Abstract
The Smg proteins Smg5, Smg6 and Smg7 are involved in nonsense-mediated RNA decay (NMD) in metazoans, but no orthologs have been found in the budding yeast Saccharomyces cerevisiae. Sequence alignments reveal that yeast Ebs1p is similar in structure to the human Smg5-7, with highest homology to Smg7. We demonstrate here that Ebs1p is involved in NMD and behaves similarly to human Smg proteins. Indeed, both loss and overexpression of Ebs1p results in stabilization of NMD targets. However, Ebs1-loss in yeast or Smg7-depletion in human cells only partially disrupts NMD and in the latter, Smg7-depletion is partially compensated for by Smg6. Ebs1p physically interacts with the NMD helicase Upf1p and overexpressed Ebs1p leads to recruitment of Upf1p into cytoplasmic P-bodies. Furthermore, Ebs1p localizes to P-bodies upon glucose starvation along with Upf1p. Overall our findings suggest that NMD is more conserved in evolution than previously thought, and that at least one of the Smg5-7 proteins is conserved in budding yeast.
INTRODUCTION
The cellular production of RNA is kept under surveillance through elaborate quality control machineries able to cope with errors that may occur during synthesis. Mutations that give rise to premature termination codons in mRNAs either due to mutations in DNA or to errors that occur during transcription or pre-mRNA splicing are handled at least in part by an intricate RNA surveillance system named nonsense-mediated RNA decay (NMD) (1). NMD detects and targets ‘nonsense’ mRNAs containing premature termination codons (PTCs) for degradation, thereby preventing their translation. Loss of functional NMD seems to have different consequences in different organisms. Yeast and worm NMD mutants as well as human cells siRNA-depleted for the NMD factor Upf2 accumulate PTC-containing mRNAs without apparent loss in viability (2–6); on the contrary, impairment of NMD in flies leads to cell cycle arrest (7). In any case, one can speculate that translation of PTC-containing transcripts could lead to the production of truncated proteins with potential deleterious effects for the cell. It has been estimated that approximately one-third of all human genetic syndromes and many types of cancer arise from mRNAs containing a PTC derived from mutations occurring in the template gene. NMD might alleviate the severity of a number of genetic disorders by decreasing the abundance of disease-associated nonsense transcripts (8).
Upf1, Upf2 and Upf3 (for UP-Frameshift; also named Smg2, Smg3 and Smg4, respectively for Suppressor with Morphogenetic effects on Genitalia) proteins play a central role in NMD. They were first discovered in yeast as the result of a genetic screen looking for mutations that suppressed an auxotrophic marker containing a PTC stemming from a frameshift (5,6). Successively, homologs of these three genes were also found in other organisms including worms, flies and humans. Upf1 is a phosphoprotein with ATPase and 5′–3′ helicase activities (9–12). It is envisioned that the helicase activity of Upf1 somehow allows the NMD surveillance complex to scan RNA molecules. Consistently, ectopically expressed helicase-dead variants of Upf1 exert a dominant negative effect on NMD in different organisms (10,13). Upf2 and Upf3 proteins seem to interact with Upf1 on the nonsense mRNA (14). Once the Upf complex has recognized an mRNA as nonsense, this mRNA is degraded via contributions from both the 5′–3′ decapping-dependent exonuclease pathway, and the deadenylation dependent 3′–5′ exosome pathway (15–17). NMD most likely takes place in cytoplasmic foci referred to as processing bodies (P-bodies) as many of the proteins that are involved in decapping and general RNA decay reside in P-bodies (18–20). Further supporting this hypothesis, yeast PTC-containing mRNAs are targeted to P-bodies in an Upf1p-dependent manner (14).
In higher eukaryotes including worms and mammals, the core of the NMD machinery also comprises four factors named Smg1, Smg5, Smg6 and Smg7. All these factors influence the phosphorylation state of Upf1. Smg1 belongs to the phosphoinositide 3-kinase-related kinase (PIKK) family and directly phosphorylates Upf1 at S/T-Q residues during NMD cycles (21). Although Saccharomyces cerevisiae Upf1 has been recently demonstrated to be a phosphoprotein (22), no obvious Smg1 ortholog has yet been identified in yeast. In contrary to Smg1, the other Smg proteins (herein referred to as Smg5-7) promote Upf1 dephosphorylation, which is likely carried out by protein phosphatase 2A (PP2A) (4,23,24). Consistently, Smg5-7 interact with PP2A and at the same time directly bind to Upf1 through highly conserved residues within their 14-3-3 domains (25). As is the case for Smg1, no obvious Smg5-7 ortholog has been found in S. cerevisiae, leading to the assumption that these factors do not exist in yeast (1,26).
The S. cerevisiae Ebs1 protein was first characterized based on its sequence similarity with yeast Est1p, a subunit of the telomerase holoenzyme, which adds newly synthesized TG-rich repeats to the termini of yeast chromosomes, the telomeres (27). Deletion of the EST1 gene leads to a continuous shortening of telomeres, which ultimately causes loss of the telomeric cap and cellular senescence (28). ebs1Δ cells also have short telomeres compared to wild-type cells, however the length of the telomeric tract is stable and cells do not experience senescence (27). These observations suggest that the telomere shortening phenotypes detected in est1Δ and in ebs1Δ cells stem from different mechanisms. More recently, it has been reported that Ebs1p functions as a ‘global inhibitor of translation’ (29).
We and others have previously demonstrated that the human genome contains three genes encoding for polypeptides with sequence similarity to yeast Est1p and Ebs1p. The three putative orthologs were named EST1A, EST1B and EST1C (30,31). Similarly to yeast Est1p, EST1A and EST1B interact with human telomerase. Furthermore, overexpression of EST1A causes telomeric dysfunctions (30,31). Strikingly EST1A, EST1B and EST1C are identical to Smg6, Smg5 and Smg7, respectively, raising the attractive possibility that Est1p and/or Ebs1p might also function in NMD.
Here we demonstrate that EBS1 (but not EST1) deletion leads to a slight but highly reproducible stabilization of NMD targets. Moreover, Ebs1p possesses several features typical of the Smg5-7 proteins: (i) Ebs1p, as well as Upf1p, localizes to P-bodies upon glucose starvation and (ii) overexpression of Ebs1p leads to accumulation Upf1p in P-bodies. Altogether, we provide evidence that Ebs1p is a new component of the yeast NMD machinery and we propose that at least one of the Smg5-7 proteins is conserved throughout evolution from yeast to man. We also demonstrate that ebs1Δ mutants and all the other tested NMD mutants are resistant to the immunosuppressive drug rapamycin, implicating the NMD machinery in cell growth control pathways.
MATERIALS AND METHODS
Antibodies
Rabbit anti-human Smg7 polyclonal antibody was generated using a synthetic 40-amino acid long peptide corresponding to a sequence close to the C-terminus of Smg7 (N-PAMGGFGIDYLSATSSSESSWHQASTPSG TWTGHGPSMED-C). Two rabbits were immunized with the peptide at Eurogentec Bel S.A. (Belgium). The peptide–antigen was coupled to NHS-activated Sepharose 4 Fast Flow (Pharmacia) and used to affinity-purify anti-Smg7 antibodies according to standard procedures. Rabbit polyclonal anti-human Upf2, Upf3 and Smg6 were kind gifts from H.M. Jack, L. Maquat and L. Harrington, respectively; goat polyclonal anti-human Upf1 antibody (BL411G) was purchased from Bethyl Laboratories; mouse monoclonal anti-Flag (M2) and anti-human αTubulin (DM1A) antibodies were from Sigma; rabbit polyclonal anti-myc (CM-100) was from Gramsch Laboratories; mouse monoclonal anti-GFP (11814460001) was from Roche; mouse monoclonal anti-HA.11 (MMS-101R-500) was from Covance; rabbit polyclonal anti-human histone H3 (1791-100) was from Abcam.
Strain constructions and genetic manipulations
Yeast strains are described in Table 1. The genotypes of the yeast strains are derived from either HFY1200 (32) (ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15, ura3-1) or S288C (his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). All haploid strains in the S288C background were derived from heterozygous diploids (Euroscarf deletion consortium) and subsequently micromanipulated to obtain the indicated haploid strain. Standard yeast growth conditions and genetic manipulations were used as described (33). Yeast transformations were performed by the lithium acetate procedure (34). YBL108 (ebs1::EBS1-MYC13) was endogenously tagged using a PCR-based method described previously (35). Strains expressing endogenously tagged Upf1p-GFP and Dcp2p-GFP were obtained from the Invitrogen GFP clone collection. Ebs1p-dsRFP was endogenously tagged using previously described methods (36). To construct YBL79, the pMX6 NAT cassette was linearized with EcoRI and transformed into YBL35. Transformants were selected on YPD medium containing 100 μg/ml nourseothricin.
Table 1.
Yeast strains used in this work
| Strain | Genotype | Background | Source |
|---|---|---|---|
| HFY1200 | Mat a | HFY | (32) |
| HFY870 | Mat a upf1::HIS3 | HFY | (32) |
| YCA1 | Mat a ebs1::TRP1 | HFY | This study |
| YCA2 | Mat a est1::HIS3 | HFY | This study |
| YCA3 | Mat a ebs1::TRP1 est1::HIS3 | HFY | This study |
| YBL40 | Mat a | S288c | This study |
| YBL35 | Mat a ebs1::KAN | S288c | This study |
| YBL38 | Mat a upf1::KAN | S288c | This study |
| YBL89 | Mat a upf1::NAT upf1::KAN | S288c | This study |
| YBL125 | Mat a Ebs1-RFP URA3 Upf1-GFP HIS3 | S288c | This study |
| YBL130 | Mat a Ebs1-RFP URA3 Dcp2-GFP HIS3 | S288c | This study |
| YBL11 | Mat a Upf1-GFP HIS3 | S288c | This study |
| YBL102 | Mat a ebs1::NAT xrn1::NAT | S288c | This study |
| YBL107 | Mat alpha ebs1::EBS1-13-myc KAN Upf1-GFP HIS3 | S288c | This study |
| YBL87 | Mat a ski2::KAN | S288c | This study |
| YBL99 | Mat a ski2::KAN ebs1::NAT | S288c | This study |
| YBL43 | Mat a xrn1::KAN | S288c | This study |
| YBL59 | Mat a upf2::KAN | S288c | This study |
| YBL47 | Mat a upf3::KAN | S288c | This study |
| YBL54 | Mat a lsm1::KAN | S288c | This study |
| YBL45 | Mat a lsm7::KAN | S288c | This study |
| YBL56 | Mat a pat1::KAN | S288c | This study |
| YBL50 | Mat a dhh1::KAN | S288c | This study |
| YBL79 | Mat a ebs1::NAT | S288c | This study |
| YBL108 | Mat a ebs1::EBS1-13-myc KAN | S288c | This study |
DNA manipulations and plasmids
Standard procedures were used for recombinant DNA manipulations (37). PCR was performed with the High Fidelity polymerase kit as recommended by the manufacturer (Roche) and verified by sequencing. Yeast genomic DNA was prepared using the Wizard genomic DNA purification kit (Promega). Telomere PCR was performed as previously described (38). Details of yeast plasmid constructions i.e. pADH-HA-Ebs1 (pBL75) and pADH-Smg7 are available upon request. All yeast galactose overexpression plasmids were ordered from Open-biosystems. The Est1-18myc plasmid was a kind gift from V. Lundblad. The centromeric DCP2-RFP plasmid (pTG003) was a kind gift from Mark Schmitt. The human Smg7 cDNA was obtained from the Kazusa DNA research Institute (clone KIAA0250). The Flag-Smg7 plasmid encodes for a N-terminally tagged Smg7 variant cloned under the control of the CMV promoter.
Immunoprecipitations and western blots
Yeast cultures measuring 50 ml were grown until mid-log phase, centrifuged washed in 10 ml of ice-cold PBS 1× and re-centrifuged. Cell pellets were resuspended in 200 μl of IP buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2% NP-40, protease inhibitor cocktail from Roche) and broken 2 × 20 s using the Bio101 disruptor. A total of 800 μl of IP buffer was then added and lysates were centrifuged at 13 000 r.p.m. for 10 min at 4°C. For IPs, 3 μl of polyclonal anti-myc antibody was added to 1 mg of cell lysate and gently mixed at 4°C for 2 h in the presence of 30 U of DNase I (Roche). Fifty microliters of pre-washed protein G beads were added and mixed for 2 h. Beads were washed 5 × 5 min and immunocomplexes were eluted in 50 μl of 3× Laemmli buffer. For human cells, protein extractions and IP conditions were previously described (3). Western blot experiments were performed according to standard procedures.
RNA isolation and northern blotting
For RNA isolation from yeast, cells were grown in 10 ml of medium until approximately OD600 0.35. For RNA half-life experiments, thiolutin (Biotrend GmbH) was directly added to the culture medium to a final concentration of 3 μg/ml. Cells were then harvested and treated with Zymolyase for 30 min at 30°C in order to produce spheroblasts. Human and yeast RNA was isolated using the RNeasy mini-kit from Qiagen. For northern blots, 5–20 μg of RNA was loaded onto formaldehyde agarose gels, separated by electrophoresis and transferred to Hybond N+ membranes (Amersham). Blots were blocked in Church buffer for 1 h at 60°C and then incubated with 32P-labeled DNA probes. Hybridizations were performed for 18 h at 50–60°C and washes were in 0.2 × SSC, 0.5% SDS at 50–60°. Signals were detected and quantified with a phosphoimager (Fuji) and the AIDA software (Raytest). The β-globin probe is a PCR fragment corresponding to exon 1 of the human gene; the human β-actin probe and the yeast 7S probe are 5′ end-labeled oligonucleotide (β-actin: 5′-GTGAGGA TCTTCATGAGGTAGTCAGTCAGGT-3′; 5′-GTCTA GCCGCGAGGAAGG-3′); the CYH2 and the ade2-1 probe are DNA fragments prepared from the pGEM4Z-CYH2 plasmid and an xrn1: ADE2 disruption plasmid, respectively (kind gifts of A. Jacobson); the URA5 and STE2 probes are PCR products of the entire ORFs and RP51B pre-mRNA is a PCR fragment of the intron.
Fluorescence microscopy
Yeast cells were washed twice with complete minimal medium and resuspended in the same. Cell suspensions were spotted onto clean slides and rapidly observed. Images were acquired using the LSM510 confocal microscope (Zeiss) equipped with Argon (wavelengths 457/488/514) and HeNe (wavelength 543) lasers.
Short hairpin (ShRNA)-mediated depletion of human proteins
ShRNA plasmids were generated as described previously (3) using a variant of the pSUPER plasmid containing the puromycin-resistance gene. The 19 nt target sequences were as follows. For human Upf1: 5′-GAGAATCGC CTACTTCACT-3′ (shUpf1); for human Smg6: 5′-GGGT CACAGTGCTGAAGTA-3′ (sh6); for human Smg7: 5′-G TATTAGTGCGACACCACT-3′ (sh7A) and 5′-CGCTTG ATCTCTTATTAGGCA-3′ (sh7B). HeLa cell lines with a stably integrated PTC-containing human β-globin reporter gene (HeLa β-globinNS39) or the wild-type β-globin gene (HeLa β-globinWT) were a kind gift from O. Mühlemann. A total of 5 × 105 cells were plated onto 30 mm dishes, grown for 15 h and transfected with 4–8 μg of plasmid DNA using the Lipofectamine 2000 reagent (Invitrogen). Twenty-four hours after transfection, cells were selected in medium containing 1.5 μg/ml puromycin (Fluka) until the mock-transfected cells were dead (1–2 days).
35S incorporation following rapamycin treatment
Yeast cultures measuring 10 ml were grown to logarithmic phase in SD medium lacking methionine. Rapamycin was added to a final concentration of 100 nM. Rapamycin treatment was for 45 min preceding the addition of labeled methionine. Cells were pelleted, and resuspended in 1 l of the same medium, followed by 10 min labeling with 7 μCi of [35S]methionine (Amersham). Incorporation was stopped by addition of cycloheximide (100 μg/ml, Sigma), followed by cell-lysis and TCA precipitation of proteins. Equal amounts of proteins were loaded on a polyacrylamide gels, and analyzed by SDS–PAGE, CoomassieR (Acros Organics) staining and autoradiography. Staining intensities were measured by an Imaging Densiometer (Bio-rad) and quantified by using the QuantityOne software (Bio-rad). Dried gels were exposed to a phosphoscreen for 4 h and analyzed using a phosphoimager (Storm 860, Amersham) and the ImageJ software (http://rsb.info.nih.gov/ij/).
Polysome analysis
Yeast cultures (200 ml) were grown to exponential phase. Rapamycin-treated samples were grown in 100 nM rapamycin for 30 min. All cultures were treated with 100 μg/ml cycloheximide for 10 min on ice. After centrifugation, pellets were washed twice and resuspended in 700 μl lysis buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 30 mM MgCl2, 100 μg/ml cycloheximide, 0.2 mg/ml heparin, 0.2 μl/ml DEPC). Subsequently cells were lyzed by bead beating. Extracts [8 absorption units at 260 nm (A260)] were loaded on 12.2 ml 15–50% linear sucrose gradients prepared in 50 mM Tris–acetate pH 7.5, 50 mM NH4Cl, 12 mM MgCl2). The gradients were centrifuged at 39 000 r.p.m. in a Beckman SW41 rotor at 4°C for 2 h 45 min followed by analysis with an ISCO UV-6 gradient collector.
RESULTS AND DISCUSSION
Sequence analysis of the Est1/Ebs1/Smg5-7 protein family
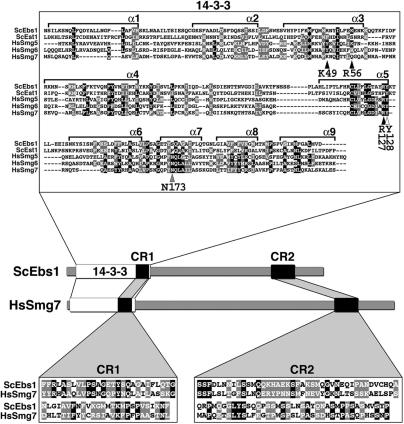
Yeast Ebs1p and Est1p are paralogous proteins that share 27% sequence identity and 48% sequence similarity. Whereas Ebs1p has been implicated in both telomere length regulation (27) and translation inhibition (29), the only proposed role of Est1p is to promote the recruitment of telomerase to chromosome ends (39). Interestingly, the N-terminal regions of both Ebs1p and Est1p comprise a 14-3-3 domain that is highly similar to that found in the higher eukaryotic NMD proteins Smg5, Smg6 and Smg7 (25). The putative 14-3-3 domain of the human Smg proteins forms a classic phosphoserine-binding pocket, which was shown to interact with phosphorylated Upf1 (25). We generated a multiple sequence alignment of the 14-3-3 domains of the human Smg proteins and the N-terminal region of both Ebs1p and Est1p (Figure 1). Careful inspection of the 14-3-3 sequences of Ebs1p and Est1p revealed differences between the two proteins with respect to key residues implicated in phosphoserine binding (40), suggesting different binding specificities. The human 14-3-3 zeta isoform (HZI) (40) residue Tyr128 is conserved in both Est1p and Ebs1p. HZI Asn173 is not conserved in either of the two yeast proteins while conservation is maintained in the human Smg proteins (Figure 1). Finally, HZI Lys49, Arg56 and Arg127 are conserved in Ebs1p and the other Smg proteins but not in Est1p (Figure 1). Pairwise alignments of Ebs1p with each of the three human Smg5-7 proteins revealed that yeast Ebs1p and human Smg7 have two highly conserved regions that do not exist in either Smg5 or Smg6. We have named these regions conserved region 1 (CR1) and conserved region 2 (CR2) (Figure 1, see insets). Furthermore, while both Smg5 and Smg6 have C-terminal PilT-amino-terminal (PIN) domains, a domain often associated with nuclease activity, (26,30,41,42), Smg7 does not contain a PIN domain, as is the case for both Est1p and Ebs1p. This sequence analysis suggests that Ebs1p has a domain organization similar to human Smg7 and further predicts that Ebs1p and/or Est1p may play a role in NMD as do the Smg proteins.
Figure 1.
The Smg5-7 14-3-3 phosphoserine-binding residues are conserved in Ebs1p. Multiple sequence alignment of the 14-3-3 domains of human Smg5 (amino acids 19–266), Smg6 (576–818) and Smg7 (1–236) proteins with the N-terminal regions of yeast Ebs1 (23–291) and Est1 (22–291) proteins. The alignment was produced using the ClustalW program. Alpha helices 1–9 (α1–α 9) are shown. Conserved amino acids are indicated as white letters on a black background while similar amino acids are white letters on a gray background. Arrowheads point to consensus 14-3-3 residues (indicated below each arrowhead) derived from the human 14-3-3 zeta isoform that have been implicated in phosphoserine binding for 14-3-3 domains across species. Black arrowheads indicate 14-3-3 residues that are conserved with respect to side-chain charge content in Ebs1p but not in Est1p, the white arrowhead points to a residue conserved in both Ebs1p and Est1p, the gray arrowhead highlights a residue, which is not conserved in either of the two yeast proteins. The cartoon depicts the domain architecture of yeast Ebs1 and human Smg7 proteins. Two conserved regions named CR1 and CR2 are indicated as black boxes and their sequence is shown in the lower insets. CR1: amino acids 231–283 in Ebs1p and 174–226 in Smg7; CR2: amino acids 710–789 in Ebs1p and 933–1013 in Smg7.
Deletion of EBS1 but not EST1 results in NMD defects similar to Smg depletion
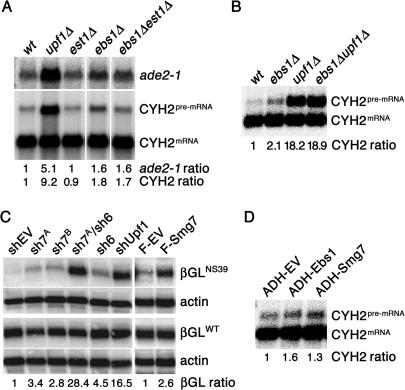
To determine if Ebs1p and/or Est1p are involved in NMD, we deleted the complete EBS1 and EST1 ORFs and determined if the two well-characterized NMD targets CYH2 pre-mRNA and a PTC containing ade2-1 allele were stabilized. While deletion of the EST1 gene did not lead to stabilization of the analyzed NMD target mRNAs, ebs1Δ cells showed a slight but very consistent accumulation of both CYH2 pre-mRNA and ade2-1 mRNA [1.52 ± 0.12 (n = 3) fold increase for ade2-1 and 1.78 ± 0.23 (n = 5) fold increase for CYH2] (Figure 2A and B). Moreover, est1Δ ebs1Δ double mutants did not show increased stabilization levels above that of the ebs1Δ single mutant, indicating that Est1p is not able to compensate for Ebs1p depletion (Figure 2A). The fact that the stabilization of the NMD targets in ebs1Δ cells was lower than the one observed in upf1Δ, upf2Δ and upf3Δ cells (Figure 2A and B and data not shown) suggests that Ebs1p plays a non-essential role in canonical NMD, explaining why EBS1 was not found in the UPF screen which identified UPF1, UPF2 and UPF3 (5,6).
Figure 2.
Ebs1 participates in NMD in a way similar to human Smg proteins. (A and B) Total RNA from log phase yeast grown in YPD was prepared from wt, upf1Δ, ebs1Δ, est1Δ, ebs1Δest1Δ and ebs1Δupf1Δ cells and subjected to northern blot analysis using a probe which detects both the CYH2 pre-mRNA and CYH2 mRNA. In (A) the filter was stripped and re-probed to detect the ade2-1 mRNA. CYH2 pre-mRNA levels (CYH2 ratio) and ade2-1 mRNA levels (ade2-1 ratio) are expressed relative to wild-type cells, after normalization using the CYH2 mRNA signal. (C) HeLaβGWT and HeLaβGNS39 cells (see Materials and Methods section) were transfected with shRNA vectors against human Smg7 (sh7A and sh7B), human Smg6 (sh6) and human Upf1 and with empty vector controls (shEV). Five days after transfection, total mRNA was prepared and the levels of β-globin mRNA were determined on a northern blot. Alternatively, the same cell lines were transfected with vectors expressing a Flag-tagged version of human Smg7 (F-Smg7) or empty vector controls (F-EV) and processed 48 h after transfection. The signal of βGNS39 (βGL ratio) is expressed relative to empty vector-transfected cells, after normalization using the actin signal (loading control) and the signal of βGWT. (D) Wild-type yeasts were transformed with centromeric plasmids harboring EBS1 and Smg7 under the constitutive ADH promoter, or with empty vector alone. Total RNA was probed using the CYH2 probe as in A–C.
In order to directly measure the stability of NMD targets, we treated wt and ebs1Δ cells with the transcription inhibitor thiolutin (43) and measured the relative amounts of CYH2 pre-mRNA and mRNA, as well as the pre-mRNA of another NMD target, RP51B at different times after addition of the inhibitor. In our experimental settings, the half-life (t1/2) of the CYH2 pre-mRNA was ∼2.7 min in ebs1Δ cells, while it was 1.2 min in wild-type cells (Supplementary Figure S1). Similarly, the t1/2 of RP51B pre mRNA was ∼6.5 min in ebs1Δ cells, while it was 3.3 min in wild-type cells. On the contrary, the t1/2 of the short-lived non-NMD targets URA5 (Wt t1/2 8.6 min, ebs1Δ t1/2 7.8 min) and STE2 (Wt t1/2 8.4 min, ebs1Δ t1/2 7 min) were not stabilized in ebsΔ cells (Supplementary Figure S1). These observations suggest that EBS1 deletion specifically impairs with the degradation of NMD targets. We then generated ebs1Δ upf1Δ double mutants and found that there was not a synergistic or an obvious additive effect with regards to CYH2 pre-mRNA stabilization (Figure 2B), suggesting that EBS1 lies in the same genetic pathway as the rest of the canonical genes involved in NMD.
Similar to the conservative effects on NMD, we noticed that the short telomere phenotype of ebs1Δ cells (27) is also less severe than that seen in upf1Δ cells (Supplementary Figure S2). This is consistent with the proposed model that the short telomere defect in upf mutants is an indirect effect of incomplete NMD (44). Nonetheless, we have recently shown that mammalian SMG proteins localize to telomeres and promote dissociation of telomeric repeat containing RNA (TERRA) from telomeres (45). It is possible that TERRA is also conserved in yeast and that the telomere shortening observed in yeast NMD mutants could derive at least in part from loss of proper TERRA regulation at chromosome ends.
To analyze the consequences of depletion of Ebs1-related human factors, we generated shRNAs directed against human Smg6 or Smg7. ShRNAs were validated by western blots, which confirmed that the levels of targeted proteins were strongly decreased (Supplementary Figure S3). As previously documented (26), shRNA-mediated depletion of Smg6 and Smg7 induced stabilization of a PTC containing β-globin mRNA (βGLNS39) when compared to empty vector transfected cells (Figure 2C). Remarkably, as is the case for yeast ebs1Δ mutants, knockdown of Smg7 did not stabilize βGLNS39 mRNA to the extent achieved using shRNA against human Upf1. However, simultaneous shRNA-mediated depletion of Smg6 and Smg7 resulted in a dramatic increase in βGLNS39 transcript that was comparable to the one observed in cells depleted for Upf1 (Figure 2C). Similar results were obtained using different shRNA-directed against Smg7, Smg6 and Upf1 (Figure 2C and data not shown). This suggests that the human Smg6 and Smg7 proteins can substitute for each other and raises the possibility that there may be another Smg-like protein in yeast (which is not Est1p), that can partially substitute for Ebs1p. We searched the yeast database for PIN domain containing proteins, which could be potential orthologs of either Smg5 or Smg6, and identified NMD4, YKR096w and YOR166c. Analysis of NMD targets in the three single mutant strains as well as in the double mutants with ebs1Δ did not reveal any increase in NMD impairment as compared to single EBS1 mutants (data not shown), ruling out the possibility that these PIN-domain-containing proteins could substitute for Ebs1p.
We then tested if overexpressing either human Smg7 or yeast Ebs1 proteins could affect NMD. Indeed, overexpression of Smg7 in human cells resulted in stabilization of βGLNS39 (Figure 2C, compare last two lanes). Similarly, overexpression of both Ebs1 and Smg7 in yeast resulted in slight stabilization of the CYH2 pre-mRNA (Figure 2D). Because Smg7 participates in the dephosphorylation of Upf1, it is possible that the impairment of NMD function observed in human cells depleted for or overexpressing Smg7 derives from a compromised balance between phosphorylated and unphosphorylated Upf1. Although yeast Upf1p has recently been demonstrated to be a phosphoprotein (22), we were unsuccessful in our attempts to detect phosphorylated Upf1p. Furthermore, there is no evidence that phosphorylation of yeast Upf1p plays a role in NMD and the human PI3-like Upf1 kinase, Smg1, has no apparent functional homolog in yeast. Indeed, deletion of the known yeast PI3-like kinases tor1, tel1 or mec1 does not impair with NMD function (data not shown).
ebs1Δ cells are competent to inhibit global translation upon rapamycin treatment
Slightly increased steady-state levels of NMD targets in EBS1Δ cells were already reported by Ford and colleagues (29). The same study also demonstrates that ebs1 deletion leads to increased translation of a lacZ reporter gene, while overexpression of Ebs1p leads to decreased translation of the same reporter. Furthermore, yeast strains deleted for EBS1 display resistance to the immunosuppressive drug and inhibitor of the TOR kinase, rapamycin (29). These observations led the authors to propose that Ebs1p might work as a global translation inhibitor and that the NMD defects as well as the rapamycin resistance observed in ebs1Δ cells are due to a failure in proper translation regulation (29).
To verify the involvement of Ebs1p in processes of translation inhibition, we performed pulse-labeling experiments with 35S-labeled methionine following rapamycin addition, which is known to result in down-regulation of translation (46). We found that 35S incorporation was inhibited to similar extents in wild-type and ebs1Δ cells following rapamycin treatment (Supplementary Figure S4). We also analyzed polysome profiles of wild-type and ebs1Δ cells following either rapamycin addition or glucose starvation and consistently we noticed the accumulation of an 80S monosome peak in both samples (Supplementary Figure S4). On the contrary, the rapamycin insensitive fpr1Δ cells showed no noticeable profile difference from the untreated control culture following the addition of rapamycin (Supplementary Figure S4). Similarly, polysome profiles from cells overexpressing Ebs1p did not show any substantial difference as compared to cells transformed with empty vector controls (Supplementary Figure S4). We then asked if Ebs1p level alterations could affect the cellular sensitivity to different translational inhibitors, including cycloheximide, paramomycin and hygromycin B. Neither the deletion nor the overexpression of Ebs1 affected the sensitivity of the cells to the tested inhibitors (Supplementary Figure S5). We thus conclude that the NMD defect and the rapamycin resistance observed in ebs1Δ cells are not a consequence of a general lack of translation inhibition. The apparent discrepancy between the data reported here and the ones shown by Ford et al. (29) might be due to the different experimental settings used to monitor translation efficiency.
We also noticed that all tested deletion mutants involved in NMD (upf1Δ, upf2Δ, upf3Δ and xrn1Δ) were highly resistant to rapamycin (Supplementary Figure S5). Furthermore, upf1Δ ebs1Δ double mutant cells had a rapamycin resistance that was similar to the respective single mutants, suggesting that both factors may function in the same genetic pathway that determines rapamycin sensitivity (Supplementary Figure S5). The rapamycin resistance of upf1Δ and ebs1Δ cells does not exactly recapitulate the defects of the respective mutants in NMD, which are much more pronounced upon UPF1 deletion. The even more pronounced rapamycin resistance of upf2Δ and upf3Δ cells, which behave identical to upf1Δ with regard to NMD-dysfunction, combined with the fact that ebs1Δ xrn1Δ and upf1Δ xrn1Δ double mutants are additive in terms of rapamycin resistance may suggest additional NMD-independent regulatory roles for these proteins (Supplementary Figure S5). In this regard, it is interesting to note that yeast Upf1p has been implicated in the trafficking of normal mRNAs in and possibly out of P-bodies while Upf2 and Upf3 work further downstream, and only on PTC-containing RNAs (14). It will be interesting to examine if Ebs1p and Upf1p collaborate in trafficking of normal mRNAs or yet another regulatory circuit that may explain the different extent of rapamycin resistance.
Ebs1p localizes to P-bodies and interacts with Upf1p
Processing bodies or P-bodies house all of the decapping proteins, such as Dcp1p, Dcp2p and the Lsm proteins, as well as the main 5' exonuclease, Xrn1p (47,48). It is predicated that the majority of RNA degradation occurs within P-bodies and it has been demonstrated that yeast NMD substrates are also targeted in an Upf1p-dependent manner to the P-bodies (14). In yeast, Upf1p itself has been detected within P-bodies upon depletion of the decapping enzymes or XRN1, which results in RNA accumulation within P-bodies and hence increased P-body size due to the accumulation of incompletely processed RNAs (14). Yeast Upf1p can also be seen in P-bodies in upf2Δ and upf3Δ mutants where Upf2p and Upf3p also trigger nonsense mRNA decapping in wild-type cells (14). Glucose starvation has also been shown to increase P-body size as judged by scoring the size of Dcp2p-GFP-containing foci (49).
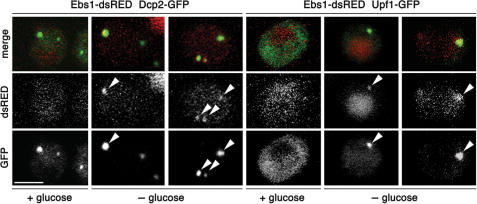
We constructed a dsRED-tagged version of Ebs1p and noticed a diffuse cytoplasmic staining in log phase cells grown in rich medium. Upon glucose starvation, Ebs1p-dsRED formed discrete cytoplasmic foci that co-localize with Dcp2p-GFP, indicating that like Upf1p, Ebs1p can localize to P-bodies (Figure 3). Furthermore, we found that Upf1p-GFP also co-localizes with Ebs1p-dsRED upon glucose starvation (Figure 3). While both Dcp2-GFP and Upf1-GFP protein levels were diminished upon glucose starvation (Supplementary Figure S6), we noticed that Ebs1p levels were not strongly affected (Supplementary Figure S6, see below). In contrary to Upf1p, only a fraction of Ebs1p-dsRED is recruited to P-bodies upon glucose starvation, with a substantial fraction remaining diffuse in the cytoplasm. This observation suggests that the P-body-associated function of Ebs1p does not require the entire cellular reservoir of Ebs1p.
Figure 3.
Ebs1p and Upf1p co-localize in P-bodies in glucose-starved cells. Yeast cells expressing Ebs1p-dsRED and Dcp2p-GFP fusions as well as cells expressing Ebs1p-dsRED and Upf1p-GFP were grown in rich synthetic media and successively resuspended in media lacking glucose while shaking at 30°C. Four hours after, confocal images of live cells were taken. Arrows point to cytoplasmic foci where a co-localization between Ebs1p-dsRED with Dcp2p-GFP and Ebs1p-dsRED with Upf1p-GFP occurred. Scale bar represents 2.5 μm.
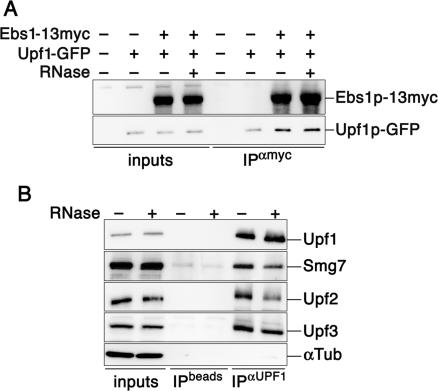
Overexpression of Smg5 and Smg7 in human cells results in the accumulation of Upf1 into P-bodies, while overexpression of Smg6 forces Upf1 into foci that do not co-localize with markers for P-bodies (50). Therefore, we overexpressed Ebs1p under the control of the ADH promoter and found that analogous to human Smg5 and 7, this also resulted in the recruitment of Upf1p-GFP into cytoplasmic foci containing an RFP-tagged version of Dcp2p (Figure 4). Importantly, overexpression of Ebs1p induces accumulation of UPF1-GFP protein as measured by western blotting experiments (Supplementary Figure S3). Because overexpression of Upf1p can increase the levels of Upf1p itself in P-bodies (14), we cannot exclude that the Upf1-containing P-bodies that we observe in cells overexpressing Ebs1p may at least in part derive from the higher intracellular levels of Upf1p. Nevertheless, co-immunoprecipitation experiments of endogenously expressed proteins show that Ebs1p-13myc (but not Est1p-18myc; data not shown) co-immunoprecipitates together with Upf1p-GFP (Figure 5A), suggesting that Ebs1p might directly interact with Upf1p in P-bodies. The amount of Upf1p that co-immunoprecipitated with Ebs1p did not increase following glucose withdrawal (Supplementary Figure 6), suggesting that glucose starvation leads to the recruitment of pre-associated Ebs1p-Upf1p molecules into P-bodies. Interestingly, the interaction between Upf1p and Ebs1p was maintained when extracts were treated with RNase prior to performing the immunoprecipitation experiments (Figure 5A). This is in partial contrast to what we observed in human cells, whereby endogenous Upf1 co-immunoprecipitates Smg7 in a partially RNA-dependent manner (Figure 5B). We speculate that the interaction between Upf1p/Upf1 and Ebs1p/Smg7 may occur through direct protein–protein interactions and in human cells also through RNA molecules simultaneously bound to both factors. The interaction of human Upf1 with human Upf2 and Upf3 is also partially lost when extracts are treated with RNase (Figure 5B), suggesting that RNA molecules stabilize the entire human NMD core complex.
Figure 4.
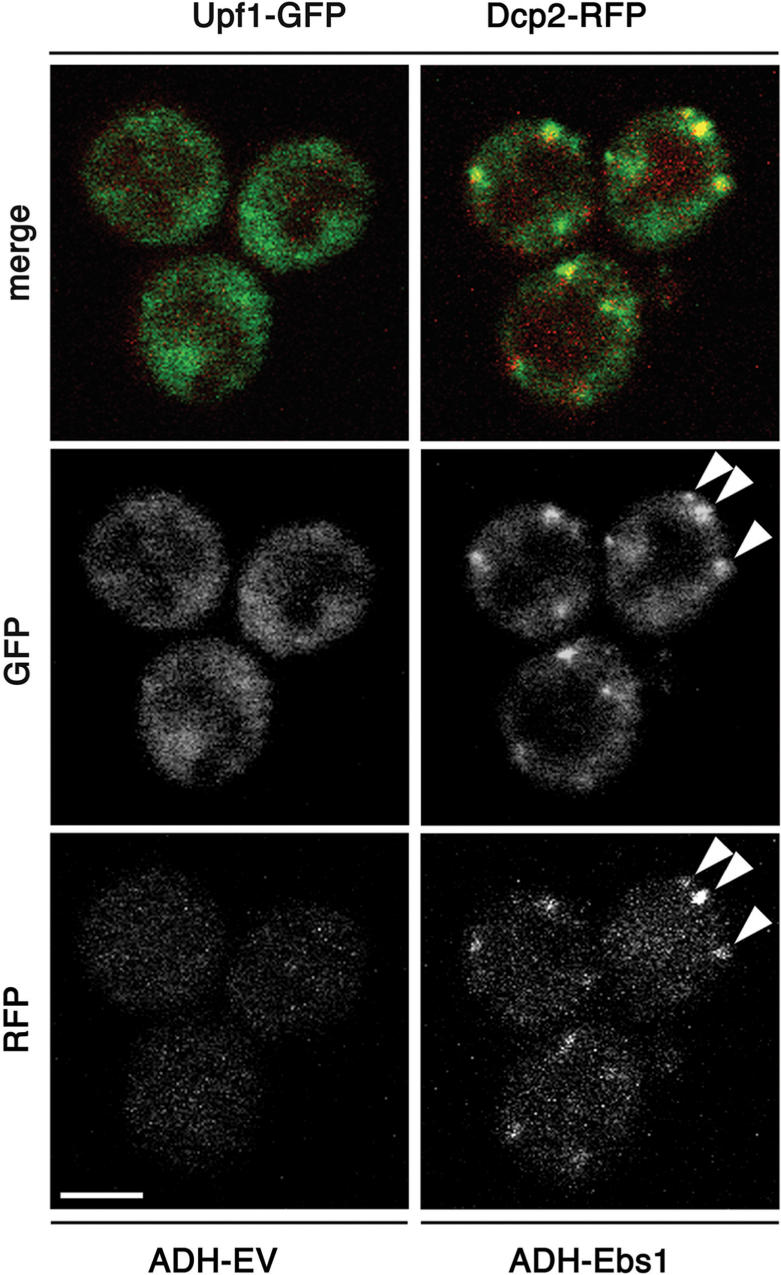
Overexpression of Ebs1 recruits Upf1p into P-bodies. Upf1p-GFP and Dcp2p-RFP cells were transformed with either empty vector (EV) or with a plasmid, which overexpressed Ebs1p under the ADH promoter (ADH-Ebs1) and subjected to confocal microscopy. Arrowheads indicate examples of cytoplasmic foci where a co-localization between Upf1p-GFP and Dcp2p-RFP occurred. At the time of imaging, 74% of the cells transformed with ADH-Ebs1 displayed 2–6 Upf1p-GFP foci and 6% of the cells transformed with empty vectors displayed 1 to 2 Upf1p-GFP foci. Scale bar represents 2.5 μm.
Figure 5.
Smg7/Ebs1p physically interacts with Upf1 proteins in a partially RNA-dependent manner. (A) Myc tagged Ebs1p was immunoprecipitated from cell lysates prepared from strains expressing Upf1p-GFP. Lysates were treated with RNase or left untreated. Western blots were performed using anti-myc and anti-GFP antibodies. (B) Protein extracts were prepared form HeLa cells and treated with RNase or left untreated. Human Upf1 was immunoprecipitated using goat polyclonal antibodies and western blot analysis was performed using rabbit polyclonal antibodies raised against human Smg7, Upf2 and Upf3 and goat polyclonal antibodies raised against Upf1. As a control for IP specificity, α-Tubulin does not co-immunoprecipitate with Upf1.
The above-described results suggest that Ebs1p could potentially be involved in either promoting Upf1p delivery or Upf1p retention in P-bodies, drawing another parallel between the Smg5-7 proteins and Ebs1p. In fact, the C-terminal region of the human Smg7 polypeptide is sufficient for recruiting Upf1 into P-bodies when overexpressed (50). The CR2 present in the C-terminus of both Smg7 and Ebs1 (Figure 1) might represent a structural domain essential for the Ebs1-induced localization of Upf1p into P-bodies. However, Upf1p-GFP is still efficiently recruited into cytoplasmic foci upon glucose starvation in ebs1Δ cells (data not shown), indicating that Ebs1p alone is not required for the Upf1p recruitment into P-bodies but possibly for downstream regulatory events, which may include Upf1p retention or release. We cannot, however, rule out the possibility that there is another protein substituting for Ebs1p in its absence, which would allow Upf1p recruitment. This second hypothesis is consistent with the fact that NMD target accumulation is incomplete in single deletion of EBS1 as well as in single knockdowns of Smg6 and Smg7 (Figure 2).
In conclusion, our data indicate that Ebs1p is a novel member of the NMD machinery in yeast and a putative ortholog of human Smg7. Thus, although it has been assumed previously that Smg proteins do not exist in yeast, our data suggest that careful screening of the yeast genome may allow identification of additional Smg-like proteins, which would further corroborate conservation of NMD throughout eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank V. Lundblad, O. Mühlemann, H.M. Jack, L. Maquat, L. Harrington, A. Jacobson, M. Schmitt and R. Tsien for sharing reagents. Work in MP's laboratory was supported by grants form the Swiss National Science Foundation and the ETHZ. Work in JL's laboratory was supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, the Human Frontier Science Program and the EU 6th Framework Programme. Funding to pay the Open Access publication charges for this article was provided by the Human Frontier Science Program grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 2.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 6.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 9.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 10.Weng Y, Czaplinski K, Peltz SW. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carastro LM, Tan CK, Selg M, Jack HM, So AG, Downey KM. Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res. 2002;30:2232–2243. doi: 10.1093/nar/30.10.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J. 2003;22:3951–3959. doi: 10.1093/emboj/cdg374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol. Cell. Biol. 2006;26:3390–3400. doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 25.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 29.Ford AS, Guan Q, Neeno-Eckwall E, Culbertson MR. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2006;5:301–312. doi: 10.1128/EC.5.2.301-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 2003;13:568–574. doi: 10.1016/s0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 31.Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 2003;13:698–704. doi: 10.1016/s0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 32.Maderazo AB, Belk JP, He F, Jacobson A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol. Cell. Biol. 2003;23:842–851. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 34.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, McKenzie A., III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 37.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (1991) Current Protocols in Molecular Biology. New York, NY: Greene Publishing Associates and Wiley-Interscience; [Google Scholar]
- 38.Forstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 40.Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 41.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 42.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parker R, Herrick D, Peltz SW, Jacobson A. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:415–423. doi: 10.1016/0076-6879(91)94032-8. [DOI] [PubMed] [Google Scholar]
- 44.Lew JE, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 1998;18:6121–6130. doi: 10.1128/mcb.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007 doi: 10.1126/science.1147182. [Epub ahead of print; Oct 4] PMID: 17916692. [DOI] [PubMed] [Google Scholar]
- 46.Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc. Natl Acad. Sci. USA. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 48.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.