Abstract
Guanine-rich DNA sequences can form G-quadruplexes stabilized by stacked G–G–G–G tetrads in monovalent cation-containing solution. The length and number of individual G-tracts and the length and sequence context of linker residues define the diverse topologies adopted by G-quadruplexes. The review highlights recent solution NMR-based G-quadruplex structures formed by the four-repeat human telomere in K+ solution and the guanine-rich strands of c-myc, c-kit and variant bcl-2 oncogenic promoters, as well as a bimolecular G-quadruplex that targets HIV-1 integrase. Such structure determinations have helped to identify unanticipated scaffolds such as interlocked G-quadruplexes, as well as novel topologies represented by double-chain-reversal and V-shaped loops, triads, mixed tetrads, adenine-mediated pentads and hexads and snap-back G-tetrad alignments. The review also highlights the recent identification of guanine-rich sequences positioned adjacent to translation start sites in 5′-untranslated regions (5′-UTRs) of RNA oncogenic sequences. The activity of the enzyme telomerase, which maintains telomere length, can be negatively regulated through G-quadruplex formation at telomeric ends. The review evaluates progress related to ongoing efforts to identify small molecule drugs that bind and stabilize distinct G-quadruplex scaffolds associated with telomeric and oncogenic sequences, and outlines progress towards identifying recognition principles based on several X-ray-based structures of ligand–G-quadruplex complexes.
INTRODUCTION
DNA can adopt structures other than the Watson–Crick duplex when actively participating in replication, transcription, recombination and damage repair. Of particular interest are guanine-rich regions, which can adopt a non-canonical four-stranded topology called the G-quadruplex. Such architectures are adopted in several key biological contexts, including DNA telomere ends, the purine-rich DNA strands of oncogenic promoter elements, and within RNA 5′-untranslated regions (UTR) in close proximity to translation start sites. Therefore, elucidation of the sequence-based diversity of G-quadruplex scaffolds could provide insights into the distinct biology of guanine-rich sequences within the genome.
Guanine-rich DNA G-quadruplexes
G-quadruplexes are built from the stacking of successive G–G–G–G tetrads (G-tetrads) and stabilized by bound monovalent Na+ and K+ cations (1). The G-tetrad is a cyclic hydrogen-bonded square planar alignment of four guanines (Figure 1a), with the guanines adopting either anti or syn alignments about glycosidic bonds (Figure 1b and c, respectively). G-quadruplexes are very stable, with their large diameter and four grooves defining a unique architecture (2) that is distinct from duplex DNA.
Figure 1.
(a) Schematic alignment of four guanines in a plane to form the G–G–G–G tetrad (G-tetrad). Each guanine uses its Watson–Crick and major groove edges to form a pair of hydrogen bonds. This leaves the minor groove edge available for further recognition. Schematic illustrating (b) anti and (c) syn guanine glycosidic torsion angle alignments. The H8 and sugar H1′ protons are closer to each other (2.5 Å) in the syn alignment compared to a longer distance (3.7 Å) in the anti alignment, a feature that can be readily monitored by NMR (256). (d) Two views of K+ cation-coordination between adjacent G-tetrad planes. Each tetrad provides four inwardly pointing guanine carbonyls, such that the dehydrated K+ cation participates in a directly coordinated tetragonal bipyramidal arrangement (10).
The backbone strands (or columns) that constitute the stacked G-tetrad core of the G-quadruplex can adopt different directionalities. Furthermore, the relative strand directionalities are geometrically related with the glycosidic conformation of the guanines. There are four possibilities: (i) Four strands are oriented in the same direction; the glycosidic angles around the G-tetrad are anti–anti–anti–anti (3–5), and occasionally syn–syn–syn–syn (6). (ii) Three strands are oriented in one direction and the fourth is oriented in the opposite direction; the glycosidic angles are syn–anti–anti–anti or anti–syn–syn–syn (7). (iii) Two neighboring strands are oriented in one direction and the two remaining strands oriented in the opposite direction (as a result of which each strand has both parallel and anti-parallel adjacent neighbors); the glycosidic angles are syn–syn–anti–anti (8–10). (iv) Each strand has adjacent anti-parallel neighbors; the glycosidic angles are syn–anti–syn–anti (11–14).
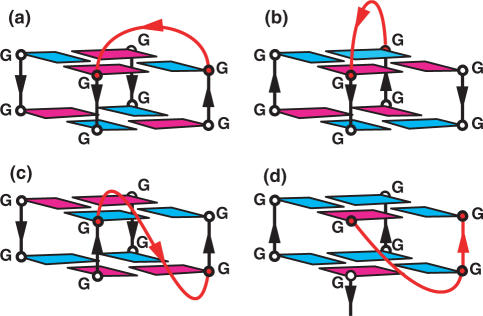
Loops in G-quadruplexes are linkers connecting G-rich tracts that support the stacked G-tetrad core. The loops can be classified into four major families that depend in part on the size and sequence of the linkers: (i) Edge-wise or lateral loops connect two adjacent anti-parallel strands (Figure 2a), and are generally composed of two or more residues (9,15). (ii) Diagonal loops connect two opposing anti-parallel strands (Figure 2b) (8–10), and are generally composed of three or more residues. (iii) Double-chain-reversal or propeller loops connect adjacent parallel strands (Figure 2c) (7,16,17), and can be as small as one and as large as six or more residues. The adenine in single-residue double-chain-reversal loops that bridge two G-tetrad planes can form hydrogen bonds with one edge of the G-tetrad resulting in A–(G–G–G–G) pentad formation (18) or two opposing edges of the G-tetrad resulting in A–(G–G–G–G)–A hexad formation (16). (iv) V-shaped loops connecting two corners of a G-tetrad core in which a support column is missing (Figure 2d) (18).
Figure 2.
Schematic illustrating (a) edge-wise, (b) diagonal, (c) double-chain-reversal or propeller and (d) V-shaped loops. The loops connect individual strands or columns bridging two G-tetrad planes. Color-coding for schematics is as follows: anti guanines in blue and syn guanines in magenta. G-rich columns in black and connecting loops in red.
Furthermore, loop residues can form base-pairing alignments, which in turn stack with the terminal G-tetrads, further stabilizing G-quadruplex structures. These include three bases in a plane, which can be classified either as base triples, where all three bases are non-contiguous in the sequence, or as base triads (19), where two adjacent bases from one strand are involved in the pairing alignment with a base from a second strand (20). Loop conformations can adopt diverse topologies (21,22) making them attractive targets for small molecule-based ligand recognition.
The G-quadruplex topology is defined by four grooves whose dimensions (depth and width) and accessibility vary based on both the overall topology and whether the loops are edge-wise or diagonal on one hand, and double-chain-reversal on the other. G-quadruplex formation requires monovalent cations, which are positioned within the central channel of stacked G-tetrads, thereby neutralizing the strong electrostatic potential associated with the inwardly pointing guanine O6 oxygen (23). The dehydrated cations are positioned either in a tetragonal bipyramidal coordination between G-tetrads planes (K+) (Figure 1d) (10), or in a range of geometries that span positioning within G-tetrad planes to out of plane alignments (Na+) (24). It has been shown that in general G-quadruplexes prefer K+ over Na+, and that this reflects in part the much greater energetic penalty for Na+ dehydration (25). Finally, the same sequence can adopt different G-quadruplex conformations in Na+ (14) and K+ (26) solution as determined by NMR, and also as monitored by fluorescently labeled oligonucleotides (27).
The subject of G-quadruplexes has been extensively reviewed in the literature (28–38). Despite a wealth of crystal and solution structures, it has proved difficult to define a comprehensive set of rules that specify the folding propensity of G-quadruplexes. Therefore, each new guanine-rich telomeric and oncogenic promoter sequence has to be individually structurally characterized as a function of monovalent cation type and, in addition, checked for conformational heterogeneity between two or more topologies in solution.
This review presents a structural biology perspective of recent advances in structures of G-quadruplexes formed by human telomeric and oncogenic promoter G-rich tracts, as well as the potential of small molecules to target-specific G-quadruplex folds, thereby setting the stage for structure-based design of new classes of cancer therapeutics. The review also highlights the increasing attention being focused on G-quadruplexes formed by G-rich RNA sequences and their role in mRNA regulation and processing.
Biology of guanine-rich genomic sequences
Guanine-rich tracts are observed in critical segments of eukaryotic and prokaryotic genomes, promoter regions, both short microsatellite and longer minisatellite repeats, ribosomal DNAs, as well as telomeres in eukaryotes and immunoglobulin heavy chain switch regions of higher vertebrates. These guanine-rich tracts have the potential to form G-quadruplexes following transient destabilization of the duplex, a process that accompanies transcription, replication and recombination. Systematic algorithmic searches of bacterial and human genomes for guanine-rich tracts (restricted to minimum of four GGG segments separated by short linkers) (39–41) have noted that such putative G-quadruplex-forming sequences are prevalent in proto-oncogenes (which promote cell proliferation) and essentially lacking in tumor-suppressor genes (which maintain genomic stability) (42).
An increasing number of proteins have been identified that bind, promote or non-catalytically disrupt G-quadruplex formation (43–46). Both the β-subunit of the Oxytricha telomere end-binding protein (βTBP) (47) and repressor activator protein 1 (RAP1) in Saccharomyces cerevisiae (48) promote intermolecular G-quadruplex formation. In addition, the MutSα protein, involved in mismatch repair, targets G-quadruplex DNA in G-loop segments and promotes synapsis of transcriptionally activated immunoglobulin switch regions (49). Activation-induced cytosine deaminase (AID) also targets G-quadruplex DNA and plays a role in immunoglobulin class switch recombination (50). On the other hand, binding of POT1, a protein conserved from fission yeast to humans (51), disrupts G-quadruplex formation at telomeric G-rich overhangs (52), thereby promoting telomere extension by telomerase (53).
In addition, helicases catalytically unwind and nucleases cleave G-quadruplexes (43–46). RecQ DNA helicase family members are associated with genomic instability and predisposition to malignancies. The Bloom and Werner syndrome RecQ helicases bind to (54) and unwind intermolecular G-quadruplex scaffolds with a 3′ to 5′ polarity in the presence of ATP and Mg cations (55,56). Furthermore, G-quadruplex-specific nucleases cut within single-stranded DNA several nucleotides upstream of the G-quadruplex using a structure-specific mode of action (57–60). Gene disruption of such nucleases can lead to cellular senescence and telomere shortening (61). Such cleavage may also be required for DNA recombination and suggests that DNA quadruplexes may play a role in the formation of interchromosomal synapsis.
Strong evidence supporting G-quadruplex formation in vivo comes from the demonstration that in vitro generated single-chain antibody fragments specific for intermolecular telomeric G-quadruplex DNA react with ciliated protozoan Stylonychia lemnae macronuclei but not corresponding micronuclei (62). Additional evidence in support of G-quadruplex formation in vivo comes from the observation that telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo (63) and that intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G-quadruplex DNA on the non-template G-rich strand, as verified from nucleolin binding and sensitivity to G-quadruplex-specific nucleases (64). In addition, attempts have been made to monitor G-quadruplex formation at telomere proximal regions of chromosomal DNA using G-quadruplex-specific fluorescent 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide (BMVC) (65). Furthermore, it has been proposed that gene function correlates with potential for G-quadruplex formation in the human genome (42). Both inter- and intramolecular G-quadruplex formation has also been demonstrated for the diabetes susceptibility locus in the promoter region of the human insulin gene (66). Finally, guanine-rich tracts containing sequences capable of G-quadruplex formation have been shown to induce apoptosis in tumor cells (67–69).
In addition, guanine-rich RNA sequences capable of G-quadruplex formation have been identified in the vicinity of polyadenylation regions (70) involved in regulating 3′-end processing of mammalian pre-mRNAs (71). Such guanine-rich motifs can interact with hnRNP H protein subfamily members, thereby potentially mediating alternative, tissue-specific splicing events. There also appears to be a combinatorial code for splicing silencing which includes a combination of RNA UAGG and GGGG motifs (72). There are several examples of RNA G-quadruplex complexes that impact on pathways ranging from RNA processing, as in the case of the exoribonuclease mXRN1p (73), to translational repression, as in the case of the fd gene 5 protein (74). Guanine-rich tracts have also been observed within neuronal RNAs that bind the RGG-rich domain of the fragile X mental retardation (FXMR) protein (75,76).
HUMAN TELOMERIC G-QUADRUPLEXES
Telomeres, nucleoprotein complexes located at the ends of eukaryotic chromosomes, are composed of tandem DNA repeats of guanine-rich sequences (77). Telomeres are essential for chromosomal stability and genomic integrity, provide sites for recombination events and transcriptional silencing, and appear to play a critical role in cellular aging and cancer (43,78–81). Telomeric DNA ends are composed of both duplex and guanine-rich 3′-overhang segments, with the former progressively decreasing in length after each round of cell division in somatic cells (82). By contrast, telomeric overhangs can be elongated by the enzyme telomerase, a ribonucleoprotein complex with reverse transcriptase activity (83), which is expressed in the majority of cancer cells, thereby helping to maintain telomere length (84).
The pairing of homologous chromatids at their telomere ends can be mediated through bimolecular quadruplex formation (11). Such quadruplex structures may also play a role in chromosome synapsis and recombination during meiosis (85).
The guanine-rich 3′-overhangs of telomeres, such as TTAGGG repeats in humans can equilibrate between single-stranded and monovalent cation-mediated G-quadruplex folds, with the latter inhibiting the activity of telomerase. The telomeric ends in a single-stranded form are maintained by hPOT1 (86), while disruption of this interaction leads to quadruplex formation. Thus, ligand-induced stabilization of telomeric G-quadruplex scaffolds in humans constitutes a promising strategy for anti-cancer drug development (87–91). Therefore, much effort has been devoted to the structural characterization of G-quadruplex topologies formed by one, two, three and four human telomeric TTAGGG repeats as a function of monovalent cation, so as to define the scaffolds for anti-cancer drug discovery.
Though extensive studies have been undertaken on both ciliate (Tetrahymena and Oxytricha) and eukaryotic (yeast and human) telomeres, the emphasis in this review will be primarily on human telomeres. Single molecule fluorescence energy transfer (FRET) studies of structure and unfolding kinetics of the intramolecular human telomere G-quadruplex revealed two stable folded conformations in both K+ and Na+ buffers (92). Both folded conformations can be opened by addition of complementary oligonucleotide, with temperature dependent studies indicating that unfolding is entropically driven in K+ buffers (ΔH = 6.4 kcal mol−1 and ΔS = −52.3 cal mol−1 K−1), while unfolding in Na+ buffers exhibits a more significant enthalpic barrier (ΔH = 14.9 kcal mol−1 and ΔS = −23.0 kcal mol−1 K−1). Single-molecule FRET spectroscopy has also been used to probe the dynamics of human telomeric DNA containing four guanine-tracts in K+ solution. Interconversion was detected between three FRET values, interpreted in terms of an unfolded and two folded G-quadruplex states, each of which was further subdivided into long- and short-lived species (93). The short-lived species were shown to determine the overall dynamics, apparently because they bridge transitions between the long-lived G-quadruplex states.
Single-repeat sequences
The earliest structural information of the human telomere focused on NMR studies of the single-repeat d(TTAGGGT) human telomere sequence in K+ cation solution (4). The NMR data established that the single-repeat human telomere sequence tetramerizes to form an all-parallel-stranded G-quadruplex composed of three stacked G-tetrads with all anti guanine glycosidic torsion angles.
Two-repeat sequences
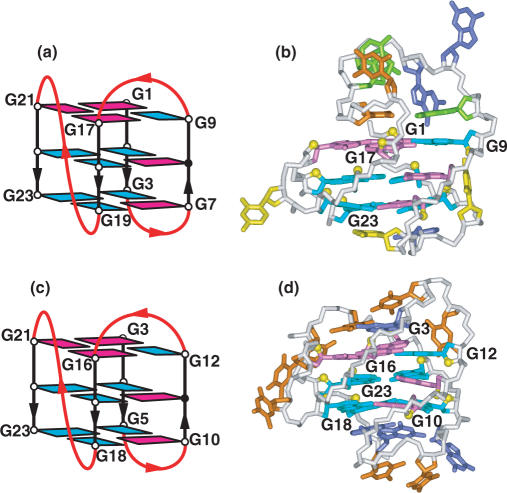
The X-ray structure of d(TAGGGTTAGGGT) crystals grown from K+-containing solution defined the architecture of the G-quadruplex formed by the two-repeat human telomere sequence (17). The structure contained an unanticipated all-parallel-stranded G-quadruplex following bimolecular association of the two-repeat human telomere sequences, with the TTA segments forming double-chain-reversal (or propeller) loops (Figure 3a). In addition, the end segments also participate in formation of a T–A–T–A tetrad, through pairing of the major groove edges of Watson–Crick A–T pairs (17).
Figure 3.
(a) X-ray structure of the two-repeat human telomere bimolecular G-quadruplex formed by the d(TAGGGTTAGGGT) sequence for crystals grown from K+ solution (coordinates deposition: 1K8P) (17). The bases are color coded as follows: guanine (blue), adenine (green) and thymine (orange). All strands are parallel in this bimolecular quadruplex and the loops are of the double-chain-reversal or propeller type. NMR-based folding topologies of interconverting (b) all parallel-stranded and (c) anti-parallel-stranded conformations of the two-repeat human telomere bimolecular G-quadruplex formed by the d(TAGGGTTAGGGT) sequence in K+ solution (94).
NMR studies on the two-repeat human telomere sequence d(TAGGGTTAGGGT) demonstrates interconversion between two dimeric G-quadruplex conformers consisting of three stacked G-tetrads in K+ solution (94). One of these conformers adopts a symmetric all-parallel-stranded G-quadruplex with double-chain-reversal loops and all anti guanines (Figure 3b), similar to that observed in the crystal structure (17). This conformer predominates for an analog containing a specific dU (in bold) for T substitution (designated U6)
5′-T1AGGG5(dU)TAGG10GT-3′
The other conformer adopts an asymmetric anti-parallel G-quadruplex with edge-wise loops composed of six syn guanines and six anti guanines (Figure 3c). This conformer predominates for an analog (designated U1,brU7) containing specific dU and dbrU (in bold) for T substitutions
5′-(dU)1AGGG5T(dbrU)AGG10GT-3′
NMR-based complementary-strand trap, concentration-jump and temperature-jump methods have been used to monitor the kinetics of interconversion and activation barriers between the parallel and anti-parallel G-quadruplex conformers (94). The equilibrium shifts towards the anti-parallel G-quadruplex (Figure 3c) at low temperature and towards the parallel G-quadruplex (Figure 3b) at high temperature for the U1,brU7 sequence, with the corresponding enthalpy being 18.5 kcal mol−1. Furthermore, the anti-parallel G-quadruplex folds faster, but unfolds slower than the parallel quadruplex at temperatures below 40°C.
A related conformational equilibrium has also been observed between a pair of bimolecular G-quadruplexes formed by the d(TGGGGTTGGGGT) two-repeat Tetrahymena sequence in Na+-containing solution (95).
Three-repeat sequences
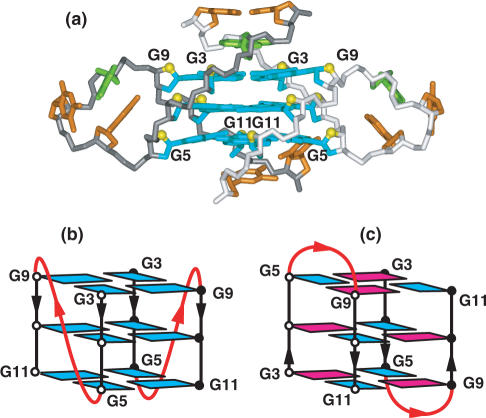
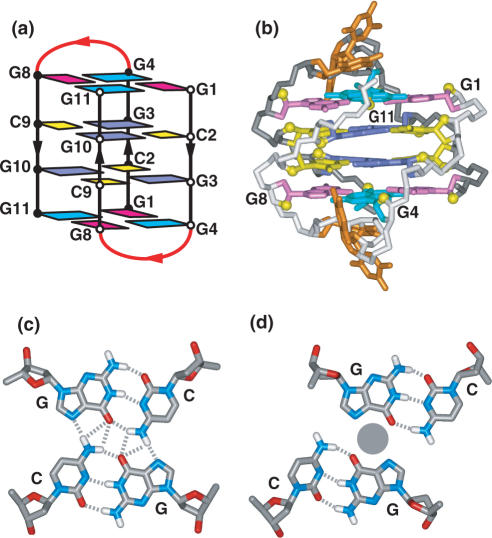
NMR-based studies have defined the folding topology (Figure 4a) and solution structure (Figure 4b) of the three-repeat human telomere sequence d[G3(T2AG3)2T]
5′-G1GGTT5AGGGT10TAGGG15T-3′
in Na+ solution (96). This sequence forms a unique asymmetric bimolecular quadruplex, in which the core composed of three stacked G-tetrads, involves all three G-tracts from one strand and only the last G-tract of the second strand. In this (3+1) G-quadruplex assembly, there is one syn–syn–syn–anti and two anti–anti–anti–syn G-tetrads, two edge-wise loops, three G-tracts oriented in one direction and the fourth oriented in the opposite direction (Figure 4a).
Figure 4.
NMR-based (a) (3 + 1) folding topology and (b) solution structure of the three-repeat human telomere bimolecular G-quadruplex formed by the d[G3(T2AG3)2T] sequence in Na+ solution (coordinates deposition: 2AQY) (96). All three G-tracts from one strand and the 3′-terminal G-tract from the partner strand are used to form the bimolecular G-quadruplex. (c) Schematic of hypothetical G-quadruplex formation when the 3′-end overhang (in red) invades the adjacent double-stranded segment of the telomere to form the so-called t-loop (97).
The (3+1) G-quadruplex topology adopted by the three-repeat human telomere sequence establishes how a segment containing three G-tracts can bind to the 3′-end G-tract of another segment. Such quadruplex formation could occur within the 3′-end overhang of human telomeres or when the 3′-end invades the adjacent double-stranded segment of the telomere to form the so-called t-loop (see schematic in Figure 4c) (97).
Earlier studies on four-repeat sequences
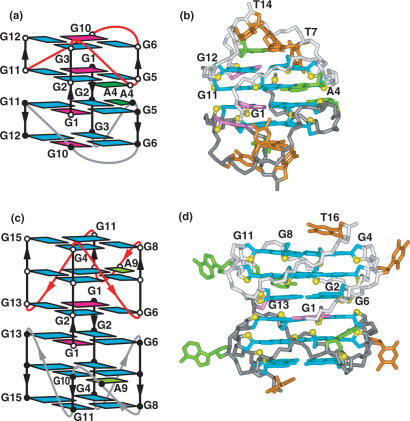
In 1993, the NMR-based folding topology (Figure 5a) and solution structure (Figure 5b) of the four-repeat human telomeric sequence d[AG3(T2AG3)3]
5′-A1GGGT5TAGGG10TTAGG15GTTAG20GG-3′
was solved in Na+ cation solution (9). The intramolecular fold contained three stacked G-tetrads connected by successive edge-wise, diagonal and edge-wise TTA loops. Each guanine-tract had both parallel and anti-parallel aligned neighboring strands around the G-quadruplex, with guanines adopting syn–syn–anti–anti glycosidic torsion alignments around each G-tetrad. The grooves were accessible for further recognition within this topology, while the connecting loops restricted access to the outward-directed faces of the terminal G-tetrads at both ends. Finally, the 5′- and 3′-terminii project toward the same ends of the G-quadruplex (Figure 5a).
Figure 5.
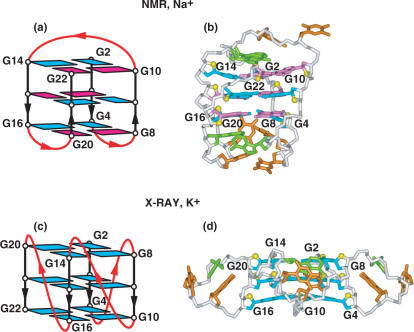
NMR-based (a) folding topology and (b) solution structure of the four-repeat human telomere unimolecular G-quadruplex formed by the d[AG3(T2AG3)3] sequence in Na+ solution (coordinates deposition: 143D) (9). The loop types starting from the 5′-end are edge-wise, diagonal and edge-wise. Individual strands have both a parallel and anti-parallel neighbor, with the G-tetrads adopting syn–syn–anti–anti alignments. X-ray-based (c) folding topology and (d) crystal structure of the four-repeat human telomere unimolecular G-quadruplex formed by the d[AG3(T2AG3)3] sequence for crystals grown from K+ solution (coordinates deposition: 1KF1) (17). All three loops are of the double-chain-reversal or propeller type, all strands are parallel and all guanines adopt anti alignments.
The X-ray structure of d[AG3(T2AG3)3] crystals grown from K+ cation solution exhibited a completely different and unanticipated fold (Figure 5c) and structure (Figure 5d) for the intramolecular G-quadruplex (17). The G-quadruplex was composed of three stacked G-tetrads, such that all strands are parallel, all guanines adopt anti conformations and all three loops are of the double-chain-reversal (or propeller) type. The double-chain-reversal loops restrict access to three of the grooves, while access is available to the outward-directed faces of the terminal G-tetrads at both ends. Finally, the 5′- and 3′-terminii project toward opposite ends of the G-quadruplex (Figure 5c), thereby facilitating potential end-to-end alignments of successive G-quadruplexes.
These very different conformers reported for the four-repeat human telomeric sequence in Na+-containing aqueous solution (9) and in K+-containing crystals (17) appear to highlight the polymorphic character of G-quadruplex scaffolds (93) as a function of medium and/or monovalent cation type. Nevertheless, accumulating evidence, including biophysical measurements (98), implied that the intramolecular parallel-stranded G-quadruplex structure of the human telomere observed in K+-containing crystals, appears unlikely to be the major form in K+-containing aqueous solution. To this end, three groups have recently systematically investigated the solution structure(s) of four guanine-repeat human telomeric sequences in K+ cation solution, while keeping in mind that the more crowded environment of the crystal may more closely reflect the crowded situation in the cell nucleus.
More recent studies on four-repeat sequences
The imino proton NMR spectrum of d[AG3(T2AG3)3] in K+ cation solution is indicative of multiple conformations in equilibrium and hence this sequence context is not readily amenable to structural characterization. Three research groups (those of Hiroshi Sugiyama, Danzhou Yang and our group) have taken somewhat different approaches to overcome this limitation and recently contributed to determination of the solution structure(s) of four-repeat human telomeres in K+ solution. Our group's approach is outlined in detail below and these results are placed in the context of independent contributions from the other two groups.
The imino proton NMR spectra corresponding to distinct predominant conformers together with one or more minor conformers were observed for the d[TAG3(T2AG3)3] sequence, where a T was added at the 5′-end (99), and for the d[TAG3(T2AG3)3TT] sequence, where a T was added at the 5′-end and a TT was added at the 3′-end (100), both in K+ cation solution, with both cases maintaining the sequence context of the TTAGG human telomere repeat.
5′-T1AGGG5TTAGG10GTTAGG15GTTAG20GG(TT)-3′
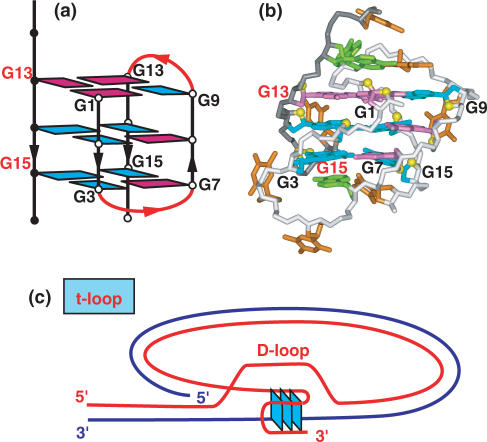
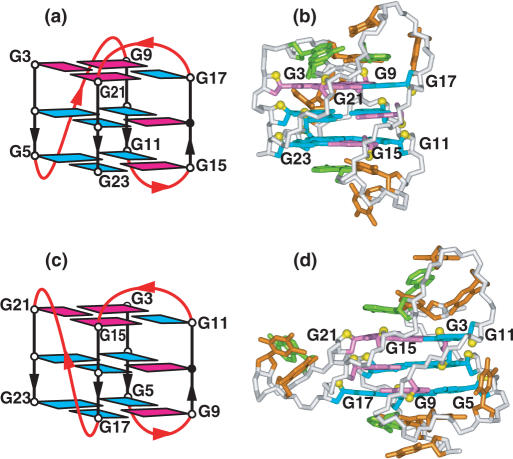
The NMR-based folding topology was determined for the predominant conformer of the d[TAG3(T2AG3)3] sequence in K+ cation solution (Figure 6a), and the solution structure determined for an analog containing terminal modifications (underlined) of this sequence, namely d[TTG3(T2AG3)3A], with the latter yielding exceptional NMR spectra reflecting a single conformer, together with the same 2D spectral characteristics of the unmodified sequence (99). Similarly, insertion of a single 8-bromoguanine at position G16 in the d[TAG3(T2AG3)3] sequence to enforce a syn glycosidic bond at this position also resulted in NMR spectra corresponding to a single conformer with all the spectral characteristics of the unmodified sequence (101). The solution structure has been determined for the d[TAG3(T2AG3)3] G-quadruplex (designated human telomere G-quadruplex form-1) (Figure 6b) (101), whose (3+1) topology differs from folds reported previously in Na+ solution (Figure 5a) (9) and K+-containing crystal (Figure 5c) (17). Instead, this G-quadruplex contains three G-tracts oriented in one direction and the fourth in the opposite direction, one anti–syn–syn–syn and two syn–anti–anti–anti G-tetrads, and a double-chain-reversal loop followed by two edge-wise loops (99).
Figure 6.
NMR-based (a) folding topology and (b) solution structure of the four-repeat human telomere unimolecular G-quadruplex formed by the d[TAG3(T2AG3)3] sequence (form-1) in K+ solution (coordinates deposition: 2JSM, 2JSK) (101). The loop types starting from the 5′-end are double-chain-reversal, edge-wise and edge-wise. Two other groups have independently investigated the same system and come to similar conclusions (102–105). NMR-based (c) folding topology and (d) solution structure of the four-repeat human telomere unimolecular G-quadruplex formed by the d[TAG3(T2AG3)3TT] sequence (form-2) in K+ solution (coordinates deposition: 2JSL, 2JSQ) (101). The loop types starting from the 5′-end are edge-wise, edge-wise and double-chain-reversal. One other group has independently investigated the same system and come to similar conclusions (106).
The same G-quadruplex folding topology (Figure 6a) has been independently reported for the four-repeat human telomere sequences in K+-containing solution by two other laboratories, one of which used NMR (102,103), while the other used both CD (104) and NMR (105). The NMR investigation by the former group focused on the sequence d[AAAG3(T2AG3)3AA], with the resulting (3+1) topology (102) stabilized by a stacked A–A–A triple (103), associated with introduction of terminal adenine modifications (underlined) at either end of the sequence. The latter groups research avoided terminal modifications and was based on judicious positioning of between four and five 8-bromoguanine substitutions, which enforce a syn guanine alignment at the corresponding guanines in the sequence (104,105).
The NMR-based folding topology has also been determined for the predominant conformer of the d[TAG3(T2AG3)3TT] sequence in K+ cation solution (100). This sequence adopts the same (3+1) G-quadruplex core topology adopted by the predominant conformer of the d[TAG3(T2AG3)3] in K+ cation solution (99) outlined in the previous paragraph, except that the first two linkers are of the edge-wise type and the last linker adopts a double-chain-reversal loop (designated human telomere G-quadruplex form-2) (Figure 6c). Insertion of a single 8-bromoguanine at position G15 in the sequence to enforce a syn glycosidic bond at this position resulted in NMR spectra corresponding to a single conformer with all the spectral characteristics of the unmodified sequence (101). The solution structure of the d[TAG3(T2AG3)3TT] G-quadruplex form-2 is shown in Figure 6d (101). An independent NMR-based study (106) has reached the same conclusions reported above regarding the folding topology (100) and solution structure (101) of form-2.
The demonstration of G-quadruplex forms 1 (Figure 6a) and 2 (Figure 6c) for the four-repeat human telomere in K+, together with the all-parallel-stranded, propeller-groove-linked G-quadruplex observed in crystals grown from K+ solution (Figure 5c) (17), support the view that multiple human telomeric G-quadruplex conformers can coexist in K+-containing solution, a conclusion reached from single molecule FRET studies of the four-repeat human telomere sequence (92). Furthermore, these studies establish that even small changes to flanking sequences perturb the equilibrium between different coexisting (3+1) G-quadruplex forms. More recent research has attempted to monitor G-quadruplex formation by the four-repeat human telomere in K+ solution under polyethylene glycol-induced crowding conditions (107) that perhaps mimic crystallization conditions.
(3 + 1) G-quadruplex fold
The (3 + 1) G-quadruplex scaffold is unique in that three stands are oriented in one direction and the fourth oriented in the opposite direction. Furthermore, two of the three G-tetrads adopt anti–anti–anti–syn alignments while the remaining G-tetrad adopts a syn–syn–syn–anti alignment. This topology was first reported in 1994 for the four-repeat Tetrahymena telomere sequence, d(T2G4)4, in Na+ solution (7) and observed a decade later for a four guanine-repeat variant bcl-2 promoter in K+ solution in which two guanines were replaced by thymines (108) (see bcl-2 sequence section).
The adaptation of the (3 + 1) core G-quadruplex by the three-repeat human telomere dimeric G-quadruplex in Na+ solution (Figure 4a) (96), as well as by the four-repeat human telomere G-quadruplexes form-1 (Figure 6a) and form-2 (Figure 6c) in K+ solution, established it to be a robust folding topology, thereby highlighting its candidacy as an important platform for structure-based drug design.
ONCOGENIC PROMOTER G-QUADRUPLEXES
Bioinformatics sequence analysis indicates that guanine-rich tracts capable of G-quadruplex formation are prevalent in the human genome (39–41). In addition, it has recently been shown that promoter regions spanning 1 kb upstream of transcription start sites of genes are significantly enriched in putative G-quadruplex-forming motifs and that these putative promoter G-quadruplex-forming regions strongly associate with nuclease hypersensitivity sites (109). It has been suggested that such promoter-based G-quadruplexes may be directly involved in gene regulation at the level of transcription (110). This has led to extensive investigations of the role of promoter-mediated G-quadruplex formation in transcriptional regulation of the oncogenic promoters of c-myc (111), VEGF (112), HIF-1α (113), bcl-2 (114) and c-kit (115,116).
Since promoter regions are part of DNA duplexes, they would be unwound during replication, prior to G-quadruplex formation. Support for this concept has emerged from single-molecule FRET studies on the c-kit promoter (117). This process could be facilitated by formation of single-stranded tracts during transcription and further stabilized through addition of G-quadruplex-stabilizing ligands (118).
Earlier studies on c-myc sequence
Human c-myc is a transcription factor that is central to regulation of cell growth, proliferation, differentiation and apoptosis (119–121). The c-myc gene that encodes this protein is tightly regulated in normal cells and its aberrant overexpression is associated with the progression of many cancers (122). c-myc can be deregulated as a result of translocation, mutation and/or amplification. An important element in the c-myc promoter region, termed the nuclease hypersensitivity element IIII (NHE IIII), controls up to 90% of total c-myc transcription (123). The 27-nt purine-rich strand of this element, which contains six guanine-tracts (underlined)
5′-T1GGGG5AGGGT10GGGGA15GGGTG20GGGAA25GG-3′
has the capacity for forming alternate G-quadruplex folds depending on which tracts participate in scaffold formation (111,124,125). Guanine to adenine mutants within the 27-nt c-myc segment that destabilize G-quadruplex formation, result in increased c-myc transcription, while ligands like the porphyrin TMPyP4 that stabilize G-quadruplex formation, result in decreased c-myc transcription (111).
The imino proton NMR spectrum of the 27-nt c-myc NHE IIII segment containing six guanine-tracts exhibited characteristics of multiple G-quadruplex folds in equilibrium, including a broad envelope characteristic of aggregated species, precluding structural characterization. Therefore, systematic NMR studies have been restricted to four and five guanine-tract sequences as part of an effort towards understanding the underlying principles contributing to c-myc G-quadruplex formation.
Initial efforts have focused on G-quadruplexes that can be generated through involvement of four of the six guanine-tracts associated with the 27-mer c-myc NHE IIII element. Over 50 sequence variants were checked prior to the identification of two that gave imino proton spectral quality reflective of distinct single conformers that justified further structural characterization (126). One of these involved the second, third, fourth and fifth guanine-tracts (designated c-myc-2345) as reflected in the sequence
TG5AGGGT10GGGGA15GGGTG20GGGAA25
while the other involved the first, second, fourth and fifth guanine-tracts (designated variant c-myc-1245), with the guanines of the third tract replaced by thymines (in bold, below), as reflected in the sequence
T1GGGG5AGGGT10TTTTA15GGGTG20GGGA
The resulting NMR-based intramolecular G-quadruplex folding topologies in K+ solution for both c-myc-2345 and thymine for guanine-containing variant c-myc-1245 sequences contain a core of three stacked G-tetrads formed by four parallel G-tracts with all anti guanines and three double–chain-reversal loops bridging G-tetrad layers (126). The c-myc-2345 fold is shown in Figure 7a, while that for variant c-myc-1245 is shown in Figure 7b. These studies establish that single-residue (A or T) double-chain-reversal loops can bridge three G-tetrad layers. Indeed, systematic studies of DNA quadruplexes with different arrangements of short and long loops confirm that single-residue loops favor parallel-stranded topologies (127). Of the two G-quadruplex folds, c-myc-2345, which has a two-residue central loop (Figure 7a), is more stable by 15° than variant c-myc-1245, which has a six-residue central loop (Figure 7b), in K+ solution. This is also reflected in the imino proton exchange lifetimes of the central G-tetrads, which are longer for the c-myc-2345 compared to variant c-myc-1245, suggesting slower unfolding kinetics for the former G-quadruplex (126).
Figure 7.
NMR-based folding topology of unimolecular G-quadruplexes formed by (a) myc-2345 and (b) thymine for guanine-containing variant myc-1245 sequences in K+ solution (126). (c) NMR-based solution structure of the thymine for guanine-containing variant myc-2345 promoter unimolecular G-quadruplex in K+ solution (coordinates deposition: 1XAV) (128).
An NMR-based solution structure has been reported for a variant c-myc-2345 sequence in which guanines G14 and G23 have been replaced by thymines (in bold, below)
TG5AGGGT10GGGTA15GGGTG20GGTAA25
The solution structure of this variant c-myc-2345 (Figure 7c) (128) adopts the topology (Figure 7a) shown previously for unmodified c-myc-2345 (126).
The NMR-based G-quadruplex topologies for myc-2345 (Figure 7a) and variant myc-1245 (Figure 7b) (126), as well as the related study of the solution structure of the variant c-myc-2345 (Figure 7c) (128) G-quadruplexes correct earlier conclusions regarding proposed c-myc folding topologies based solely on interpretation of footprinting data (111), in an otherwise highly cited contribution.
More recent studies on c-myc sequence
The variant c-myc-1245 (126) and c-myc-2345 (128) sequences replace guanines by thymines within G-rich tracts. Thymine, unlike inosine, has nothing in common with guanine, and thymine for guanine substitutions represent a significant perturbation of the wild-type c-myc sequence. Therefore, structural studies were next extended to the c-myc sequence containing five of the six guanine-tracts associated with the 27-mer c-myc NHE IIII element, while avoiding any thymine for guanine substitutions. This sequence (designated c-myc-23456)
5′-TG5AGGGT10GGGGA15GGGTG20GGGAA25GG-3′
is composed of the second, third, fourth, fifth and sixth guanine-tracts. The NMR-based folding topology (Figure 8a) and solution structure (Figure 8b) of the c-myc-23456 G-quadruplex in K+ solution is composed of three stacked guanine tetrads formed by four parallel guanine-tracts with all anti guanines and a snap-back 3′-end syn guanine (129). The guanines involved in G-tetrad formation are highlighted in bold below
5′-TG5AGGGT10GGGGA15GGGTG20GGGAA25GG-3′
and involve guanines from each of the five tracts. This snap-back configuration is facilitated by a stable diagonal loop, which contains a G–(A-G) triad, which stacks on and caps the G-tetrad core at one end of the G-quadruplex. The 5′- and 3′-ends of the sequences are at opposite ends of the snap-back c-myc-23456 G-quadruplex (Figure 8a) (129), as they are for the c-myc-2345 (Figure 7a) and variant c-myc-1245 (Figure 7b) G-quadruplexes (126).
Figure 8.
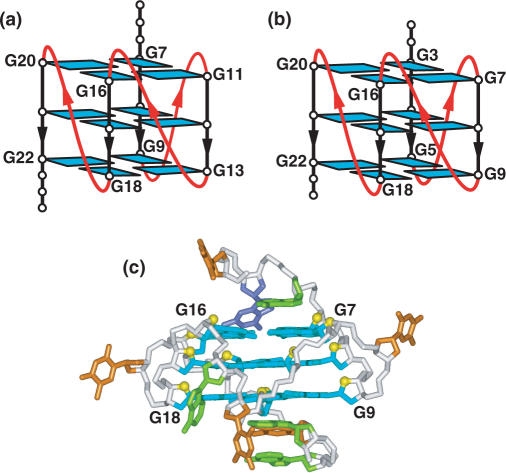
NMR-based (a) folding topology and (b) solution structure of the unimolecular G-quadruplex formed by the myc-23456 sequence in K+ solution (coordinates deposition: 2A5P) (129). NMR-based (c) folding topology and (d) solution structure of the unimolecular G-quadruplex formed by the c-kit1 sequence in K+ solution (coordinates deposition: 2O3M) (135).
c-kit sequences
The proto-oncogenic c-kit promoter encodes for a tyrosine kinase receptor, thereby regulating signal transduction cascades that control cell growth and proliferation (130). Oncogenic cellular transformations in c-kit are associated with mutations in structurally important regions, with human gastrointestinal stromal tumors (GIST) associated with mutations around the two main autophosphorylation sites in the juxtamembrane region (131), while myeloid leukemias and human germ cell tumors are associated with kinase domain mutants (132). The drug Gleevec (imatinib) is an effective in vitro and in vivo inhibitor of c-kit kinase activity and is widely used clinically against GIST (133). Like other small molecule drugs targeted against kinases, new patterns of resistance mutations within the active site, result in diminished binding and clinical effectiveness of the drug (134).
Selective gene regulation at the transcription level provides an alternate approach to c-kit inhibition. This can be achieved by induction of G-quadruplex structures within G-rich tracts of the c-kit promoter and their potential stabilization by bound ligands. Recently, imino proton NMR spectral studies established that the c-kit1 22-mer sequence
5′-A1GGGA5GGGCG10CTGGG15AGGAG20GG-3′
positioned between −87 and −109 nt upstream of the transcription start site of the human c-kit gene, forms a single G-quadruplex scaffold in K+ solution (115). Expectations that this sequence, which contains four GGG tracts (underlined, above), forms a conventional G-quadruplex, appeared unlikely when it was found that mutations within the linker segments were detrimental to G-quadruplex formation (115). It should be mentioned that a second highly conserved guanine-rich sequence has been recently identified in the c-kit gene, at a site critical for core promoter activity (116).
The NMR-based solution structure has been determined for the 22-mer c-kit1 sequence in K+ cation solution (135). The c-kit1 sequence, which exhibits an exceptionally well-resolved NMR spectrum (115), adopts a G-quadruplex topology (Figure 8c) and solution structure (Figure 8d) composed of three stacked G-tetrads and four connecting loops. The guanines involved in G-tetrad formation (in bold, below) include isolated guanine G10, but excludes G20 of the last G-tract.
5′-A1GGGA5GGGCG10CTGGG15AGGAG20GG-3′
Two single-residue linkers (A5 and C9) form two double-chain-reversal loops that bridge three G-tetrad layers, the two-residue linker connects two adjacent corners (G10 and G13), while the five-residue linker allows the terminal G21–G22 step to be inserted back into the G-quadruplex core. The loops are stabilized through formation of a Watson–Crick A–T pair that stacks over the top of the G-quadruplex and two non-canonical G–A pairs that stack over the bottom of the G-quadruplex.
This structure establishes a new folding principle that an isolated guanine (G10 in the present case) within a non-G-tract segment can participate in the formation of the structured G-quadruplex core (135). This result raises an element of caution regarding the use of programs that predict G-quadruplex folding topologies from sequence data, where they rely solely on the participation of guanines within G-tracts. Another notable feature is associated with formation of a snap-back parallel-stranded G-quadruplex core, where the last two guanines insert back into the core to complete adjacent G-tetrad alignments (Figure 8c). The 5′- and 3′-ends of the sequences are at opposite ends of the snap-back c-kit1 G-quadruplex, thereby allowing continuation of the DNA sequence in both directions without significant steric hindrance.
Both the c-myc 23456 (Figure 8a) (129) and c-kit1 (Figure 8c) (135) scaffolds contain distinct pronounced clefts, with their unique surface topologies making them attractive site-selective targets for drugs.
bcl-2 sequence
The bcl-2 gene mediates the t(14;18) chromosomal translocation associated with the onset of lymphomas (136,137). The bcl-2 gene is overexpressed in several human cancers, with the gene product functioning as an apoptosis inhibitor, thereby impacting adversely on the therapeutic action of cancer treatment regimes in the clinic (138). Thus, both the bcl-2 gene and its gene product constitute rational targets for anti-cancer therapy.
Transcriptional initiation of bcl-2 is controlled by a major promoter P1, containing a guanine-rich strand upstream of the initiation site and proximal to a nuclease hypersensitivity region (114). This bcl-2 promoter region contains six guanine-tracts containing three or more contiguous guanines (underlined)
5′-GGGGCG1GGCG5CGGGA10GGAAG15GGGGC20GGGAG25CGGGG-3′
with non-denaturing gel, footprinting and cd data interpreted in terms of a mixture of at least three G-quadruplex conformers in K+ solution. The second to fifth G-tracts (designated bcl-2 2345) forms the most stable G-quadruplex (114), and an attempt has been made to structurally investigate this sequence composed of the four central guanine-tracts. The NMR studies were undertaken on a variant in which guanines G15 and G16 were replaced by thymines (in bold, below) (108).
5′-G1GGCG5CGGGA10GGAAT15TGGGC20GGG-3′
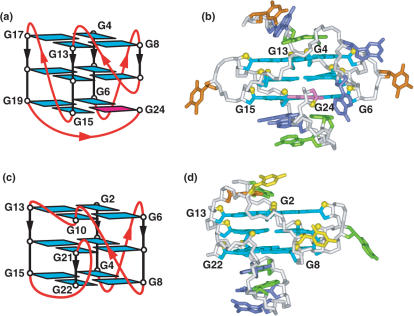
The thymine for guanine-containing variant of the bcl-2 2345 adopts a (3 + 1) G-quadruplex topology (108) (Figure 9a) and solution structure (139) (Figure 9b).
Figure 9.
NMR-based (a) (3 + 1) G-quadruplex folding topology and (b) solution structure of the thymine for guanine-containing variant bcl-2 2345 promoter unimolecular G-quadruplex in K+ solution (coordinates deposition: 2F8U) (139). NMR-based (c) (3 + 1) G-quadruplex topology and (d) solution structure adopted by the four-repeat Tetrahymena telomere unimolecular G-quadruplex formed by the d[(T2G4)4] sequence in Na+ solution (coordinates deposition: 186D) (7).
The same (3 + 1) G-quadruplex scaffold was first reported over a decade ago for the four-repeat Tetrahymena telomere sequence, d(T2G4)4,
5′-T1TGGG5GTTGG10GGTTG15GGGTT20GGGG-3′
in Na+ solution, with its unanticipated double-chain-reversal loop-containing folding topology (Figure 9c) and solution structure (Figure 9d) (7), considered to be an anomaly at that time.
Replacement of single guanines by inosines, where the exocyclic amino groups are replaced by protons, have been used previously in NMR-based studies of G-quadruplex formation in efforts to improve spectral quality (96). By contrast, replacement of two guanines by thymines in variant bcl-2 2345 constitutes a much more serious perturbation, especially for an internal guanine-tract, preventing these two guanines from potential participation in G-tetrad formation. Thus, opportunities exist for structurally investigating unperturbed bcl-2 oncogenic promoter sequences, perhaps involving five of the six guanine-tracts, as was accomplished previously for c-myc-23456 (129).
VEGF and HIF-1α sequences
Vascular endothelial growth factor (VEGF) stimulates the formation of new blood vessels, providing oxygen and nutrients to primary tumor sites, thereby facilitating the proliferation of cancer cells. VEGF-mediated tumor angiogenesis, has stimulated interest in the VEGF gene and its potential as a target for cancer therapy (140). Elevation of VEGF expression in cancer is primarily regulated at the transcription level, with the VEGF promoter containing a purine-rich strand composed of five guanine-tracts of at least three guanines each (underlined)
5′-G1GGGC5GGGCC10GGGGG15CGGGG20TCCCG25GCGGG30G-3′
that also serves as binding sites for Sp1 and Egr-1 transcription factors. The guanine-rich VEGF sequence forms G-quadruplex structures in monovalent cation solution (as monitored by cd and footprinting measurements), which are stabilized by G-quadruplex-interacting agents TMPyP4 and telomestatin (112). In addition, a DNase1 and S1 nuclease hypersensitivity site was identified to the 3′-side of the G-quadruplex forming region, but not for mutant sequences that inhibit quadruplex formation. Finally, the cd spectrum of the guanine-rich VEGF sequence in K+ is consistent with formation of a parallel-stranded G-quadruplex. Overall, the results are suggestive of the importance of structural transitions in enhancing open promoter complex formation, thereby facilitating transcriptional regulation (112).
Hypoxia inducible factor-1α (HIF-1α) is activated in many common human tumors and is associated with local invasion and metastasis (141). The HIF-1α promoter contains five guanine-rich tracts of at least three guanines each (underlined)
5′-G1GGCG5CGCGG10GGAGG15GGAGA20GGGGG25CGGG-3′
capable of all-parallel-stranded G-quadruplex formation in K+ solution, as indicated by chemical probing, cd and DNA polymerase arrest assays (113). Considerable effort has gone towards targeting HIF-1α in cancer therapy (142).
To date, no systematic structural investigations have been undertaken to determine the G-quadruplex structures adopted by the guanine-rich tracts of either the VEGF or HIF-1α promoters.
TRIPLET REPEAT DISEASE G-QUADRUPLEXES
A series of nucleotide or repeat expansion disorders caused by the dynamic intergenerational expansion of triple repeat d(CGG)n–d(CCG)n, d(CAG)n–d(CTG)n and d(GAA)n–d(TTC)n sequences are associated with neurological, neuromuscular and neurodegenerative disorders (143,144). These diseases exhibit genetic anticipation, whereby the symptoms and penetrance are manifested in subsequent generations at a decreased age of onset and increased severity. The expandable repeats are found in diverse settings ranging from coding segments, to 5′- and 3′-UTRs, promoter regions and introns. It is likely that the pathogenesis of these debilitating diseases, and their disruption of cellular replication, repair and recombination machineries, reflects unusual DNA conformations generated for long repeats, for which several secondary structural models have been proposed in the literature (145–149). These guanine-containing repeats within complementary repetitive strands of the duplex can form slip-out hairpin-like folds (150), which in turn could form higher order architectures, including quadruplex formation following bimolecular association. One of these repeat expansion models proposes that the higher order structures stall the replication fork, giving time for addition of extra repeats, prior to replication fork restart (151).
Though the early emphasis on triplet expansion diseases was focused on the DNA template, more recent analysis has brought RNA repeats to the forefront, with the emphasis on gain-of-function contributions at the RNA level (152). Thus, structural studies need to be undertaken on both triplet repeat-containing DNAs and RNAs.
CGG triplet repeats
There has been considerable interest in the molecular basis for expansion of d(CGG)n–d(CCG)n tracts in genomic DNA that results in the onset of the FXMR syndrome (153,154), the single most common inherited cause of mental retardation (155). The d(CGG)n triplet repeat (can be designated CGG, GGC or GCG repeat depending on the phase of the readout) is observed within the first exon of the FMR-1 gene with n < 30 nt in normal individuals. This number increases up to ∼200 nt in premutation carriers and further expands up to 2000 nt in individuals afflicted with fragile X syndrome. The genetic instability associated with the expansion of d(CGG)n repeats to the diseased state is facilitated by hypermethylation of cytosine residues (156) and results in suppression of FMR-1 gene transcription (154) and delay in replication in patients with the FMR-1 syndrome (157). It was initially shown that the fragile X syndrome d(CGG)n repeat forms a stable G-quadruplex in the presence of monovalent cations when n = 7, and also when n = 5, for its methylated cytosine counterpart (158). In addition, d(CGG)n repeats form structures that block DNA synthesis in vitro (159), with the block overcome by the Werner syndrome (WRN) helicase (160). Interestingly, the cationic porphyrin TMPyP4 (161) and the hnRNP-related protein CBF-A (162,163), both destabilize quadruplex formation, in contrast to their structural stabilization of the human telomere G-quadruplex.
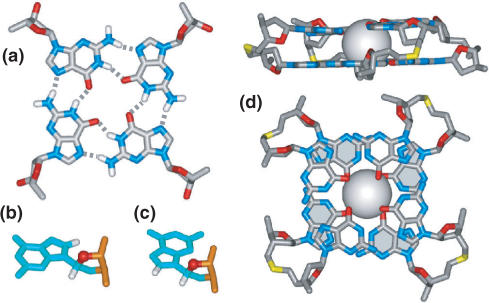
Very high-quality NMR spectra were observed for d(GCGGT3GCGG), a sequence that embeds CGG and GCG steps, in Na+ solution, thereby defining a distinct folding topology (Figure 10a) and solution structure (Figure 10b) (164).
5′-G1CGGT5TTGCG10G-3′
The sequence forms a bimolecular quadruplex containing G–C–G–C tetrads (Figure 10c) flanked by G–G–G–G tetrads in solution. The loops adopt edge-wise conformations and are aligned at opposite ends of the bimolecular quadruplex, while the strands directionalities alternate around the G-quadruplex and the G-tetrads adopt anti–syn–anti–syn alignments (Figure 10a). These studies establish the pairing alignments that can be potentially utilized by sequences containing the fragile X syndrome d(CGG)n triplet repeat to form quadruplex structures. Such quadruplex structures, stabilized by a mixture of G–C–G–C and G–G–G–G tetrads [see also, (165), for an alternate, but not structurally characterized quadruplex model], could serve as potential blockage sites for the progress of replication forks and account for the blockage at the fragile X locus observed experimentally (157).
Figure 10.
NMR-based (a) folding topology and (b) solution structure of the bimolecular G-quadruplex formed by the d(GCGGT3GCGG) sequence in Na+ solution (coordinates deposition: 1A6M) (164). Schematics illustrating mixed tetrad pairing alignments observed for (c) direct and (d) slipped G–C–G–C tetrads, associated with pairing along the major groove edges. A monovalent cation (gray ball) most likely bridges the acceptor atoms along the major groove edges of opposing guanines in the slipped G–C–G–C tetrad.
GAA triplet repeats
The d(GAA)n-repeat is of considerable biological interest since expansion of d(GAA)n–d(TTC)n triplet repeats located within the first intron of the frataxin gene contributes to Friedrich's ataxia, an autosomal recessive neurodegenerative disease (166). The non-G–C rich nature of the sequence, together with the intronic localization and the requirement of both alleles, makes Friedrich's ataxia unique amongst the triplet-repeat disease sequences. Expression of the d(GAA)n triplet repeat leads to reduced levels of frataxin mRNA transcripts, and it has been shown to reflect impediment in transcription elongation, in a length and supercoil dependent manner (167). This impediment could reflect formation of a stable nucleic acid architecture (168), and several models have been proposed ranging from triplexes (169) to parallel-stranded duplexes (170). The parallel-stranded duplex model for d(GAA)n triplet repeats is intriguing, since further bimolecular pairing could result in quadruplex formation.
NOVEL QUADRUPLEX FOLDS AND TETRAD PAIRING ALIGNMENTS
G-quadruplexes can contain pairing alignments beyond the G-tetrad and considerable effort has gone into defining these alignments. These include other homo- and mixed-tetrad pairing alignments, triads, pentads, hexads and heptads. Triads and triples are generally observed within edge-wise and diagonal loop regions, where they stack on terminal G-tetrads. By contrast, mixed tetrads, pentads and hexads are observed at both the ends and within G-quadruplexes.
Double-chain-reversal loops
Early structural studies identified edge-wise (9,15), and diagonal (8,10) loops that bridged anti-parallel-aligned columns around the G-quadruplex. An unanticipated development was the identification of double-chain-reversal loops that bridged adjacent parallel-aligned columns within the four-repeat Tetrahymena G-quadruplex (7). In this case, two thymine residues span three stacked G-tetrad planes. Next it was demonstrated that single residue double-chain-reversal loops can span two G-tetrad planes in an all parallel-stranded G-quadruplex (16). The importance of double-chain-reversal loops emerged center-stage following the structure determination of the four-repeat human telomere from crystals grown in K+ solution (17), where all three TTA loops were of the double-chain-reversal (or hairpin) type and each spanned three stacked G-tetrad planes (Figure 5c). The next discovery was that single-residue double-chain-reversal loops could span both two (16) and three (126) stacked G-tetrads. The latter result was most unexpected but was confirmed in subsequent studies on additional G-quadruplex folds (128,129,171,172).
Mixed tetrads
The standard view of G-quadruplex formation involves a scaffold stabilized by stacked G–G–G–G tetrads. Nevertheless, mixed tetrads can also stabilize G-quadruplex formation and these include major groove-aligned G–C–G–C tetrads of the direct (Figure 10c) (14,164,173,174) and slipped (Figure 10d) (26) type and major groove-aligned A–T–A–T tetrads of the direct (17) and slipped (173) type. Minor groove-aligned mixed G–G–G–G and A–T–A–T tetrads have also been structurally characterized, but the bases deviate significantly from the tetrad plane (175,176).
A–A–A–A tetrads
NMR studies on d(AGGGT) in K+ solution are consistent with formation of a parallel-stranded G-quadruplex (177). Somewhat unexpectedly, nuclear Overhauser enhancement (NOE) cross peaks were observed between the adenine amino protons and the non-exchangeable H8 and H2 protons. This has lead to the proposal of A–A–A–A tetrad formation, with rapid interconversion between N6H•••N7 and N6H•••N3 hydrogen-bonding alignments. Furthermore, the terminal adenine residues appear to adopt syn glycosidic torsion angles based on the strong H8 to H1′ NOEs observed at short mixing times, suggestive of an A(syn)–A(syn)–A(syn)–A(syn) alignment (177). A more definitive approach would have been to use 15N isotopic labeling to directly monitor scalar coupling to define hydrogen-bonding alignments (178,179); (174), thereby validating the proposed A–A–A–A tetrad formation. The NMR-based conclusions contrast with crystallographic studies of RNA sequences (discussed in more detail in topologies and tetrad alignments section), where A–A–A–A tetrads have been definitively identified, but shown to adopt A(anti)–A(anti)–A(anti)–A(anti) alignments (180,181).
Triads
The concept of an anti-parallel DNA duplex stabilized by base triads was proposed more than a decade ago (19). A base triad involves alignment of three bases in a plane, where a base from one strand interacts through hydrogen bonding with two adjacent co-planar bases from the partner strand. The coplanar-aligned adjacent bases essentially form a platform, a feature identified initially in RNA (182). A triad differs from a triple, where the three bases come from three distinct strands. There are now several examples of base triads stacked over the terminal G-tetrads of G-quadruplexes. These include A–(T-A) (Figure 11a) (101,183), G–(C-A) (Figure 11b) (20), T–(A-A) (184), T–(A-T) (101), G–(A-G) (129), and G–(T-T) (185) triads, where in each case, the co-planar adjacent bases that constitute the platform, are indicated in brackets.
Figure 11.
Schematics illustrating base triad pairing alignments for (a) A–(T-A) (183), (b) G–(C-A) (20) triads. Schematics illustrating (c) A–(G–G–G–G) pentad (18,171) and (d) A–(G–G–G–G)–A hexad (16) alignments. NMR-based (e) folding topology and (f) solution structure of end-to-end stacked tetramolecular G-quadruplexes formed by four d(GGAGGAG) strands in 150 mM Na+ solution (coordinates deposition: 1EEG) (16).
Pentads and hexads
NMR-based investigations of G-quadruplexes have also identified formation of A–(G–G–G–G) pentads (Figure 11c) (18,171), A–(G–G–G–G)–A hexads (Figure 11d) (16) and heptads (186). Such alignments essentially are composed of G–(A-G) triads, where one or more A residue(s) align(s) along one or more minor groove edge(s) of a G-tetrad.
Effect of guanine substitutions on G-quadruplex formation
A systematic and penetrating study has reported on the impact of guanine modifications on formation of parallel four-stranded G-quadruplexes (187). These authors measured G-quadruplex association and dissociation kinetics to estimate the energetic penalty associated with single-site modifications of 12 different substitutions. Modifications involving the hydrogen-bonding positions on the guanine ring (O6, N1, N2 and N7) were detrimental to G-quadruplex stability as reflected in decreased association rate constants and reduced quadruplex lifetimes. The most deleterious effects were observed for central guanine substitutions, suggestive of an important role for this position in the nucleation process. By contrast, modifications that perturb neither the central carbonyl group alignment nor the cyclic hydrogen-bonding pattern are tolerated, as are other planar bicyclic ring systems that retain such constraints. Thus, substitution of guanine by either 8-bromoguanine or 6-methyl-isoxanthopterin accelerates quadruplex formation, especially when substituted at the 5′-end of the G-tract. It is conceivable that the bromo and methyl groups in these substitutions favor hydrophobic collapse during the process of strand association. These modifications also favor a syn glycosidic torsion angle, which correlates with the observation of syn glycosidic torsion angles at the 5′-guanine positions in (3 + 1) G-quadruplex scaffolds (7,101). Finally, non-guanine tetrads are destabilizing when positioned internally within a G-quadruplex, but can be accommodated when positioned over terminal G-tetrads due to stabilizing stacking interactions (187). A systematic study has also been undertaken on the effect of G-tract length on the topology and stability of intramolecular G-quadruplexes (188).
STACKED AND INTERLOCKED G-QUADRUPLEXES
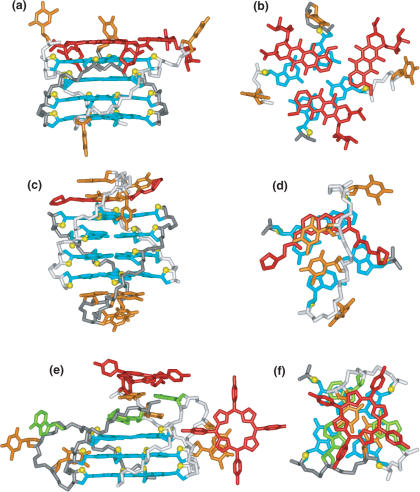
Two G-quadruplexes can interact through end-to-end stacking (16) or alternately through an interlocked configuration (18,171), where a guanine from one monomer completes the G-tetrad through interaction with three guanines from the other monomer. Such quadruplex–quadruplex interactions, especially those of the interlocked type, result in very stable topologies, and can involve the participation of junctional G–C–G–C tetrads (Figure 10c) (174), A–(G–G–G–G) pentads (Figure 11c) (18) and A–(G–G–G–G)–A hexads (Figure 11d) (16).
End-to-end stacked G-quadruplexes
Previous NMR-based studies had demonstrated that the d(GGAGGAT) sequence formed a two-stranded arrowhead motif-aligned solely through non-canonical pair formation under low (10 mM) Na+ counterion conditions (189). Further NMR-based studies of this sequence and related d(GGAGGAG) indicated a pronounced conformational change on proceeding to moderate (150 mM) Na+ counterion conditions (16). Structural characterization of the moderate salt conformer demonstrated formation of end-to-end stacked G-quadruplexes involving four strands with a unique folding topology (Figure 11e) and solution structure (Figure 11f) for the d(GGAGGAG) sequence in 150 mM Na+ solution (16). Each G-quadruplex monomer, formed by alignment of two d(GGAGGAG) strands, is composed of a junctional A–(G–G–G–G)–A hexad (Figure 11d), a G–G–G–G tetrad and an A–A non-canonical pair. The A3 residue, involved in double-chain-reversal loop formation, also participates in A–(G–G–G–G)–A hexad formation. The end-to-end stacking of G-quadruplex monomers is mediated through stacking of their junctional A–(G–G–G–G)–A hexads (Figure 11e). A combination of Brownian dynamics and molecular dynamics simulations identified several stable monovalent cation-binding sites within the end-to-end stacked G-quadruplexes scaffold (16).
V-shaped scaffold mediates interlocked G-quadruplex formation
The guanine-rich d(G3AG2T3G3AT) sequence
5′-G1GGAG5GTTTG10GGAT-3′
contains one GG and two GGG segments and therefore was not expected to form a monomeric intramolecularly folded G-quadruplex. Despite this limitation of lacking four guanine-tracts, the sequence gave exceptional NMR spectra associated with a single conformation in Na+ solution (18). The stoichiometry of two, coupled the number of resonances, established that d(G3AG2T3G3AT) folds by interaction between symmetry-related G-quadruplexes. A uniformly 13C,15N-labeled sample was prepared to facilitate resonance assignments and identify hydrogen-bonding alignments (178,179); (174). NMR-based NOE and hydrogen-bonding constraints defined the folding topology (Figure 12a) and solution structure (Figure 12b) associated with a pair of interacting G-quadruplexes (18). Each symmetry-related G-quadruplex monomer undergoes three sharp turns, with all purines involved in pairing alignments. The first turn is of the double-chain-reversal type, the second turn is of the edge-wise type and the last involves a new alignment, the V-shaped turn (Figure 2d). Each monomer contains two stacked G(anti)–G(anti)–G(anti)–G(syn) tetrads, one of which forms a A–(G–G–G–G) pentad. There is a break in one of the four G-G columns that link adjacent G-tetrads within each monomer, resulting in a V-shaped scaffold. The A–(G–G–G–G) pentad from each monomer mutually stack on each other, with each pentad containing four bases from one monomer and a syn G1 from the partner monomer, thereby resulting in interlocked G-quadruplex formation (18).
Figure 12.
NMR-based (a) folding topology and (b) solution structure of the V-shaped interlocked bimolecular G-quadruplex formed by the d(G3AG2T3G3AT) sequence in Na+ solution (coordinates deposition: 1JJP) (18). NMR-based (c) folding topology and (d) solution structure of interlocked bimolecular G-quadruplex formed by the d(G4TG3AG2AG3T) 93del sequence in K+ solution (coordinates deposition: 1Y8D) (171).
Targeting HIV-1 integrase using interlocked G-quadruplexes
HIV-1 integrase catalyzes the integration of proviral DNA into the host-cell genome, a reaction critical for efficient viral replication. NMR-based studies have solved the solution structure of an in vitro selected guanine-repeat-containing 93del DNA sequence
5′-G1GGGT5GGGAG10GAGGG15T-3′
a potent nanomolar inhibitor of both processing and strand transfer functions of HIV-1 integrase (190). This sequence forms an unusually stable interlocked G-quadruplex architecture in K+ solution with a fold shown schematically in Figure 12c and solution structure shown in Figure 12d (171). Within each monomer subunit, one A–(G–G–G–G) pentad is sandwiched between two G–G–G–G tetrads, all G-stretches are parallel, are linked by three double-chain-reversal loops, and all guanines are anti, except for G1, which is syn. Interlocked G-quadruplexes formation is achieved through mutual pairing of G1 of one monomer, with three other guanines of the other monomer, to complete junctional G-tetrad formation.
The interlocked G-quadruplexes scaffold with its distinct surface architecture could be shape-specifically targeted by ligands, or in turn serve as a ligand that targets interfacial channels on multimeric proteins. Indeed, molecular docking approaches suggest that the 93del interlocked DNA G-quadruplex could potentially be positioned within a basic canyon formed between subunits of the tetrameric HIV-1 integrase (171).
Snap-back G-quadruplexes
The snap-back parallel-stranded G-tetrad core scaffolds have been observed for both the c-myc-23456 (Figure 8a) (129) and the c-kit1 (Figure 8c) (135) G-quadruplexes. In both cases, there is an interruption of the G-tetrad core, with base-pairing alignments in the last connecting loop important in stabilizing the snap-back scaffold. The c-myc-23456 (Figure 8a) and c-kit1 (Figure 8c) G-quadruplexes differ in that the former involves insertion of a single syn guanine (129), while the latter involves insertion of two anti guanines (135). The snap-back feature allows for continuation of the DNA sequence in both directions without significant steric hindrance.
RNA QUADRUPLEXES
The majority of attention has focused on DNA quadruplexes, their diversity of scaffolds and their potential role in biology. By contrast, RNA quadruplexes have received less attention, despite implications of their involvement at sites of RNA packaging (191), endonucleolytic cleavage activity (192), translational control (193) and mRNA turnover (73).
Topologies and tetrad alignments
RNA quadruplex formation was initially established from NMR studies on r(UGGGGU) (194) and subsequently by a 0.61 Å crystal structure of the same sequence for crystals grown in Sr2+-containing solution (195). This sequence forms a parallel four-stranded RNA quadruplex with all anti guanines in both solution and the crystalline state, with Sr2+ sandwiched between every other G-tetrad plane in the crystal. These studies also identified formation of U–U–U–U tetrads, as well as G- and U-containing octads in the crystal, where uracils pair with the minor groove edges of guanines of the G-tetrad. Additional aspects of quadruplex structure have emerged from the 1.4 Å crystal structure of d(brU)-r(GAGGU) (181) and 1.5 Å crystal structure of r(U)-d(brG)-r(AGGU) (180). These studies unequivocally demonstrate formation of Na+ cation-coordinated all anti A–A–A–A tetrads involving either N6H•••N7 (former quadruplex) or N6H•••N3 (latter quadruplex) hydrogen-bonding alignments. More recently, a crystal structure of r(UGGUGU) established an even higher order architecture associated with a dimer of quadruplexes scaffold (6).
Oncogenic 5′-UTR regions
Bioinformatic searches for guanine-rich sequences in 5′-UTRs of the human genome has recently identified up to 3000 putative G-quadruplex-forming elements (196). One of these sequences, an 18-mer containing four guanine-tracts
5′-G1GGAG5GGGCG10GGUCU15GGG-3′
is associated with the 5′-UTR of the oncogenic N-ras sequence, located 14-nucleotides downstream of the 5′-cap and 222-nucleotides upstream of the translation start site. This sequence, which contains four guanine-tracts is highly conserved across species, both within the guanine-rich segments and its position relative to the translation start site, and forms a G-quadruplex (as monitored by cd) as a function of monovalent cation. The measured tm was 63°C in 1 mM K cation, and the stabilization decreased in the order K+ > Na+ > Li+ (196). The RNA G-quadruplex was very stable since unfolding was not observed even at 95 C in K+ solution. The CD spectrum exhibited characteristics of a parallel-stranded G-quadruplex with a positive peak at 263 nm and a negative peak at 241 nm. The authors used a cell-free translation system coupled to a reporter gene assay to demonstrate that the N-ras G-quadruplex inhibits gene expression at the translational level (196). This seminal result opens opportunities for the identification of small molecule therapeutic agents with the potential for stabilizing 5′-UTR RNA G-quadruplex formation, thereby inhibiting translation of oncogenes.
FUTURE OPPORTUNITIES: QUADRUPLEX STRUCTURE
There remain many structural challenges associated with G-quadruplex architecture, as well as the impact of structure on function. Several of these are outlined below.
Quadruplexes containing G–A–G–A tetrads
To date there is no evidence for formation of all-purine major groove-aligned G–A–G–A tetrads within a quadruplex scaffold. The design and identification of G–A–G–A tetrads would expand the repertoire of sequences that can form quadruplex scaffolds. There are several possibilities for non-canonical G–A pairing alignments stabilized by two hydrogen bonds. These include G(anti)–A(anti) pairing along their Watson–Crick edges (197–199), sheared G(anti)–A(anti) pairing using the minor groove edge of G and major groove edge of A (200,201), G(anti)–A(syn) pairing using the major groove edge of A (202), and G+(syn)–A(anti) pairing using the major groove edge of protonated G (203,204). Such G–A non-canonical pairs could potentially align along their major groove edges to form G–A–G–A tetrads. It remains to be demonstrated whether G–A–G–A tetrad alignments can be accommodated in an otherwise G–G–G–G tetrad-containing G-quadruplex. The G–A–G–A tetrad has only two inwardly pointing carbonyls in contrast to four inwardly pointing carbonyls in the G–G–G–G tetrad available for monovalent cation coordination. Thus, the role of monovalent cations in stabilization of G–A–G–A tetrads is less clear at this time. It should be noted that ethanol can substitute for monovalent cations in facilitating quadruplex formation (205).
It is conceivable that the Friedrich's ataxia d(GAA)n triplet-repeat sequence (166) could adopt a quadruplex scaffold stabilized by G–A–G–A and A–A–A–A tetrads. It remains to be established whether such a quadruplex forms in solution, and if so, what the strand directionalities and which tetrad alignments define the topology.
Quadruplex–quadruplex junctions
Published structural efforts to date have focused on human telomeres containing one (4), two (17,94), three (96) and four repeats (9,17,99–106). However, there are interesting questions concerning (TTAGGG)n repeats, where n is >4, especially regarding the issue of whether adjacent human telomeric G-quadruplexes stack on each other (17), thereby adopting a beads-on-a-string architecture (206). In addition, in a particularly innovatively designed experiment, it has been shown that chiral cyclic-helicene molecules of a particular size, which are capable of wedge formation, can target quadruplex–quadruplex junctions and stabilize higher order human telomere structures (207). Both the architecture of quadruplex–quadruplex junctions and their complexes with ligands such as chiral cyclic-helicene constitute a significant challenge for the future.
Quadruplex–duplex junctions
In contrast to significant progress on the structures of the G-quadruplex folds of the c-myc (126,128,129) and c-kit (135) sequences in K+ solution, and the human telomere sequence in Na+ (9) and K+ (96,99–106) solution, little is known about the architecture and stability of quadruplex–duplex junctions, as would occur in a natural context, where telomeric G-quadruplexes could either cap the ends of the telomere or where telomeric and oncogenic promoter G-quadruplexes extrude out of a duplex segment. Studies of G-quadruplex–duplex junctions and their stability represent a significant structural challenge that will require a systematic investigation involving variations in the length and sequence of junctional residues.
Telomere t-loops
Electron microscopy studies demonstrate that the telomeric G-overhang segment of chromosomal termini may adopt a non-linear configuration through formation of a lariat-shaped t-loop structure, where the overhang segment invades an adjacent duplex region through formation of a displacement D-loop (Figure 4c) (97). It should be noted that such a t-loop architecture has the potential for sequestering and protecting the 3′-terminii of telomere overhangs. Structural studies have the opportunity to discriminate between proposed alternate models of t-loop formation.
Oncogenic VEGF and HIF-1α DNA promoters
Both the VEGF and HIF-α promoter sequences (see earlier VEGF and HIF-1α sequences section) contain five guanine-tracts and hence may adopt more than one conformation in solution. Perhaps, through judicious choice of single inosine for guanine substitutions, high-quality NMR spectra corresponding to a single G-quadruplex conformation may be achievable for each of these oncogenic promoter sequences, as was successfully achieved previously for the five guanine-repeat c-myc-23456 G-quadruplex (129).
It should be noted that there are two examples of single bases separating G-tracts in both the VEGF and HIF-1α promoters, and these are potential sites for double-chain-reversal loops (7,16,126). Indeed, there are some similarities in the sequence between VEGF-1234 and HIF-1α–2345 in that they have an approximate consensus element
GGGGNGGG(G)NN(N)GGGGGNGGG(G)
with footprinting studies indicating that this represents a common structural motif (112,113).
Oncogenic 5′-UTR RNAs
To date, no structures have been reported for oncogenic 5′-UTR RNA sequences. Potential candidates containing four guanine-tracts that impact on oncogenic events (196), include the 5′-UTR of N-ras (see earlier Oncogenic 5′-UTR regions section for sequence), as well as the 5′-UTR of Friend leukemia integration protein 1 (fli1), which exhibits the sequence
5′-GGGAGGGCCCAGGGCGCCAGGG-3′
the 5′-UTR of apoptosis regulator (bcl-2), which exhibits the sequence
5′-GGGGGCCGUGGGGUGGGAGCUGGGG-3′
and the 5′-UTR of transcription factor AP1 (jun), which exhibits the sequence
5′-GGGGAGGGGACCGGGGAAGAGAGGG-3′
Each of these sequences contains single-residue linkers with the potential for formation of double-chain-reversal loops. One can anticipate that several unanticipated topologies are likely to emerge for RNA quadruplexes, as reported previously for DNA quadruplexes (35). Furthermore, unlike oncogenic promoter DNA quadruplexes whose formation requires prior melting of the duplex segment, no such constraint exists for the primarily unstructured 5′-UTR RNA sequences.
Quadruplexes in living cells
The ultimate challenge would be to develop methods for probing structures and conformational transitions involving G-quadruplexes in living cells. Given the rich diversity of G-quadruplex scaffolds and their propensity to interconvert, it will be a challenge to identify small molecules that exhibit recognition selectivity for distinct scaffolds at the cellular level. Clearly, one anticipates further developments, for instance of fluorescent dyes, along the lines of the carbazole BMVC (65), to further address this problem.
DRUGS TARGETED TO G-QUADRUPLEX SCAFFOLDS
The ribonucleoprotein enzyme telomerase, composed of an endogenous RNA template and a reverse transcriptase, can maintain telomere length by adding TTAGGG repeats to the 3′-ends of chromosomes (79,208). Telomerase is active in the majority of human tumor cells and requires single-stranded telomere ends as a primer for its activity (84,209). In this regard, telomerase levels correlate with cancer progression and the metastatic state. Telomerase activity can be negatively regulated in vivo through monovalent cation-mediated G-quadruplex formation at telomeric ends (210), and hence small molecules that bind and stabilize G-quadruplex structures constitute potent telomerase inhibitors. This concept was first validated when it was demonstrated that 2,6-diamidodianthraquinone inhibits the activity of telomerase by interacting with and stabilizing G-quadruplex structures (211). Some ligands could also act as molecular chaperones by increasing the association constant for G-quadruplex formation (212). G-quadruplex formation denies access of telomerase and telomeric DNA-binding proteins to telomere overhangs, thereby selectively interfering with telomere maintenance in tumor cells (87,88,213–218). Overall, telomerase inhibition leads to telomere-length reduction, tumor-cell senescence and ultimately apoptosis.
Diverse families of compounds have been identified that exhibit selectivity for G-quadruplexes over their duplex counterparts and inhibit telomerase action in human tumor cell lines with IC50 values in the sub-μM range. Many of these compounds contain polyaromatic heterocyclic ring systems, including anthraquinones, acridines, perylenes and porphyrins (215,216,219,220), capable of extensive π-stacking interactions with terminal G-tetrads (221,222), and in some cases containing at least two side chains directed towards the G-quadruplex grooves. Some insights into the principles of ligand-G-quadruplex recognition have emerged from the few published NMR and X-ray structures of complexes.
Polyaromatic heterocyclic rings
NMR studies of complexes formed between the single-repeat human telomere sequence d(TTAGGGT) and dicationic perylene tetracarboxylic diimide (223) and fluorinated pentacyclic quino[4,3,2-kl]acridinium cation (224) ligands, establish that these polycyclic ring systems interact with the all-parallel tetramolecular G-quadruplex through end-stacking over terminal G-tetrads. To date, despite occasional claims, there is no definitive spectroscopic and structural evidence that supports intercalation of polycyclic ring system-containing chromophores between G-tetrads of G-quadruplexes.
An X-ray structure has been reported for the antitumor drug daunomycin (Figure 13a) bound to the all-parallel-stranded tetramolecular G-quadruplex formed by d(TGGGGT) (Figure 14a) (222). Daunomycin aligns in a trimeric arrangement, with its anthracycline chromophores optimally end-stacked on the terminal G-tetrad (Figure 14b). In addition, the daunosamine sugar rings are positioned in the grooves and anchored through intermolecular hydrogen bonds.
Figure 13.
Chemical formulas of (a) daunomycin (222), (b) 9-benzylamino-substituted acridine (227), (c) bisquinolinium-substituted phenanthroline (X=NCH3 +, Y=CH; X=CH, Y= NCH3+) (229), (d) 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)porphyrin (TMPyP4), (e) Mn(III) porphyrin with flexible cationic arms (234), (f) telomestatin (235), (g) oxazole-containing 24-membered macrocycle (HXDV) (240), (h) steroid diamine funtumine substituted by a guanylhydrazone moiety (244) and (i) left-handed chiral cyclic-helicene with a short linker (207).
Figure 14.
(a) X-ray structure of the complex of three molecules of daunomycin (in red) bound to the tetramolecular d(TGGGGT) G-quadruplex (coordinates deposition: 1O0K) (222). (b) Overlap of daunomycin molecules over the terminal G-tetrad. (c) X-ray structure of the complex of disubstituted aminoalkylamido acridine (in red) bound to the bimolecular d(G4T4G4) G-quadruplex (coordinates deposition: 1L1H) (221). (d) Overlap of the substituted acridine over the terminal G-tetrad. (e) X-ray structure of the porphyrin TMPyP4 (in red) bound to the d(TAG3T2AG3) bimolecular G-quadruplex (coordinates deposition: 2HRI) (233). (f) Overlap of the porphyrin ring of TMPyP4 with a terminal base pair.
One of the best-characterized telomerase inhibitors belongs to the family of 3,6,9-trisubstituted acridine molecules. The first-generation compound, BRACO-19, exhibits cell growth arrest, chromosomal end-to-end fusions (225) and antitumor activity in tumor xenografts (226), all within a short exposure time. BRACO-19 also induces G-quadruplex formation by competing with hPOT1 for binding to single-stranded telomeric overhangs. Such uncapping of telomerase from telomere ends, induces a rapid DNA damage response and selective cell death. More recently, the key anilino substitutent in BRACO-19 has been replaced by a benzylamino substitutent (Figure 13b) resulting in enhanced quadruplex interaction and superior telomerase inhibitor activity (227).
The X-ray structure of a disubstituted aminoalkylamido acridine bound to two-repeat Oxytricha d(GGGGTTTTGGGG) sequence establishes that the acridine ring system end-stacks with the terminal G-tetrad of the bimolecular G-quadruplex (Figure 14c) (221). The acridine ring threads through the diagonal loop of the bimolecular G-quadruplex, with stacking and intermolecular hydrogen-bonding interactions stabilizing complex formation (Figure 14d).
Malignant glioblastomas are very aggressive and invasive tumors of the central nervous system that are highly refractive to surgery, radiotherapy and chemotherapy. A family of bisquinolinium-substituted 2,6-pyridine-dicarboxamide derivatives were shown to inhibit cell proliferation at low doses and induce massive apoptosis in cultures of glioma cell lines (228). The apoptosis was preceded by multiple cell cycle alterations associated with telomere end fusion and anaphase bridge formation, suggesting that these pyridine-based G-quadruplex ligands could serve as promising agents against malignant gliomas. Furthermore, recent improvements within this family of G-quadruplex-binding ligands have resulted following replacement of the central pyridine-based core by a phenanthroline core (229). These bisquinolinium-substituted phenanthroline compounds (Figure 13c), which also adopt a planar crescent-shaped alignment due to internally organized hydrogen-bonded syn–syn conformation, exhibit high affinity and excellent selectivity for the four-repeat human telomere G-quadruplex. The current model of complex formation involves stacking of the planar ligands on terminal G-tetrads of G-quadruplexes, given that the crescent-shaped ligand exhibits excellent geometric complementarity with the dimensions of the G-tetrad.
Porphyrins
Porphyrins have been used successfully as ligands for targeting G-quadruplexes (214,230,231). The most extensively studied cationic porphyrin has been 5,10,15,20-tetrakis-(N-methyl-4-pyridyl)porphyrin (TMPyP4) (Figure 13d), which induces telomerase inhibition upon targeting telomeric G-quadruplexes (232) and down-regulates the expression of the c-myc oncogene (111).
NMR-based approaches have been used to investigate the complex of TMPyP4 with the five guanine-tract-containing c-myc-23456 G-quadruplex (129). Large upfield shifts are observed for a subset of imino proton resonances on complex formation, with slow exchange between free and bound forms. Exchange cross peaks observed in the NOESY spectrum of a sample containing equal amounts of free and bound forms, allowed assignments of the imino protons of the complex based on the known assignments in the free form. The TMPyP4 porphyrin ring stacks towards one end of the G-quadruplex in the solution structure of the complex (129).
Recently, an X-ray structure has been solved of TMPyP4 bound to the all-parallel-stranded bimolecular G-quadruplex formed by the two-repeat human telomere d(TAGGGTTAGGG) sequence in K+ solution (Figure 14e) (233). Somewhat unexpectedly, the porphyrin rings do not stack on the terminal G-tetrads, but rather stack on the TTA nucleotides, both on base pairs formed at the 5′-ends of the G-quadruplex (Figure 14f), as well as the double-chain-reversal or propeller loops that span the grooves of the structure. In addition, the propeller loops undergo a conformational transition on complex formation.
A limitation of first-generation cationic porphyrins such as TMPyP4 is that they exhibit poor selectivity between G-quadruplex and duplex DNA. To overcome this limitation, studies were extended to a Mn(III)-coordinated porphyrin containing a central aromatic core and four relatively flexible arms carrying cationic end groups (Figure 13e). The binding of this Mn(III) porphyrin to the four-repeat human telomere DNA established that it targets the human telomere G-quadruplex by four orders of magnitude over duplex DNA (234). Furthermore, telomerase inhibition occurred with IC50 = 580 nM. A working model has been put forward for this remarkable selectivity, where the porphyrin is proposed to stack on terminal tetrads and the flexible cationic arms are likely to be positioned in the grooves. Since the substituted Mn(III) porphyrin has two axial ligands, one of these would have to be replaced by a Mn-bound water molecule that in turn could potentially insert into the central channel in the complex (234).
Macrocyclic torands
One of the most promising telomerase inhibitor candidates is telomestatin (Figure 13f), a macrocyclic torand natural product isolated from Streptomyces anulatus, consisting of seven oxazole rings and one thiazole ring that targets G-quadruplexes with high specificity (IC50 = 5 nM) (235), causing growth arrest, apoptosis and telomere dysfunction (236). Furthermore, telomestatin shows selectivity for cancer cell lines over normal cells, activates key components of the DNA damage-response pathway and sensitizes tumor cells to chemotherapeutic agents (237). Telomestatin may also exhibit selectivity, since it has been suggested that telomestatin and related macrocycle Se2SAP bind preferentially to different folds of the human telomere G-quadruplex (238). These results suggest that the equilibrium between conformational states of the human telomere G-quadruplex can potentially be shifted on complex formation with specific macrocyclic ligands.
Oxazole-based peptide macrocycles represent a new class of chemically synthesized G-quadruplex-binding ligands (239,240) as analogs of telomestatin. One of these, an oxazole-containing 24-membered macrocycles consisting of a hexazole designated HXDV (Figure 13g), inhibits the growth of human lymphoblastoma cells with an IC50 of 0.4 μM (240). HXDV binds and thermally stabilizes the structure of the four-repeat human telomere in K+ solution, but not to duplex or triplex DNA (241). The binding stoichiometry is two HXDV molecules per G-quadruplex, presumably consistent with stacking of the cyclic hexazoles on the terminal G-tetrads at either end of the G-quadruplex. Thermodynamic and mobility studies demonstrate that the binding of HXDV is entropically driven, with the entropic driving force reflecting contributions from favorable drug-induced alteration in the configurational entropy of the DNA (241). A challenge in all these studies is linking quadruplex binding to biological effects at the cellular level, as has been documented to date for BRACO-19 and telomestatin.
Shape-selective recognition
Many of the drug–G-quadruplex complexes solved to date emphasize recognition principles highlighting the contributions of intermolecular stacking, hydrogen-bonding and hydrophobic interactions at the expense of shape-complementarity to recognition. Nevertheless, shape-selective recognition represents a promising area for future growth, with some very elegant demonstrations attesting to its potential for G-quadruplex recognition.
The non-planar and non-aromatic steroid diamines have long been of interest as potential nucleic acid-binding ligands since they have been postulated to bind to and stabilize kink sites in DNA (242). Experimental support for this hypothesis emerged following NMR demonstration of partial insertion of the steroid diamine, dipyrandium, between unstacked base pairs of poly (dA–dT) (243). Most importantly, a temperature melting fluorescence-based screen of natural and synthetic molecules identified two steroid diamines, malouetine and funtumine that induce G-quadruplex stabilization (244). Of the two, funtumine substituted by a guanylhydrazone moiety (Figure 13h), is more promising, since it interacted selectively in vitro with human telomeric G-quadruplex. Funtumine-induced senescence and telomere shortening, as well as rapid telomeric G-overhang degradation and anaphase bridge formation, associated with uncapping of telomeric ends. These new results on first-generation steroid diamines hold promise for the future, given that they can be easily synthesized and modifications readily incorporated, in efforts to increase the selectivity and potency for human telomere G-quadruplex targets.
Recently, chiral cyclic-helicene molecules have been shown to exhibit chiral and selective binding to higher order structures by wedging between two adjacent four-repeat intramolecular human telomere G-quadruplexes connected by a TTA linker (207). A left-handed chiral cyclic-helicene with a short linker (Figure 13i) appears to be sandwiched within a chiral cleft formed by two human telomere G-quadruplexes stacked 3′-to-5′ with a connecting TTA loop in d[AGGG(TTAGGG)7], as monitored by cd and fluorescence (helicenes are strongly fluorescent) studies.
FUTURE OPPORTUNITIES: QUADRUPLEX RECOGNITION
The current literature on drugs targeted to G-quadruplexes is primarily restricted to planar aromatic chromophores involved in end-stacking on terminal G-tetrads in G-quadruplexes (215,216). There remain many structural challenges associated with G-quadruplex recognition, as well as their impact on function. Several of these are outlined below.
Structure-based drug design targeted to G-quadruplex scaffolds
The Watson–Crick and major groove edges of guanines are involved in hydrogen bonding within the G-tetrad alignment (Figure 1a), leaving the minor groove edge available for further recognition. Indeed, it has been shown that adenines can pair with G-tetrads to form A–(G–G–G–G) pentads (Figure 11c) (18) and A–(G–G–G–G)–A hexads (Figure 11d) (16), as a result of non-canonical G–A pair formation. It is thus conceivable that successive G-tetrad base edges can be targeted by (A)n-containing sequence segments. An alternate strategy has been to prepare conjugates containing quadruplex-stabilizing acridines linked to oligonucleotides that are complementary to the human telomere sequence (245).
The four grooves can adopt distinct dimensions based on the strand directionalities around the G-quadruplex, thereby offering the possibility of discriminating between distinct quadruplex types. The grooves are accessible for edge-wise and diagonal loops but occluded for double-chain-reversal loops. At this time, the current understanding of quadruplex groove-specific recognition is restricted to the interaction between the aglycone sugar ring and grooves in the daunomycin–G-quadruplex complex (222). Other promising developments include identification of a group of structurally related compounds that selectively target quadruplex grooves as monitored by circular dichroism binding measurements (246).
The diversity of loops linking G-rich strands in G-quadruplexes range from edge-wise to diagonal and double-chain-reversal types. These loops can also vary in length and sequence (21,22,247,248) and can adopt distinct conformations stabilized by non-canonical pairs, triples, triads and mixed tetrads. Thus, loops projecting from G-quadruplexes serve as promising distinct targets, as yet unexploited by drug design approaches.
To date, there has been less emphasis on the contribution of shape-complementarity between ligand and G-quadruplex to molecular recognition. It has been demonstrated that shape-specific complementarity between ligand and three-helical junction targets is key to both ribozyme-based catalysis (249) and metallosupramolecular helicate recognition (250). We anticipate that this will represent a challenging area for future investigation.
Combinatorial approaches to identification of G-quadruplex-binding drugs
A library-based approach is eventually needed for unbiased and selectivity-driven identification of ligands that target unique G-quadruplex topologies and discriminate against closely related counterparts. To this end, the earliest combinatorial selection approaches generated carbocyanine–peptide conjugate libraries (251). Furthermore, ribosomal display has yielded antibody fragment libraries (62) that exhibit specificity for different G-quadruplex fold families. Recently, click chemistry has been used to generate bistriazole ligands to generate pharmacophores capable of π-stacking interactions with G-tetrads (252).
Library-based approaches can also be applied to identify proteins that target G-quadruplexes in a sequence and structure-specific manner. To this end, selection approaches have been used to engineer tandem zinc finger proteins that bind G-quadruplex scaffolds and effectively inhibit the activity of telomerase (253,254).
Further development of such library-based approaches should provide opportunities for modulating processes from DNA recombination to maintenance of telomere length and integrity.
Protein–DNA/RNA quadruplex complexes
Despite over a decade of structural research on G-quadruplexes, there is still no structure for a protein–G-quadruplex complex. This is unfortunate since there is an extensive literature on proteins that bind G-quadruplexes (44–46), including proteins that either facilitate or non-catalytically disrupt G-quadruplex formation, as well as helicases that catalytically unwind G-quadruplexes in an ATP-dependent manner and nucleases that cleave at or adjacent to G-quadruplex scaffolds. Many of these proteins bind specific G-quadruplex scaffolds. Therefore, there is a pressing need to devote efforts at structurally characterizing complexes of proteins that target DNA and/or RNA families of G-quadruplexes and G-quadruplex–duplex junctions.
G-quadruplex forming sequences have also been identified in alternatively spliced pre-mRNA sequences (71). One of the most important challenges in the future centers on determination of the structures of RNA G-quadruplex scaffolds and RNA G-quadruplex–duplex junctions adopted by alternatively spliced pre-mRNA sequences on complex formation with bound proteins.
Addendum
A recent paper monitored the conformational transition of the four-repeat human telomere sequence d[G3(T2AG3)3] in 150 mM K+ solution on addition of PEG 200, a mediator of molecular crowding conditions (255). The CD spectrum changed from one typical of a (3+1) G-quadruplex to one typical of an all parallel-stranded G-quadruplex at 40% (w/v) of added PEG 200. The human telomere G-quadruplex in K+ solution containing 40% PEG 200 exhibited unusual stability and negatively impacted on polymerase processivity. These data provide strong support for formation of an all parallel-stranded G-quadruplex for the four-repeat human telomere in K+ solution under molecular crowding conditions (255).
ACKNOWLEDGEMENTS
Research in the Patel laboratory on the structure and recognition of G-quadruplexes is funded by NIH grant GM034504-22. The earlier contributions of Serge Bouaziz, Natalya Chernichenko, Andrey Gorin, Abdelali Kettani, Kim Ngoc Luu, Ananya Majumdar, Eugene Skripkin, Yong Wang and Na Zhang, former members of the Patel laboratory, are gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by GM034504-22.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman SB, Cohen GH, Davies DR. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 3.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Patel DJ. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992;31:8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- 5.Laughlan G, Murchie AI, Norman DG, Moore MH, Moody PC, Lilley DM, Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 6.Pan B, Shi K, Sundaralingam M. Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex. Proc. Natl Acad. Sci. USA. 2006;103:3130–3134. doi: 10.1073/pnas.0507730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 8.Smith FW, Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 10.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 11.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 12.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 13.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 14.Kettani A, Bouaziz S, Gorin A, Zhao H, Jones RA, Patel DJ. Solution structure of a Na cation stabilized DNA quadruplex containing G•G•G•G and G•C•G•C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 15.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. A dimeric DNA interface stabilized by stacked A•(G•G•G•G)•A hexads and coordinated monovalent cations. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 17.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. V-shaped scaffold: a new architectural motif identified in an A•(G•G•G•G) pentad-containing dimeric DNA quadruplex involving stacked G(anti)•G(anti)•G(anti)•G(syn) tetrads. J. Mol. Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]
- 19.Kuryavyi VV, Jovin TM. Triad-DNA: a model for trinucleotide repeats. Nat. Genet. 1995;9:339–341. doi: 10.1038/ng0495-339. [DOI] [PubMed] [Google Scholar]
- 20.Kettani A, Basu G, Gorin A, Majumdar A, Skripkin E, Patel DJ. A two-stranded template-based approach to G•(C-A) triad formation: designing novel structural elements into an existing DNA framework. J. Mol. Biol. 2000;301:129–146. doi: 10.1006/jmbi.2000.3932. [DOI] [PubMed] [Google Scholar]
- 21.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 22.Hazel P, Parkinson GN, Neidle S. Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex. J. Am. Chem. Soc. 2006;128:5480–5487. doi: 10.1021/ja058577+. [DOI] [PubMed] [Google Scholar]
- 23.Hud NV, Plavec J. The role of cations in determining quadruplex structure and stability. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 100–130. [Google Scholar]
- 24.Phillips K, Dauter Z, Murchie AI, Lilley DM, Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 25.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 26.Bouaziz S, Kettani A, Patel DJ. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998;282:637–652. doi: 10.1006/jmbi.1998.2031. [DOI] [PubMed] [Google Scholar]
- 27.Risitano A, Fox KR. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 28.Guschlbauer W, Chantot JF, Thiele D. Four-stranded nucleic acid structures 25 years later: from guanosine gels to telomere DNA. J. Biomol. Struct. Dyn. 1990;8:491–511. doi: 10.1080/07391102.1990.10507825. [DOI] [PubMed] [Google Scholar]
- 29.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert DE, Feigon J. Multistranded DNA structures. Curr. Opin. Struct. Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 31.Patel DJ, Bouaziz S, Kettani A, Wang Y. Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric, and triplet repeat disease DNA sequences. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. pp. 389–453. [Google Scholar]
- 32.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 33.Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 34.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 35.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structural diversity of G-quadruplex scaffolds. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 81–99. [Google Scholar]
- 36.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkinson GN. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 1–30. [Google Scholar]
- 39.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 44.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 45.Maizels N. Quadruplexes and the biology of G-rich genomic regions. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 228–252. [Google Scholar]
- 46.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 47.Fang G, Cech TR. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993;74:875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- 48.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 50.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 51.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 52.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 53.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 55.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 56.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 58.Sun H, Yabuki A, Maizels N. A human nuclease specific for G4 DNA. Proc. Natl Acad. Sci. USA. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghosal G, Muniyappa K. Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res. 2005;33:4692–4703. doi: 10.1093/nar/gki777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang MY, Yarborough OH, III, Conyers GB, McPhie P, Owens RA. Stable secondary structure near the nicking site for the adeno-associated virus type 2 Rep proteins on human chromosome 19. J. Virol. 2005;79:3544–3556. doi: 10.1128/JVI.79.6.3544-3556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z, Lee A, Gilbert W. Gene disruption of a G4-DNA-dependent nuclease in yeast leads to cellular senescence and telomere shortening. Proc. Natl Acad. Sci. USA. 1995;92:6002–6006. doi: 10.1073/pnas.92.13.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 64.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 66.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 68.Qi H, Lin CP, Fu X, Wood LM, Liu AA, Tsai YC, Chen Y, Barbieri CM, Pilch DS, et al. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 69.Tsai YC, Qi H, Liu LF. Protection of DNA ends by telomeric 3′ G-tail sequences. J. Biol. Chem. 2007;282:18786–18792. doi: 10.1074/jbc.M702171200. [DOI] [PubMed] [Google Scholar]
- 70.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliver AW, Bogdarina I, Schroeder E, Taylor IA, Kneale GG. Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol. 2000;301:575–584. doi: 10.1006/jmbi.2000.3991. [DOI] [PubMed] [Google Scholar]
- 75.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 76.Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 77.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greider CW. Mammalian telomere dynamics: healing, fragmentation shortening and stabilization. Curr. Opin. Genet. Dev. 1994;4:203–211. doi: 10.1016/s0959-437x(05)80046-2. [DOI] [PubMed] [Google Scholar]
- 79.Blackburn EH. Telomeres: no end in sight. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 80.Rhodes D, Giraldo R. Telomere structure and function. Curr. Opin. Struct. Biol. 1995;5:311–322. doi: 10.1016/0959-440x(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 81.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 82.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 83.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 84.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 85.Anuradha S, Muniyappa K. Molecular aspects of meiotic chromosome synapsis and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:49–132. doi: 10.1016/S0079-6603(04)79002-9. [DOI] [PubMed] [Google Scholar]
- 86.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 87.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 88.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 89.Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cairns D, Anderson RJ, Perry PJ, Jenkins TC. Design of telomerase inhibitors for the treatment of cancer. Curr. Pharm. Des. 2002;8:2491–2504. doi: 10.2174/1381612023392720. [DOI] [PubMed] [Google Scholar]
- 91.Pendino F, Tarkanyi I, Dudognon C, Hillion J, Lanotte M, Aradi J, Segal-Bendirdjian E. Telomeres and telomerase: pharmacological targets for new anticancer strategies? Curr. Cancer Drug Targets. 2006;6:147–180. doi: 10.2174/156800906776056482. [DOI] [PubMed] [Google Scholar]
- 92.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Phan AT, Patel DJ. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and anti-parallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Phan AT, Modi YS, Patel DJ. Two-repeat Tetrahymena telomeric d(TGGGGTTGGGGT) Sequence interconverts between asymmetric dimeric G-quadruplexes in solution. J. Mol. Biol. 2004;338:93–102. doi: 10.1016/j.jmb.2004.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang N, Phan AT, Patel DJ. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 98.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm706. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 105.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 106.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;15:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kan ZY, Lin Y, Wang F, Zhuang XY, Zhao Y, Pang DW, Hao YH, Tan Z. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007;35:3646–3653. doi: 10.1093/nar/gkm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dexheimer TS, Fry M, Hurley LH. DNA quadruplexes and gene regulation. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 180–207. [Google Scholar]
- 111.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 114.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shirude PS, Okumus B, Ying L, Ha T, Balasubramanian S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene c-kit. J. Am. Chem. Soc. 2007;129:7484–7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rangan A, Fedoroff OY, Hurley LH. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 119.Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu. Rev. Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 120.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jaattela M. Multiple cell death pathways as regulators of tumour initiation and progression. Oncogene. 2004;23:2746–2756. doi: 10.1038/sj.onc.1207513. [DOI] [PubMed] [Google Scholar]
- 122.Slamon DJ, deKernion JB, Verma IM, Cline MJ. Expression of cellular oncogenes in human malignancies. Science. 1984;224:256–262. doi: 10.1126/science.6538699. [DOI] [PubMed] [Google Scholar]
- 123.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 124.Boles TC, Hogan ME. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987;26:367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- 125.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rachwal PA, Findlow IS, Werner JM, Brown T, Fox KR. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 129.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, et al. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 132.Kitamura Y, Hirota S, Nishida T. A loss-of-function mutation of c-kit results in depletion of mast cells and interstitial cells of Cajal, while its gain-of-function mutation results in their oncogenesis. Mutat. Res. 2001;477:165–171. doi: 10.1016/s0027-5107(01)00117-8. [DOI] [PubMed] [Google Scholar]
- 133.Tuveson DA, Willis NA, Jacks T, Griffin JD, Singer S, Fletcher CD, Fletcher JA, Demetri GD. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene. 2001;20:5054–5058. doi: 10.1038/sj.onc.1204704. [DOI] [PubMed] [Google Scholar]
- 134.Schittenhelm MM, Shiraga S, Schroeder A, Corbin AS, Griffith D, Lee FY, Bokemeyer C, Deininger MW, Druker BJ, et al. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer Res. 2006;66:473–481. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 135.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 137.Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 138.Reed JC, Kitada S, Takayama S, Miyashita T. Regulation of chemoresistance by the bcl-2 oncoprotein in non-Hodgkin's lymphoma and lymphocytic leukemia cell lines. Ann. Oncol. 1994;5:61–65. doi: 10.1093/annonc/5.suppl_1.s61. [DOI] [PubMed] [Google Scholar]
- 139.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martiny-Baron G, Marme D. VEGF-mediated tumour angiogenesis: a new target for cancer therapy. Curr. Opin. Biotechnol. 1995;6:675–680. doi: 10.1016/0958-1669(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 141.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 142.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 143.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 144.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 145.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 147.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 149.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 150.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 151.Mirkin SM. Molecular models for repeat expansions. Chemtracts Biochem. Mol. Biol. 2004;17:639–662. [Google Scholar]
- 152.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 153.Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence d(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 154.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 155.Richards RI, Sutherland GR. Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem. Sci. 1997;22:432–436. doi: 10.1016/s0968-0004(97)01108-0. [DOI] [PubMed] [Google Scholar]
- 156.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 157.Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 158.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 161.Weisman-Shomer P, Cohen E, Hershco I, Khateb S, Wolfovitz-Barchad O, Hurley LH, Fry M. The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res. 2003;31:3963–3970. doi: 10.1093/nar/gkg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weisman-Shomer P, Naot Y, Fry M. Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)n are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem. 2000;275:2231–2238. doi: 10.1074/jbc.275.3.2231. [DOI] [PubMed] [Google Scholar]
- 164.Kettani A, Kumar RA, Patel DJ. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 165.Usdin K. NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucleic Acids Res. 1998;26:4078–4085. doi: 10.1093/nar/26.17.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 167.Grabczyk E, Usdin K. The GAA•TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, et al. GAA instability in Friedreich's ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 169.Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD. Sticky DNA: self-association properties of long GAA•TTC repeats in R.R.Y triplex structures from Friedreich's ataxia. Mol. Cell. 1999;3:465–475. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 170.LeProust EM, Pearson CE, Sinden RR, Gao X. Unexpected formation of parallel duplex in GAA and TTC trinucleotide repeats of Friedreich's ataxia. J. Mol. Biol. 2000;302:1063–1080. doi: 10.1006/jmbi.2000.4073. [DOI] [PubMed] [Google Scholar]
- 171.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 173.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. Dimeric DNA quadruplex containing major groove-aligned A•T•A•T and G•C•G•C tetrads stabilized by inter-subunit Watson-Crick A•T and G•C pairs. J. Mol. Biol. 2001;312:1073–1088. doi: 10.1006/jmbi.2001.5002. [DOI] [PubMed] [Google Scholar]
- 174.Majumdar A, Patel DJ. Identifying hydrogen bond alignments in multistranded DNA architectures by NMR. Acc. Chem. Res. 2002;35:1–11. doi: 10.1021/ar010097+. [DOI] [PubMed] [Google Scholar]
- 175.Leonard GA, Zhang S, Peterson MR, Harrop SJ, Helliwell JR, Cruse WB, d'Estaintot BL, Kennard O, Brown T, et al. Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 Å resolution. Structure. 1995;3:335–340. doi: 10.1016/s0969-2126(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 176.Salisbury SA, Wilson SE, Powell HR, Kennard O, Lubini P, Sheldrick GM, Escaja N, Alazzouzi E, Grandas A, et al. The bi-loop, a new general four-stranded DNA motif. Proc. Natl Acad. Sci. USA. 1997;94:5515–5518. doi: 10.1073/pnas.94.11.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Patel PK, Koti AS, Hosur RV. NMR studies on truncated sequences of human telomeric DNA: observation of a novel A-tetrad. Nucleic Acids Res. 1999;27:3836–3843. doi: 10.1093/nar/27.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2Jnn couplings. J. Am. Chem Soc. 1998;120:8293–8297. [Google Scholar]
- 179.Pervushin K, Ono A, Fernandez C, Szyperski T, Kainosho M, Wuthrich K. NMR scalar couplings across Watson-Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc. Natl Acad. Sci. USA. 1998;95:14147–14151. doi: 10.1073/pnas.95.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pan B, Xiong Y, Shi K, Sundaralingam M. An eight-stranded helical fragment in RNA crystal structure: implications for tetraplex interaction. Structure. 2003;11:825–831. doi: 10.1016/s0969-2126(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 181.Pan B, Xiong Y, Shi K, Deng J, Sundaralingam M. Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads: implications for specific binding in RNA tetraplexes. Structure. 2003;11:815–823. doi: 10.1016/s0969-2126(03)00107-2. [DOI] [PubMed] [Google Scholar]
- 182.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 183.Kettani A, Bouaziz S, Wang W, Jones RA, Patel DJ. Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nat. Struct. Biol. 1997;4:382–389. doi: 10.1038/nsb0597-382. [DOI] [PubMed] [Google Scholar]
- 184.Kuryavyi V, Kettani A, Wang W, Jones R, Patel DJ. A diamond-shaped zipper-like DNA architecture containing triads sandwiched between mismatches and tetrads. J. Mol. Biol. 2000;295:455–469. doi: 10.1006/jmbi.1999.3345. [DOI] [PubMed] [Google Scholar]
- 185.Kuryavyi V, Majumdar A, Shallop A, Chernichenko N, Skripkin E, Jones R, Patel DJ. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)•G(syn)•G(anti)•G(anti) tetrads flanked by a G•(T-T) triad and a T•T•T triple. J. Mol. Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
- 186.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)4 triplet repeat DNA with a G•G•G•G tetrad and a G(A)•G(A)•G(A)•G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 187.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny JL. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- 189.Kettani A, Bouaziz S, Skripkin E, Majumdar A, Wang W, Jones RA, Patel DJ. Interlocked mismatch-aligned arrowhead DNA motifs. Structure. 1999;7:803–815. doi: 10.1016/s0969-2126(99)80104-x. [DOI] [PubMed] [Google Scholar]
- 190.de Soultrait VR, Lozach PY, Altmeyer R, Tarrago-Litvak L, Litvak S, Andreola ML. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 191.Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Horsburgh BC, Kollmus H, Hauser H, Coen DM. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell. 1996;86:949–959. doi: 10.1016/S0092-8674(00)80170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cheong C, Moore PB. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 195.Deng J, Xiong Y, Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)4 with divalent Sr2+ ions at subatomic resolution (0.61 A) Proc. Natl Acad. Sci. USA. 2001;98:13665–13670. doi: 10.1073/pnas.241374798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the N-RAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Patel DJ, Kozlowski SA, Ikuta S, Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG•dA mismatch site. Biochemistry. 1984;23:3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- 198.Prive GG, Heinemann U, Chandrasegaran S, Kan LS, Kopka ML, Dickerson RE. Helix geometry, hydration, and G•A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 199.Leonard GA, McAuley-Hecht KE, Ebel S, Lough DM, Brown T, Hunter WN. Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti)•A(anti) base pairs. Structure. 1994;2:483–494. doi: 10.1016/S0969-2126(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 200.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 201.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 202.Brown T, Hunter WN, Kneale G, Kennard O. Molecular structure of the G•A base pair in DNA and its implications for the mechanism of transversion mutations. Proc. Natl Acad. Sci. USA. 1986;83:2402–2406. doi: 10.1073/pnas.83.8.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gao X, Patel DJ. The G(syn)•A(anti) mismatch formation in DNA dodecamers at acidic pH: pH dependent conformational transition of G•A mispairs detected by proton NMR. J. Am. Chem. Soc. 1988;110:5178–5182. [Google Scholar]
- 204.Brown T, Leonard GA, Booth ED, Chambers J. Crystal structure and stability of a DNA duplex containing A(anti)•G(syn) base-pairs. J. Mol. Biol. 1989;207:455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- 205.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 207.Xu Y, Yamazaki S, Osuga H, Sugiyama H. The recognition of higher-order G-quadruplex by chiral cyclic-helicene molecules. Nucleic Acids Symp. Ser. 2006;50:183–184. doi: 10.1093/nass/nrl091. [DOI] [PubMed] [Google Scholar]
- 208.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 209.de Lange T. Activation of telomerase in a human tumor. Proc. Natl Acad. Sci. USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 211.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 212.De Cian A, Mergny JL. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mergny JL, Helene C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 214.Arthanari H, Bolton PH. Porphyrins can catalyze the interconversion of DNA quadruplex structural types. Anticancer Drug Des. 1999;14:317–326. [PubMed] [Google Scholar]
- 215.Searle MS, Balkwill GD. DNA quadruplex-ligand recognition: structure and dynamics. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC publishing; 2006. pp. 131–153. [Google Scholar]
- 216.Riou JF, Gomez D, Morjani H, Trentesaux C. Quadruplex ligand recognition: biological aspects. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 154–179. [Google Scholar]
- 217.Mergny JL, Gros J, De Cian A, Bourdoncle A, Rosu F, Sacca B, Guittat L, Amrane S, Mills M, et al. Energetics, kinetics and dynamics of quadruplex folding. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 31–80. [Google Scholar]
- 218.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat. Rev. Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 219.Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 220.Kerwin SM. G-Quadruplex DNA as a target for drug design. Curr. Pharm. Des. 2000;6:441–478. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 221.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 222.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 223.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-Based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 224.Gavathiotis E, Heald RA, Stevens MF, Searle MS. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003;334:25–36. doi: 10.1016/j.jmb.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 225.Incles CM, Schultes CM, Kempski H, Koehler H, Kelland LR, Neidle S. A G-quadruplex telomere targeting agent produces p16-associated senescence and chromosomal fusions in human prostate cancer cells. Mol. Cancer Ther. 2004;3:1201–1206. [PubMed] [Google Scholar]
- 226.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 227.Martins C, Gunaratnam M, Stuart J, Makwana V, Greciano O, Reszka AP, Kelland LR, Neidle S. Structure-based design of benzylamino-acridine compounds as G-quadruplex DNA telomere targeting agents. Bioorg. Med. Chem. Lett. 2007;17:2293–2298. doi: 10.1016/j.bmcl.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 228.Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, et al. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 229.De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 230.Yamashita T, Uno T, Ishikawa Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005;13:2423–2430. doi: 10.1016/j.bmc.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 231.Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-Ya K, White E, et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J. Am. Chem. Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 232.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- 233.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 234.Dixon IM, Lopez F, Tejera AM, Esteve JP, Blasco MA, Pratviel G, Meunier B. A G-quadruplex ligand with 10,000-fold selectivity over duplex DNA. J. Am. Chem. Soc. 2007;129:1502–1503. doi: 10.1021/ja065591t. [DOI] [PubMed] [Google Scholar]
- 235.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 236.Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K. Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 237.Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 238.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 239.Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. Oxazole-based peptide macrocycles: a new class of G-quadruplex binding ligands. J. Am. Chem. Soc. 2006;128:13662–13663. doi: 10.1021/ja064713e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Minhas GS, Pilch DS, Kerrigan JE, LaVoie EJ, Rice JE. Synthesis and G-quadruplex stabilizing properties of a series of oxazole-containing macrocycles. Bioorg. Med. Chem. Lett. 2006;16:3891–3895. doi: 10.1016/j.bmcl.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 241.Barbieri CM, Srinivasan AR, Rzuczek SG, Rice JE, LaVoie EJ, Pilch DS. Defining the mode, energetics and specificity with which a macrocyclic hexaoxazole binds to human telomeric G-quadruplex DNA. Nucleic Acids Res. 2007;35:3272–3286. doi: 10.1093/nar/gkm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sobell HM, Tsai CC, Gilbert SG, Jain SC, Sakore TD. Organization of DNA in chromatin. Proc. Natl Acad. Sci. USA. 1976;73:3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Patel DJ, Canuel LL. Steroid diamine-nucleic acid interactions: partial insertion of dipyrandium between unstacked base pairs of the poly(dA-dT) duplex in solution. Proc. Natl Acad. Sci. USA. 1979;76:24–28. doi: 10.1073/pnas.76.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Brassart B, Gomez D, De Cian A, Paterski P, Montagnac A, Qui KH, Temime-Smaali N, Trentesaux C, Mergny J-L, et al. A new steroid derivative stabilizes G-quadruplexes and induces telomere uncapping in human tumor cells. Mol. Pharm. 2007;72:631–640. doi: 10.1124/mol.107.036574. [DOI] [PubMed] [Google Scholar]
- 245.Casals J, Debethune L, Alvarez K, Risitano A, Fox KR, Grandas A, Pedroso E. Directing quadruplex-stabilizing drugs to the telomere: synthesis and properties of acridine-oligonucleotide conjugates. Bioconjug. Chem. 2006;17:1351–1359. doi: 10.1021/bc060194t. [DOI] [PubMed] [Google Scholar]
- 246.White EW, Tanious F, Ismail MA, Reszka AP, Neidle S, Boykin DW, Wilson WD. Structure-specific recognition of quadruplex DNA by organic cations: influence of shape, substituents and charge. Biophys. Chem. 2007;126:140–153. doi: 10.1016/j.bpc.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 247.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cevec M, Plavec J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry. 2005;44:15238–15246. doi: 10.1021/bi0514414. [DOI] [PubMed] [Google Scholar]
- 249.Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Hobartner C, Polonskaia A, Phan AT, Wombacher R, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat. Struct. Mol. Biol. 2005;12:218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Oleksy A, Blanco AG, Boer R, Uson I, Aymami J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. Int. Ed. Engl. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 251.Schouten JA, Ladame S, Mason SJ, Cooper MA, Balasubramanian S. G-quadruplex-specific peptide-hemicyanine ligands by partial combinatorial selection. J. Am. Chem. Soc. 2003;125:5594–5595. doi: 10.1021/ja029356w. [DOI] [PubMed] [Google Scholar]
- 252.Moorhouse AD, Santos AM, Gunaratnam M, Moore M, Neidle S, Moses JE. Stabilization of G-quadruplex DNA by highly selective ligands via click chemistry. J. Am. Chem. Soc. 2006;128:15972–15973. doi: 10.1021/ja0661919. [DOI] [PubMed] [Google Scholar]
- 253.Isalan M, Patel SD, Balasubramanian S, Choo Y. Selection of zinc fingers that bind single-stranded telomeric DNA in the G-quadruplex conformation. Biochemistry. 2001;40:830–836. doi: 10.1021/bi001728v. [DOI] [PubMed] [Google Scholar]
- 254.Patel SD, Isalan M, Gavory G, Ladame S, Choo Y, Balasubramanian S. Inhibition of human telomerase activity by an engineered zinc finger protein that binds G-quadruplexes. Biochemistry. 2004;43:13452–13458. doi: 10.1021/bi048892t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Xue Y, Kan Z-Y, Wang Q, Yao Y, Liu J, Hao Y-H, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 256.Patel DJ, Kozlowski SA, Nordheim A, Rich A. Right-handed and left-handed DNA: studies of B- and Z-DNA by using proton nuclear Overhauser effect and P NMR. Proc. Natl Acad. Sci. USA. 1982;79:1413–1417. doi: 10.1073/pnas.79.5.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]