total of 0.75 fmol of the 3′-end-labelled (*, asterisk) oligonucleotide containing an AP site or in cluster with 8-oxoG following incubation for 30 min at 37°C with the indicated proteins and further incubated for 10 min with 0.1 M NaBH4. The reaction products were separated by electrophoresis in a 20% denaturating polyacrylamide gel.
8-oxoG retards the efficiency of ligase I but not pol β during short-patch repair of an AP site at position +1
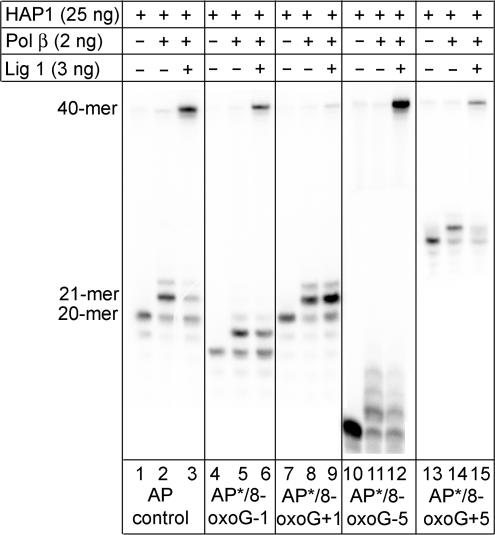
To confirm whether the retarded processing of an AP site in a cluster containing 8-oxoG at position +1 using EM7 nuclear extracts deficient in XRCC1 is indeed due to reduced efficiency of ligase I as a backup during short-patch repair, the influence of 8-oxoG on ligation was studied in more detail using purified proteins. Purified HAP1, pol β and ligase I were used to repair an AP site in the presence and absence of 8-oxoG in the various bistranded clusters. The AP control is initially incised by HAP1 (Figure 4, lane 1) followed by addition of a single nucleotide by pol β (Figure 4, lane 2) and finally restoration of the oligonucleotide through ligation by ligase I (Figure 4, lane 3). That ligation occurs infers that pol β has removed the 5′-dRP arising from HAP1 incision of the AP site (39). Kinetic data showed that rejoining is achieved rapidly after gap filling in the presence of excess DNA ligase I. If 8-oxoG is present in the clusters at positions −1, +5 and −5 to the AP site, the sequence of events shown in Figure 4 (lanes 4–6 and 10–15) for repair of the HAP1 incised AP is similar to that of the AP control. pol β incorporates a single nucleotide with similar efficiency to that of the control AP site (Figure 4, lanes 5, 11, 14) although the ligation step involving ligase1 is slightly retarded (1.4-fold reduction). In contrast, when 8-oxoG is present in the cluster at position +1, the efficiency of repair of the AP site is reduced 7-fold compared with that of the control AP site (Figure 4 lanes 7–9). Although incision of the AP site by HAP1 and incorporation of a single nucleotide by pol β occur with similar efficiencies, ligation by ligase1 is greatly retarded compared with that in the control AP site.
Figure 4.
Effect of 8-oxoG on the efficiency of repair of an AP site using purified proteins to re-construct the short-patch repair pathway. A total of 0.75 fmol of 5′-end-labelled substrate containing an AP site alone or in the presence of 8-oxoG on the complementary strand were incubated for 30 min at 37°C with the indicated amounts of purified enzyme as described in the Materials and Methods section and the intermediates separated by electrophoresis on a 20% denaturating polyacrylamide gel.
To verify that the efficiency of ligase1 is drastically reduced and not due to inhibition of the dRP lyase (dRPase) activity of pol β within the cluster containing 8-oxoG at position +1, we therefore studied the effect of 8-oxoG on removal of dRP by pol β. We constructed a 3′-end-labelled substrate containing 8-oxoG opposite the pre-incised AP site at the various positions (Figure 5). In all cases a band representing dRP is seen following HAP1 treatment with the position of the band depending on the position of the AP site in the various oligonucleotides (see Table 1 and Figure 5 lanes 6, 10, 14 and 18). Addition of pol β results in a faster migrating band as a result of excision of the dRP terminus (Figure 5 lanes 7, 11, 15 and 19). We found no inhibitory effect of 8-oxoG in the clusters or of the AP control on the excision of dRP by pol β. Consistent with the findings using 5′-32P-labelled substrates, the efficiency of the ligation step is slightly reduced (1.5-fold) for all the clusters with the exception of 8-oxoG at position +1 where a dramatic reduction (∼8-fold) in the rate of ligation was seen relative to that of the AP control (Figure 5, lanes 4 and 16). Therefore ligase1 may substitute for XRCC1/ligase III in the repair of an AP site in a cluster containing 8-oxoG except when in position +1.
8-oxoguanine does not affect the efficiency of flap removal by purified FEN1 and ligation by ligase I during long-patch repair
During long-patch repair of a HAP1-SSB and to a lesser extent an AP site when present in a bistranded cluster containing 8-oxoG, incorporation of >1 base occurs but rejoining is severely retarded as shown earlier. During long-patch repair, FEN1 is required to excise the generated flap prior to ligation by ligase1. Retarded rejoining especially with a HAP1-SSB could be due to reduced efficiency of either FEN1 to remove the flap or ligase1. To address this we have 3′-32P-end-labelled a substrate containing a reduced AP site in a cluster with 8-oxoG at position +1 to direct repair of the AP site through long-patch repair. The efficiency of excision by the FEN1 of the pol β-generated flap was determined using a reconstituted system containing HAP1, pol β and FEN1 mixed prior to addition of the oligonucleotides. Figure 6 shows that after HAP1 cleavage of the reduced AP site (lanes 1 and 5), the flap generated by pol β is readily removed from the AP control (lane 3) and (+1) cluster containing 8-oxoG (lane 7). Therefore removal of the flap overhang by FEN1 is not retarded by 8-oxoG. Retardation of FEN1 was also not observed using the −1, −5 and +5 cluster containing 8-oxoG and a reduced AP site (data not shown).
Figure 6.
Effect of 8-oxoG on the efficiency of repair of a reduced AP site using purified proteins to re-construct the long-patch repair pathway. A total of 0.75 fmol of 5′-end-labelled substrate containing an AP-site control or when in the presence of 8-oxoG on the complementary strand at position +1 were incubated for 30 min at 37°C with the indicated amounts of purified enzyme as described in the Materials and Methods section. The intermediates were separated by electrophoresis on a 20% denaturating polyacrylamide.
Ligation of the repair intermediates, following removal of the flap, by ligase1 occurs with the AP control and (+1) cluster containing 8-oxoG (Figure 6, lanes 4 and 8, respectively). Since ligation during long-patch repair of a HAP1-SSB is retarded in the presence of 8-oxoG containing clusters, namely −5, −1 and +1 (Figure S4), the substrates were first incubated with HAP1 to convert the reduced AP site into a SSB, before the sequential addition of the other proteins in the order predicted for the long-patch pathway. The removal of the flap by FEN1 is not inhibited with the HAP1-SSB substrates (control or the clusters containing 8-oxoG). However, the ligation step by ligase1 still occurs with the control and +1 cluster as described earlier but is severely retarded with the −5 cluster (data not shown).
DISCUSSION
Building on our previous findings using xrs5 extracts (22,26), we have verified using AA8 nuclear extracts that an AP site is repaired ≥4× more efficiently than a SSB when in a bistranded cluster containing 8-oxoG and that both the efficiency and mechanism of repair depends on the orientation and interlesion distance to 8-oxoG. We had previously shown that 8-oxoG retards the efficiency of XRCC1/ligase III but not pol β during the repair of a HAP1-SSB (22). Since differences in the efficiency of repair between an AP site, which is converted into a HAP1-SSB in the extracts, and a HAP1-SSB when in a bistranded cluster containing 8-oxoG are as yet not known, we have further investigated the influence of other potential candidate proteins involved in BER, namely PARP-1, XRCC1 and ligase1, a known substitute for XRCC1-ligase III complex during short-patch BER of a single AP site (36).
Using PARP-1 deficient cells and/or PARP-1 inhibitors, we probed whether PARP-1 is involved through prolonging the lifetime of intermediate(s) formed during processing of a HAP1-SSB in a bistranded cluster with 8-oxoG. However the respective rates of repair of a HAP1-SSB or AP site in the 8-oxoG containing clusters are independent of the presence or absence of PARP-1 (Bellon, unpublished data). Therefore PARP-1 does not appear to play a major role in contributing to the observed difference in repair efficiency of an AP site relative to a HAP1-SSB when in the clusters, even though we and others have shown that PARP-1 may bind to a sub-set of radiation-induced SSB and inhibit their repair, if NAD+, a co-factor for PARP-1, is absent (40,41).
An alternative candidate is XRCC1 which is a scaffold protein with no obvious enzymatic activity but is known to form a complex with ligase III (28) as well as interacting with other proteins involved in BER of both an AP site and HAP1-SSB (35). To establish whether XRCC1 contributes to the difference in repair efficiency of an AP site relative to a HAP1-SSB when in the clusters, we explored whether any of the individual steps in BER and SSBR are dependent upon the presence of XRCC1. The major findings with an AP site in a bistranded cluster with 8-oxoG are: (i) the overall rate of repair of an AP site control and when in the clusters is essentially independent of the presence of XRCC1 except when positioned at +1 to 8-oxoG where a 2–3-fold retardation was seen, (ii) ligase1 acts with a slightly reduced efficiency as a backup to XRCC1/ligase III for short-patch repair except when positioned at +1 to 8-oxoG when ligation is drastically retarded and (iii) the efficiency of FEN1 is not effected by the presence of 8-oxoG. The major findings with a HAP1-SSB in a bistranded cluster with 8-oxoG are: (i) the low rate of repair of HAP1-SSB when in the clusters is essentially independent of the presence of XRCC1, although we had previously shown that the repair of a HAP1-SSB is already very inefficient in XRCC1 containing extracts (22), (ii) both short-patch and long-patch rejoining of the HAP1 SSB are inefficient even though repair intermediates representing addition of one or more bases are formed and (iii) ligase1 is retarded for some clusters during long-patch repair of the HAP1-SSB.
The BER pathways of an AP site in a bistranded cluster containing 8-oxoG, using the −1 cluster as an example, are shown in Figure 7 based on our findings with the various cell extracts and purified proteins and the known pathways previously proposed for an individual AP site (18,19). If we concentrate initially on the repair of the AP site in the cluster when XRCC1 is present, its repair proceeds via both short- and long-patch BER in the ratio of ∼4 : 1, respectively based on the findings with ddNTP (Figures 1 and S3). The incision of the AP site by HAP1 and subsequent addition of the first base (steps 1 and 2) are not affected by the presence of 8-oxoG (Figure S1). In the presence of XRCC1, the dRP is then removed by pol β (step 3) in competition with addition of additional bases by polymerases (step 4) and commitment to the long-patch BER pathway (Figure S1). Alternatively the intermediate arising from removal of dRP in step 3 is ligated by XRCC1/ligase III (step 5) in competition with addition of additional bases by polymerases (step 6a) and commitment to the long-patch BER pathway. Once committed to long-patch BER, more than one base is added (Figure S1) and the resulting flap removed by FEN1 followed by ligation with ligase1 (Figure 6). In the absence of XRCC1, ligase1 is able to substitute for XRCC1/ligase III (steps 9 and 10) (Figure 4), albeit with a slightly reduced efficiency (1.4-fold reduced efficiency), with the exception of when 8-oxoG is at position +1 (Figure 4). That ligase1 may substitute for XRCC1/ligase III had previously been demonstrated in the repair of an individual AP site (36). As the percentage of repair of the AP site in the absence of XRCC1 is also reduced 2- to 3-fold (Figure 2B), this reduction would be consistent with an increase in the probability of commitment to long-patch repair by step 6b competing with the lowered efficiency of ligation in step 10 and to a lesser extent at the earlier stage when step 4 would compete with step 9, involving removal of dRP. As this latter step is not retarded in the presence of 8-oxoG (Figure 5), its importance in shifting the balance from short-patch to long-patch repair is predicted to be minimal. Long-patch repair would then proceed as described earlier following step 6a or step 4.
Figure 7.
General scheme of the effect of XRCC1 on the repair of an AP site in a clustered DNA damage containing 8-oxoG at position −1 as an example.
When 8-oxoG is in the +1 position, repair of the AP site is retarded in the absence of XRCC1 (Figure 3). To obtain a more detailed understanding of how cells deal with such complex lesions we analysed in more detail the efficiency of repair depending on the sub-pathways taken (42). The inhibition of ligase1 in XRCC1 deficient cells line for +1 cluster causes a significant shift from short-patch to long-patch BER as discussed earlier. Using purified enzymes (39) to re-construct short-patch repair of the AP site in the +1 cluster, we confirmed that the stalled short-patch repair of the SSB arising from incision of the AP site is due to inhibition of DNA ligase1 and not due to reduced efficiency of dRP lyase activity of pol β.
For a HAP1-SSB within clustered DNA damage site containing 8-oxoG, the low level of repair occurs via a mix of short- and long-patch BER (Figures 1 and S2). Having shown that 8-oxoG retards the rate of ligation by XRCC1/ligaseIII during short-patch repair of HAP1-SSB (22), a low level of rejoining still occurs when long-patch repair is prevented using ddNTP. This observation is consistent with XRCC1/ligase III complex being stalled (Figure S4). It is therefore suggested that repair occurs predominantly via long-patch repair as the ligation step during short-patch repair is inefficient and repair intermediates, in which >1 base has been incorporated, are formed (Figure S3). We additionally confirmed this inhibition of short-patch ligation since the efficiency of repair of a HAP1-SSB is essentially independent of the presence of XRCC1 and ligase1 does not act as an effective backup during the ligation step of a HAP1-SSB (data not shown). In contrast, the absence of XRCC1 does not strongly affect the processing of an AP site when in the corresponding clusters. To identify which step might be responsible for the low rejoining efficiency during long-patch repair of a HAP1-SSB in the presence of 8-oxoG (using a reduced AP site to ensure long-patch repair pathway), we have reconstituted the pathway using purified enzymes. The efficiency of removal of the flap by FEN1 is not inhibited by the presence of 8-oxoG, whereas ligation by ligase1 when 8-oxoG at position −5 is retarded (data not shown). It is important to note that under the conditions and the extract concentrations used for repair, 8-oxoG is not removed when opposite an AP site or a HAP1-SSB in any of the tested positions (<5%) and that DSB are not formed (<2%) as previously shown (22,26).
The main impact of 8-oxoG is to reduce greatly the efficiency of the ligation steps for both short- and long-patch BER with a HAP1-SSB when compared with an AP site. We suggest that distortions in the structure of DNA around the HAP1-SSB and to a lesser extent the AP site caused by 8-oxoG may reduce recognition by DNA ligase1 to the ligatable termini. That the AP site in a cluster is repaired more efficiently and proceeds mainly by short-patch repair infers that the appropriate conformation/structure of the ligatable intermediate is retained relative to that of a HAP1-SSB. DNA ligases are known to be selective to the conformation/structure and chemical characteristics of base pairs around the DNA termini (43,44). For instance, the fidelity of ligation strongly discriminates against mismatch base pairs at the 3′-OH-end of the nick and when in upstream positions. As a general rule, mismatched base pairs are poorly tolerated upstream of the 3′-OH-ends but are better tolerated downstream of the 5′-P-end of a nicked DNA substrate.
In conclusion, clustered DNA damage sites induced by ionizing radiation have been suggested to have serious consequences to organisms, including cancer induction. The major difference between the repair of an AP site and a HAP1-SSB when in a cluster is the greater efficiency of the short-patch pathway for repair of the AP site compared with that for the corresponding cluster containing a HAP1-SSB. Additionally ligase1 acts as a backup for XRCC1/ligase III in the short-patch repair pathway for the AP site but only weakly if at all for a HAP1-SSB when in a cluster. As a consequence, the reduced ability of lesions within a cluster to be processed by the repair machinery in cycling cell may result in extension of the lifetime of lesions within a cluster. This extension may lead to enhance mutation frequencies or additional DSB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
We are grateful to Prof. John Thacker (MRC, Harwell) for providing us AA8 and EM7 cell lines. We thank Prof. Alan Tomkinson for the generous gift of DNA ligase1. This work was partly funded by the European Commission (CLUSTOXDNA, MCRTN-CT-2003-505086). Funding to pay the Open Access publication charges for this article was provided by Medical Research Council (UK) and the European Commission.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ward JF. Biochemistry of DNA lesions. Radiat. Res. Suppl. 1985;8:S103–S111. [PubMed] [Google Scholar]
- 2.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 3.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenner TJ, Fulford J, O'Neill P. Contribution of base lesions to radiation-induced clustered DNA damage: implication for models of radiation response. Radiat. Res. 2001;156:590–593. doi: 10.1667/0033-7587(2001)156[0590:cobltr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA damages induced by x rays in human cells. Radiat. Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 7.Gulston M, de Lara C, Jenner T, Davis E, O'Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Souza DI, Harrison L. Repair of clustered uracil DNA damages in Escherichia coli. Nucleic Acids Res. 2003;31:4573–4581. doi: 10.1093/nar/gkg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malyarchuk S, Youngblood R, Landry AM, Quillin E, Harrison L. The mutation frequency of 8-oxo-7,8-dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Repair. 2003;2:695–705. doi: 10.1016/s1568-7864(03)00040-5. [DOI] [PubMed] [Google Scholar]
- 10.Malyarchuk S, Brame KL, Youngblood R, Shi R, Harrison L. Two clustered 8-oxo-7,8-dihydroguanine (8-oxodG) lesions increase the point mutation frequency of 8-oxodG, but do not result in double strand breaks or deletions in Escherichia coli. Nucleic Acids Res. 2004;32:5721–5731. doi: 10.1093/nar/gkh911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shikazono N, Pearson C, O'Neill P, Thacker J. The roles of specific glycosylases in determining the mutagenic consequences of clustered DNA base damage. Nucleic Acids Res. 2006;34:3722–3730. doi: 10.1093/nar/gkl503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison L, Brame KL, Geltz LE, Landry AM. Closely opposed apurinic/apyrimidinic sites are converted to double strand breaks in Escherichia coli even in the absence of exonuclease III, endonuclease IV, nucleotide excision repair and AP lyase cleavage. DNA Repair. 2006;5:324–335. doi: 10.1016/j.dnarep.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearson CG, Shikazono N, Thacker J, O’Neill P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic Acids Res. 2004;32:263–270. doi: 10.1093/nar/gkh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair. 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat. Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat. Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 17.Lomax ME, Gulston MK, O’Neill P. Chemical aspects of clustered DNA damage induction by ionising radiation. Radiat. Prot. Dosimetry. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 18.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhry MA, Weinfeld M. The action of Escherichia coli endonuclease III on multiply damaged sites in DNA. J. Mol. Biol. 1995;249:914–922. doi: 10.1006/jmbi.1995.0348. [DOI] [PubMed] [Google Scholar]
- 20.David-Cordonnier MH, Boiteux S, O’Neill P. Efficiency of excision of 8-oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 21.David-Cordonnier MH, Cunniffe SM, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 22.Lomax ME, Cunniffe S, O’Neill P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair. 2004;3:289–299. doi: 10.1016/j.dnarep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Chaudhry MA, Weinfeld M. Reactivity of human apurinic/apyrimidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem. 1997;272:15650–15655. doi: 10.1074/jbc.272.25.15650. [DOI] [PubMed] [Google Scholar]
- 24.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 25.Eot-Houllier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Interplay between DNA N-glycosylases/AP lyases at multiply damaged sites and biological consequences. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 27.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 30.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson D.M., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 34.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 35.Beernink PT, Hwang M, Ramirez M, Murphy MB, Doyle SA, Thelen MP. Specificity of protein interactions mediated by BRCT domains of the XRCC1 DNA repair protein. J. Biol. Chem. 2005;280:30206–30213. doi: 10.1074/jbc.M502155200. [DOI] [PubMed] [Google Scholar]
- 36.Sleeth KM, Robson RL, Dianov GL. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry. 2004;43:12924–12930. doi: 10.1021/bi0492612. [DOI] [PubMed] [Google Scholar]
- 37.David-Cordonnier MH, Boiteux S, O'Neill P. Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein. Nucleic Acids Res. 2001;29:1107–1113. doi: 10.1093/nar/29.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 40.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 41.Hodgkins PS, Fairman MP, O'Neill P. Rejoining of gamma-radiation-induced single-strand breaks in plasmid DNA by human cell extracts: dependence on the concentration of the hydroxyl radical scavenger, Tris. Radiat. Res. 1996;145:24–30. [PubMed] [Google Scholar]
- 42.Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair. 2007;6:254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Fujimoto H, Pinak M, Nemoto T, O'Neill P, Kume E, Saito K, Maekawa H. Molecular dynamics simulation of clustered DNA damage sites containing 8-oxoguanine and abasic site. J. Comput. Chem. 2005;26:788–798. doi: 10.1002/jcc.20184. [DOI] [PubMed] [Google Scholar]
- 44.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.