Abstract
Caloric restriction (CR) reduces the incidence and progression of spontaneous and induced tumors in laboratory rodents while increasing mean and maximum life spans. It has been suggested that CR extends longevity and reduces age-related pathologies by reducing the levels of DNA damage and mutations that accumulate with age. This hypothesis is attractive because the integrity of the genome is essential to a cell/organism and because it is supported by observations that both cancer and immunological defects, which increase significantly with age and are delayed by CR, are associated with changes in DNA damage and/or DNA repair. Over the last three decades, numerous laboratories have examined the effects of CR on the integrity of the genome and the ability of cells to repair DNA. The majority of studies performed indicate that the age-related increase in oxidative damage to DNA is significantly reduced by CR. Early studies suggest that CR reduces DNA damage by enhancing DNA repair. With the advent of genomic technology and our increased understanding of specific repair pathways, CR has been shown to have a significant effect on major DNA repair pathways, such as NER, BER and double-strand break repair.
INTRODUCTION
More than 70 years ago, McCay and his colleagues demonstrated that a reduction in total food intake after weaning significantly increased both mean and maximum life spans of laboratory rats (1). Over the last seven decades, numerous laboratories have successfully repeated McCay's findings using various strains of rats and mice as well as non-mammalian species, such as fish and flies (2–6). Thus, food restriction has been established as a powerful experimental tool, and the anti-aging action of food restriction has become one of the most active areas of research in the realm of biogerontology (6). While life span extension by food restriction appears to be due to alterations in aging processes, the underlying mechanism(s) by which food restriction exerts its anti-aging effects remain elusive. Identification of important anti-aging and anti-tumor targets of food restriction and elucidating the molecular mechanisms by which food restriction exerts its beneficial effects could eventually provide targets for intervention in humans.
Extensive studies by Masoro's (7) and Good's (8) laboratories conclusively demonstrate that the increase in the survival of rodents by food restriction is due to a decreased intake of calories, i.e. caloric restriction (CR). Interestingly, the increase in survival is attained along with retardation in disease and pathology, e.g. cancer; thus, it is often proposed that CR increases the survival of rodents simply by retarding disease processes rather than altering aging and senescence (4). Although the distinction between senescence and disease processes is difficult to interpret, research over the last three decades demonstrates that CR retards the changes that occur with age in most physiological processes and that these changes generally precede any alterations observed in pathology and disease (4,8–10). While restricting the dietary intake of proteins and restriction of dietary methionine have been shown by some laboratories to increase both mean and maximum life spans of rats (11,12), other laboratories have reported that methionine restriction is not the key to life span extension by food restriction (13,14). To date, CR is the best described experimental strategy known to increase the survival of mammals by retarding aging. Thus, CR offers scientists a powerful tool for studying the biological and molecular mechanism(s) involved in aging and senescence.
Although it is well established that CR (without malnutrition) increases survival of rodents and reduces the pathology and disease associated with aging, the molecular and cellular event(s) underlying these various outcomes are currently unknown. Initially, several hypotheses were advanced to explain the mechanistic basis by which CR prolonged survival of laboratory rodents (4,15–19). Originally, it was proposed that CR increased survival by retarding growth and development. Later, reduction in body fat content was proposed as the physiological basis for the increased survival rates of rodents fed a CR diet (20). Subsequently, Sacher (21) proposed that CR increased survival by reducing the metabolic rate of the rodents. However, studies over the last three decades have refuted these hypotheses (22,23).
Recent efforts on dissecting the mechanism(s) by which CR extends life span lean towards the impact of CR on stress response and signaling mechanisms. Currently, the major hypotheses proposed are the oxidative damage attenuation hypothesis, the altered glucose–insulin system hypothesis, the alteration of the growth hormone-IGF-1 axis hypothesis and the hormesis hypothesis (14). In the oxidative damage attenuation hypothesis, CR has been demonstrated to reduce the accumulation of oxidative damage to biological molecules including DNA, protein and lipids in rodents (24,25). This reduction in oxidative damage is proposed to be due to a decline in the generation of reactive oxygen species (ROS) (26), an enhancement of protective mechanisms (27–31), an increase in repair capacity (32–34) or a combination of all the aforementioned. However, recent findings suggest that oxidative stress, in at least some studies, does not alter aging processes; thus, knowing whether CR extends life span via a reduction in oxidative damage remains elusive (14). The premise of the glucose–insulin hypothesis is that CR reduces the levels of plasma glucose and insulin/insulin signaling. Ultimately, CR-mediated life extension could be a result of increased glucose effectiveness, insulin responsiveness or both (35–39). This hypothesis remains exciting in light of data that show loss of the insulin signaling system results in life extension in nematodes, fruit flies and mice (14). Moreover, CR has the potential to reduce the IGF-1 signaling, which has been observed in mammalian models with a resultant increase life span (40,41).
The hormesis hypothesis proposes a benefit to health from low-intensity stressors (14). CR may occur through a hormesis mechanism in that it could act as a mild stressor eliciting an adaptive response namely, enhanced maintenance and repair systems. CR, as part of the hormetic effect, has been found to enhance the expression of one or more stress response genes (33,42–46). In line with CR as a low-intensity stressor, it is relevant to this article to suggest that particular enzymes involved in DNA repair pathways may function as stress response genes when exposed to low intensity stress, such as CR.
Caloric restriction and genomic stability
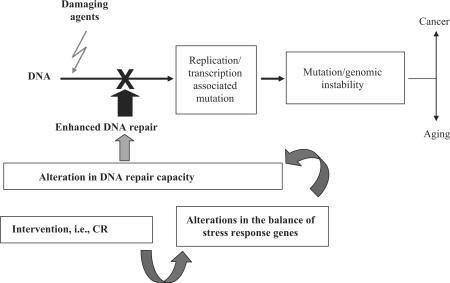
The importance of preventing an accumulation of damage to genetic material cannot be overstated. Protection of DNA is believed to be a key element in preventing cancer as well as in delaying or preventing the aged phenotype. When DNA damage persists in the genome, through replicative processes and/or through transcription-associated mutagenesis, this damage becomes permanent, in the form of mutations and/or chromosomal breakage and instability. This increased genomic instability promotes both carcinogenic and aging processes, as outlined in Figure 1. A question that recently has been addressed by numerous laboratories is whether DNA repair capacity can be manipulated, i.e. whether key enzymes in DNA repair pathways are stress response genes and whether these enzymes could be upregulated in response to environmental stimuli, and are there environmental factors that play a role in determining the effectiveness of these critical pathways? In support of a role for the environment in modulating the efficiency of DNA repair capacity, numerous laboratories have addressed the question of whether CR improves genomic stability via enhancing the expression/activity of enzymes in DNA repair pathways (47). In other words, it is rational to propose that an age-related decline in the ability of a cell/organism to maintain the integrity of its genome is a fundamental mechanism underlying the aging process with CR being at the core of life extension through maintenance of genomic integrity (48). This hypothesis is attractive because the integrity of the genome is very important to a cell/organism and because DNA is constantly exposed to insults from both endogenous and exogenous factors and because DNA damage has been associated with various biological endpoints, including mutations, cancer and many of the age-associated diseases (49–53). Furthermore, DNA integrity, as maintained by a number of DNA repair systems, is essential for the survival of cells and organisms (53). The accumulation of DNA damage in somatic cells was first proposed as a basic mechanism in the aging process (54), and it was hypothesized that the accumulation of DNA damage eventually results in the inactivation of genes and cell death (54). In 1967, Alexander (55) extended this theory by proposing that the age-related accumulation of un-repaired DNA damage and subsequent mutations in cells would alter DNA replication and transcription resulting in an aged phenotype. Accordingly, if expression of essential proteins is reduced or inhibited through alterations in genetic materials, a cell may lose function and/or viability, and this could be a primary cause of aging. Studies conducted over the past 50 years support these hypotheses by showing age-related alterations in DNA damage, mutation and metabolism (56,57). In addition, many of these alterations are accelerated in Werner's syndrome, Hutchinson Gilford Progeria and Cockayne syndromes, genetic diseases that show many clinical symptoms of premature aging and involve mutations in DNA repair genes. Interestingly, CR has been shown to reverse the age-related alterations in DNA damage/repair and mutations. In this article, we review the proposed mechanisms of DNA damage and repair while providing insight into current research that may assist in further understanding the molecular and cellular events involved in aging and the life-prolonging effect of CR. However, there are a variety of mechanisms by which genomic integrity can be affected. For example, the level of DNA damage and/or mutations can be affected by changes in carcinogen activation (55), increased detoxification of carcinogens, increased ability to repair DNA or a combination of these factors. CR is a well-established model for preventing the onset and delaying the progression of spontaneous and chemically induced tumors. It is inviting to suggest that an upregulation in DNA repair capacity provides a mechanistic explanation for the improved genomic stability observed in CR animals. We suggest that CR is an ‘intervention’ that alters the activation of specific ‘stress response genes’, key enzymes in DNA repair pathways, which then results in upregulation of ‘DNA repair' capacity’. Enhanced DNA repair reduces the levels of DNA damage, consequently reducing mutation frequency, which would result in maintenance of genomic stability.
Figure 1.
Modulation of genomic stability.
Mechanisms of DNA repair
The ultimate effect of any DNA alteration on cell function is dependent upon the speed and fidelity with which the damage is repaired. In replicating cells, the lasting effect of a particular damage will depend upon whether the damage is repaired before replication of the damaged area occurs. During the life span of any organism, the genome is constantly exposed to a variety of agents (58). The various insults to genomic DNA can be classified as endogenous and exogenous damages. ‘Endogenous damages’ include mainly replication errors, alkylating damage, and oxidative and hydrolytic damage arising from the surrounding water and cellular metabolism. Some of the hydrolytic damages encountered by the DNA are, (i) depurination/depyrimidination of nucleotides and (ii) deamination of nitrogenous bases (e.g. cytosine deamination to uracil) (59). Oxidative damage to DNA includes (i) ring-saturated pyrimidines like thymine glycols and cytosine hydrates, (ii) ring-opened purines forming FaPy products (formamidopyrimidine), (iii) 8-hydroxydeoxyguanosine and (iv) damage arising from lipid peroxidation products like M1G and etheno adduct (59). The various ‘exogenous damages’ to DNA are introduced by (i) physical agents like ionizing radiations (e.g. oxidative damage), ultraviolet rays (e.g. cyclobutane dimers), and heat, and (ii) chemical agents (59). These chemical agents can be direct acting, such as alkylating agents, or indirect acting, requiring activation, e.g. benzopyrene (59).
Despite the large amount of DNA damage that occurs every day because of body temperature, metabolism and environmental insults, only a few stable changes, i.e. mutations, accumulate in the DNA sequence; the rest of the damage is eliminated with remarkable efficiency by elaborate DNA repair systems that have evolved in cells/organisms to remove all types of genomic instability (58). In mammalian cells, there is a variety of repair mechanisms—namely, direct removal of lesions, excision repair, mismatch repair and non-homologous end joining, which are each catalyzed by a different set of enzymes (60–63). Reversal of damage is the simplest repair mechanism involving a single enzyme that catalyzes the direct reversal of the damage, e.g. reversal of alkylating damage by O6-methylguanine DNA methyltransferase (MGMT). This repair pathway repairs both single- and double-strand DNA and requires no knowledge of the genetic information (59). Excision repair is the mechanism responsible for the repair of most DNA damage. In this repair pathway, damaged or inappropriate bases are excised from DNA and replaced by the correct nucleotide. Base excision repair pathway (BER) is believed to repair small, non-helix-distorting lesions in the DNA. The BER pathway has been estimated to be responsible for the repair of as many as one million nucleotides per cell per day (64), stressing its importance in the maintenance of genomic stability. It also is involved in the repair of the endogenous damage that arises spontaneously in the living cell. This pathway repairs nucleotides with small base adducts like oxidized bases (e.g. 8-oxoG), hydrolytically deaminated bases (e.g. cytosine to uracil), and alkylated bases (e.g. 3-methyl adenine) (65). BER usually replaces a single-damaged nucleotide and sometimes can replace up to 6 nt, hence is classified as short-patch and long-patch BER (59,66–68). Nucleotide excision repair (NER) pathway repairs large and bulky helix distorting adducts to DNA like the ones created by carcinogens (e.g. benzopyrene, cis-platin), UV (e.g. cyclobutane pyrimidines dimers) and some oxidative adducts. NER in human cells requires about 25 proteins, including 7 associated with the genetic disease Xeroderma Pigmentosum (XP) (69). The enzymes involved in BER have strict substrate specificity, whereas the enzymes in NER have a very broad specificity, accommodating a large number of substrates. Mismatch repair pathway (MMR) predominantly repairs replication errors and also when there is insertion or deletion of nucleotides in DNA (63). In addition to the repair of damaged nucleotides and/or single-strand breaks, double-stand breaks (DSB) created by DNA damaging agents can be repaired by a specialized pathway like non-homologous end joining (NHEJ), homologous recombination, synthesis-dependent strand annealing, break-initiated repair, and single strand annealing (59).
Effect of age on DNA repair
Research over the past decades suggest that many steps in DNA metabolism are altered with age in a variety of tissues and animal models (56,57). The relation of DNA repair to aging has been studied by measuring the ability of cells from organisms of various life spans to repair DNA damage and by experiments that have compared the ability of cells from young and old organisms to repair DNA damage. Interest was peaked by the original study conducted by Hart and Setlow (70) who demonstrated a positive correlation between UV-induced DNA repair by fibroblasts with species’ life span because it appeared that increased DNA repair was associated with a longer life span. A similar correlation has been shown with lymphocytes (71). However, not all studies in this area have shown an analogous relation between DNA repair capacity and species’ life span (72). These conflicting results could be accounted for by the relatively crude assays often used in measuring DNA repair and the many pathways accessible to a cell for repair of damaged DNA. With the utilization of specially designed systems for evaluating DNA repair, more recent studies tend to suggest that particular types of DNA damage are not restored as efficiently in cells from old organisms.
With the advent of transgenic and knockout models, it has become possible to study complex processes by genetically manipulating individual components of the pathway or process (73). Theoretically, DNA repair pathways could be altered in two ways: repair could be reduced or enhanced. In order to enhance a specific DNA repair pathway, a systematic investigation of each component of the DNA repair pathway is necessary to determine the rate-limiting component of these repair pathways. However, this approach has been proven difficult because components of most DNA repair pathways need to work in concert to enhance the repair capacity. Currently, the simplest approach of altering DNA repair in animal models is to utilize ‘knockout’ animal models. These models would potentially display a repair-deficient phenotype by reducing the activity of one essential component of the pathway. In such a transgenic model, the deficient gene product would become limiting and DNA repair reduced; thus, such animal models could potentially be used to accelerate the accumulation of DNA damage/mutations and to more directly test the Somatic Mutation Theory of Aging, i.e. these animals should exhibit accelerated aging (i.e. reduced life span), earlier incidence of disease process(es), increased changes in biomarkers of aging, and reduced lifespan. Despite difficulties, many mammalian animal models have been produced that are deficient in DNA repair genes, e.g. DNA polymerase β, XPA, XPC genes (73,74), and the impact of aging on these models has been examined. For example, DNA polymerase β heterozygous knockout mice displaying haploinsufficiency in the BER pathway, show accelerated mortality and tumor development (75). In addition, XPA homozygous knockout mice with functional NER deficiency display accelerated mutations in liver (76) and increased hepatocellular adenomas with age (77), while, homozygous XPC knockout animals show accelerated spontaneous mutagenesis with age in lymphocytes (78) and increased susceptibility to skin, liver and lung cancer in response to carcinogens (79). Interestingly, neither XPA nor XPC knockout models display accelerated aging. However, patients with trichothiodystrophy, a heritable disorder arising from point mutations in the XPD gene creating functional NER deficiency, show accelerated aging (80), while Xpdm/m mice exhibit prominent premature aging features and a significant reduction (20%) in life span (81).
Effect of caloric restriction on DNA repair/damage
Over the past three decades, a number of investigators have compared the ability of cells to repair a variety of types of DNA damage in CR rodents. Originally, these studies were designed to determine the impact of CR on DNA repair capacity as measured by unscheduled DNA synthesis (UDS). This reputable assay measures the integration of radioactive thymidine into DNA by either autoradiography or scintillation counting after exposure of cells to a DNA damaging agent in an environment where replication is inhibited or minimal. Findings from Licastro et al. (82) and Weraarchakul et al. (53) showed that irrespective of age, dietary restriction significantly increased UDS levels after UV-irradiation in lymphocytes obtained from spleens of mice, as well as in the hepatocytes and kidney cells obtained from rats. Furthermore, using the same technique, Srivastava and Busbee (83) and Tilley et al. (84) measured UDS levels in hepatocytes and lung fibroblasts obtained from CR rats, supporting the view that CR enhances the ability of cells to repair the UV-induced DNA damage. In addition to UV-irradiation, Lipman et al. (85) observed higher levels of UDS induced by methylmethanesulfonate in cultured skin fibroblasts obtained from CR rats as compared to animals fed ad libitum. Moreover, Asakura et al. (86) showed that cultured hepatocytes obtained from young rats treated with hepatocarcinogens, i.e. dimethylnitrosamine (DMN), on a short-term food restriction modulated the response of UDS. In contrast to the above findings, Shaddock et al. (87) did not observe any beneficial effect of CR on DNA repair. Contradictions in findings by Shaddock's laboratory could be due to the nature of carcinogens used by their lab. While carcinogens used by Shaddock's group required metabolic activation, other investigators used direct-acting mutagens. Inconsistencies also could arise from the use of relatively crude assays in measuring DNA repair, i.e. while UDS in combination with UV is a rather specific measure of the removal of 6–4 photoproducts by NER, UDS itself neither directly nor selectively measures the removal of a particular type of damage.
Over the last decade, research on the impact of CR on DNA repair has focused on specific DNA repair pathways and the expression of key enzymes within these pathways (Table 1). As such, understanding the mechanism of DNA repair, as well as the function of key enzymes both dependently and independently of these repair pathways has become the key to dissecting the possible beneficial effects of CR on these pathways. Originally, Hanawalt's laboratory reported heterogeneity in the repair of the dihydrofolate reductase (DHFR) gene, its upstream DNA fragments and the whole genome in response to UV-induced DNA damage (88,89). Subsequently, studies showed that the removal of cyclobutane pyrimidine dimers (CPDs) from the transcriptionally active genes/DNA fragments was more efficient than the removal of CPDs from the non-transcribed DNA in variety of organisms, ranging from E. coli (90) and yeast (91) to cells from rodents (89,92,93) and humans (94,95). Thus, it was established that NER preferentially repairs transcriptionally active genes, and the transcribed strand of the transcriptionally active gene is restored more efficiently than the non-transcribed strands (88). Utilizing Hanawalt's methodology, Guo et al. (92) characterized how aging affects the ability of cultured hepatocytes obtained from rats to repair UV-induced DNA fragments/genes (32,92). In line with Bohr's findings, Guo et al. (32,92) demonstrated heterogeneity in the removal of CPDs from various DNA fragments studied. The transcriptionally active albumin gene was shown to be repaired more efficiently than the transcriptionally inactive embryonic myosin heavy chain (MHCemb) gene, H-ras fragments and the genome overall in cultured hepatocytes (32,92). Interestingly, an age-related decline in the removal of CPD's in unexpressed DNA (i.e. MHCemb), H-ras fragments and the genome overall was observed 24 h post-UV treatment, which was reversed by CR (32). While, Guo et al. (32) did not observe an age-related decline in the repair of albumin fragment 24 h post UV-treatment, an age-related decline in the rate of repair of the transcriptionally active albumin fragment was observed 12 h post UV-treatment. Intriguingly, the age-related decline in repair of albumin fragment at 12 h post UV-treatment was also reversed by CR (32). Thus, CR appears to alter the age-related decline in the rate of repair of specific genes. Also, the effect of age and CR on the DNA repair capacity of cells by the NER pathway appears to be heterogeneous in nature, i.e. the effect of age and CR is gene- as well as strand-dependent.
Table 1.
Effect of caloric restriction on DNA repair pathways
| DNA repair pathway | Species tissue/cells | CR Diet (% of ad libitum) | DNA-damaging agent | Ages studied (in months) | Changes with CR | Reference |
|---|---|---|---|---|---|---|
| Base excision repair | Brain, liver, spleen, testes (Male Fischer 344 rats) | 60 | 24 | CR reverses age-related decline in BER by up-regulating β-pol | Cabelof et al. (33) | |
| Brain, liver, spleen, testes (Male Fischer 344 rats) | 60 | DMS (50 mg/kg body weight) | 6 | CR up-regulates β-pol and BER when exposed to oxidizing agents | Cabelof et al. (33) | |
| 2-NP (50 mg/kg body weight) | ||||||
| Brain, liver, heart, kidney (C57BL/6 mice) | 60 | NA | 12 | CR enhances uracil-initiated BER in a tissue-specific manner | Stuart et al. (34) | |
| Nucleotide excision repair | Hepatocytes (Male Fischer 344 rats) | 60 | UV (5–30 J/m2) | 6, 12 and 24 | CR reverses the age-related decline in repair of transcriptionally active strand | Guo et al. (32) |
| Mismatch repair | Small and Large Intestine (Mlh ± C57BL/6 mice) | 70 | NA | Entire life span | NS difference in adenoma number but small increase in life- span seen with CR | Tsao et al. (99) |
| NHEJa repair | Kidney, lung, testes, liver | 60 | NA | 6, 12, 18 and 24 | CR reverses age-related decline in ku protein in a tissue-specific manner | Um et al. (46) |
UV = ultraviolet irradiation; DMS = dimethyl sulfate, 2-NP = 2-nitropropane, NA = not applicable and NS = not significant.
aNon homologous end joining.
In addition to the aforementioned studies on the NER pathway, CR was shown to reverse the age-related decrease in DNA polymerase α-specific activity and fidelity (96). Furthermore, Prapurna and Rao (97) discovered that long-term CR enhances the activity of total DNA polymerases in various regions of the brain in rats. Thus, CR appears to impact fidelity of both the DNA replication machinery as well as the component of the DNA repair pathways in rodents. More recently, Cabelof et al. (33) showed that CR reverses the age-related decline in BER. They demonstrated that DNA polymerase β message, protein and activity are up-regulated by CR, indicating that DNA polymerase β, the rate-limiting enzyme in the BER pathway and a stress response gene, is the specific polymerase altered in response to CR. They also showed that CR resulted in a significant up-regulation in BER and expression of DNA polymerase β in young animals as well providing them with resistance against two carcinogens: Dimethyl sulfate and 2-nitropropane. Their observations on the effect of CR on BER during aging provide an explanation for the anti-aging action of CR, supporting the hormesis theory of aging and CR.
In addition, Stuart et al. (34) provided evidence showing the differential effect of CR on mitochondrial and nuclear DNA-initiated BER. They observed 30% lower mitochondrial DNA damage (8-hydroxydeoxyguanosine), accompanied by a decrease in the uracil-initiated BER capacity in mitochondria isolated from brain and kidney. However, they showed a 46% decrease in mitochondrial DNA damage (8-hydroxydeoxyguanosine) accompanied by an increase in the uracil-initiated BER capacity of liver mitochondria. The reduced mitochondrial BER observed in brain and kidney could be attributed to the reduced AP endonuclease and polymerase γ activity observed during CR. They concluded that the mitochondrial genomic stability enhanced by CR was due to decreased ROS production by mitochondria rather than changes in the repair of mitochondrial DNA damage. However, Bohr's laboratory observed different effects of CR on nuclear BER (34). Uracil-initiated BER capacity in nuclei from kidney or liver was upregulated by CR. They did not observe any significant differences in the activities of DNA glycosylases (OGG1, UDG, NTH1) and AP endonuclease, supporting the observation of Cabelof et al. (33) that DNA polymerase β is the polymerase enhanced during CR.
Um et al. (46) investigated the tissue-specific changes of the non-homologous end joining (NHEJ) repair protein, Ku and the mitochondrial HSP70 in aging rats and the effects of CR on these proteins. They showed that CR reverses the age-related increase in mtHSP70 (age-related apoptotic sensitive marker) and the age-related decrease in Ku in a tissue-specific manner. Ku80/70, the regulatory component of the DNA double-strand break recognition protein DNA-PK, is altered during aging and CR prevents this alteration. CR reverses the age-related decline in Ku80/70 in the kidney and lung with no significant difference seen in liver between old ad libitum-fed animals and CR animals. CR restored the Ku expression in the testis of the 18-month-old rats but was unable to restore the Ku expression in the testis of the 24-month-old rats. Moreover, Sir2, which has been suggested to play a central role in CR-induced longevity in lower organisms, (e.g. yeast), has been shown to repair DNA double-strand breaks by NHEJ, along with other proteins responsible for repairing breaks created from the recombinational repair pathway (98).
Tsao et al. (99) studied Mlh1 deficient mice (MMR) prone to lymphomas, intestinal adenomas and carcinomas on different diets, e.g. high, fat low calcium diet, (HFLC) and a CR diet. They observed no significant difference in the incidence of adenocarcinoma and lymphoma with CR; however, the number of adenomas was significantly greater in the HFLC-fed mice. They suggest that diet may have an enhancing effect on tumorigenesis before the onset of genomic instability caused by MMR deficiency.
Above and beyond DNA repair pathways, the levels of DNA mutation and DNA damage in tissues of rodents fed either ad libitum or CR diets have been ascertained by several investigators (Table 2). DNA mutations are indicative of damaged DNA that has escaped repair processes, and analysis of the frequency of mutations in the hypoxanthine phosphoribosyl transferase (hprt) gene is a well-established assay to quantify DNA mutation in vivo. Dempsey et al. (100) and Casciano et al. (101) observed significant reduction in mutations in the hprt gene in lymphocytes obtained from CR-fed mice and rats as compared to ad libitum fed animals. In addition to analysis of mutation frequency, the accumulation of DNA damage in various tissues of laboratory rodents has been shown to be reduced by CR by several laboratories. Yu's laboratory demonstrated that CR lowered the level 8-hydroxydeoxyguanosine levels (the major type of endogenous oxidative damage) in liver DNA of rats (102), and Djuric et al. (103) showed a significant reduction in the level of 5-hydroxymethyluracil (another oxidative product) in DNA from liver and mammary gland of rats fed a CR diet. Sohal et al. (104) also observed reduced levels of 8-hydroxydeoxyguanosine with CR in all tissues studied in mice, e.g. skeletal, muscle, brain, heart, liver and kidney. Kaneko et al. (105) also found that CR reduced the onset of the age-related increase in 8-hydroxydeoxyguanosine levels. Moreover, CR has been shown to decrease DNA damage products excreted in the urine of human subjects (106,107). In contrast to above findings, Fu et al. (108) showed no significant effect of CR on the accumulation of single-strand breaks in DNA in a variety of tissues obtained from mice fed a CR diet. In their analysis, Fu et al. (108) fed their CR mice 53% of the calories consumed by their ad libitum-fed controls, and the level of DNA damage was determined against control mice that consumed 80% of the calories consumed by the mice fed ad libitum. In their study, the control animals were not ad libiutm fed; thus, the effect of 20% restriction of calorie intake in ‘control’ animals may have masked the beneficial impact of CR.
Table 2.
Effect of caloric restriction on DNA damage
| DNA damage | Species tissue/cells | CR diet (% of ad libitum) | Ages studied (in months) | Changes with CR (% reduction) | Reference |
|---|---|---|---|---|---|
| 8-hydroxydeoxy-guanine | Brain (Male C57BL/6N mice) | 60 | 15 | 34 | Sohal et al. (104) |
| Heart (Male C57BL/6N mice) | 60 | 15 | 12 | Sohal et al. (104) | |
| Kidney (Male C57BL/6N mice) | 60 | 15 | NS | Sohal et al. (104) | |
| Liver (Male C57BL/6N mice) | 60 | 9–17 | NS | Sohal et al. (104) | |
| 23 | 35 | ||||
| Skeletal muscle (Male C57BL/6N mice) | 60 | 15 | 20 | Sohal et al. (104) | |
| Brain (Male B6D2F1 mice) | 60 | 24–26 | ∼22 | Hamilton et al. (111) | |
| Heart (Male B6D2F1 mice) | 60 | 24–26 | ∼12 | Hamilton et al. (111) | |
| Liver (Nuclear) (Male B6D2F1 mice) | 60 | 24–26 | ∼19 | Hamilton et al. (111) | |
| Liver (Mitochondrial) (Male B6D2F1 mice) | 60 | 24–26 | ∼42 | Hamilton et al. (111) | |
| Kidney (Male B6D2F1 mice) | 60 | 24–26 | ∼12 | Hamilton et al. (111) | |
| Brain (Male Fischer 344 rats) | NA | 6–24 | NS | Kaneko et al. (105) | |
| 30 | 30 | ||||
| 60 | 24–26 | ∼32 | Hamilton et al. (111) | ||
| Heart (Male Fischer 344 rats) | NA | 6–24 | NS | Kaneko et al. (105) | |
| 30 | 34 | ||||
| 60 | 24–26 | ∼50 | Hamilton et al. (111) | ||
| Kidney (Male Fischer 44 rats) | NA | 6–24 | NS | Kaneko et al. (105) | |
| 30 | 45 | ||||
| 60 | 24–26 | NS | Hamilton et al. (111) | ||
| 60 | 22–24 | ∼27 | Ward et al. (112) | ||
| Liver (Male Fischer 344 rats) | NA | 6–24 | NS | Kaneko et al. (105) | |
| 30 | 31 | ||||
| Liver (Nuclei) (Male Fischer 344 rats) | 60 | 3 | 30 | Chung et al. (102) | |
| 24 | 30 | ||||
| 60 | 24–26 | NS | Hamilton et al. (111) | ||
| 60 | 22–24 | ∼30 | Ward et al. (112) | ||
| Liver (Mitochondrial) (Male Fischer 344 rats) | 60 | 24 | 20 | Chung et al. (102) | |
| 60 | 24–26 | ∼50 | Hamilton et al. (111) | ||
| Muscle (Male Fischer 344 rats) | 60 | 24–26 | ∼55 | Hamilton et al. (111) | |
| Brain (Sprague-Dawley rats) | 60 | 24 | ∼40 | Wolf et al. (109) | |
| Heart (Sprague-Dawley ats) | 60 | 24 | ∼50 | Wolf et al. (109) | |
| Liver (Sprague-Dawley rats) | 60 | 24 | ∼40 | Wolf et al. (109) | |
| Skeletal muscle (Sprague-Dawley rats) | 60 | 24 | ∼30 | Wolf et al. (109) | |
| Intestine (Sprague-Dawley rats) | 60 | 24 | ∼30 | Wolf et al. (109) | |
| Lymphocytes (Sprague-Dawley rats) | 60 | 24 | ∼55 | Wolf et al. (109) | |
| Liver (Mitochondrial) (Male Wistar rats) | 60 | 24 | ∼46 | Lopez-Torres et al. (115) | |
| 5-hydroxymethyl uracil | Liver (Female Fischer 344 rats) | 60 | 2 | 43 | Djuric et al. (103) |
| Mammary gland (Female Fischer 344 rats) | 60 | 2 | 38 | Djuric et al. (103) | |
| Fpg and Endo III sensitive oxidized bases | Aorta (Male B6D2F1 mice) | 60 | 26 | ∼26 | Guo et al. (29) |
| Single-strand breaks | Brain (Male C57BL/6N mice) | 53 | 6-25 | NS | Fu et al. (108) |
| Kidney (Male C57BL/6N mice) | 60 | 6–25 | NS | Fu et al. (108) | |
| Liver (Female C57BL/6N mice) | 53 | 6–25 | NS | Fu et al. (108) | |
| mtDNA deletions | Brain (Male Fischer 344 rats) | 60 | 6–24 | NS | Kang et al. (114) |
| Liver (Mitochondrial) (Male Fischer 344 rats) | 60 | 6 | NS | Kang et al. (114) | |
| 18 | 73 | ||||
| 24 | 71 | ||||
| 24 | 20 | ||||
| Mutations (hypoxanthine Phosphoribosyl Transferase locus) | Splenic lymphocytes (Female HALH/0) | 60 | 6 | NS | Dempsey et al. (100) |
*NA = not applicable, NS = not significant and mt = mitochondrial.
More recently, Guo et al. (29) demonstrated that CR decreased an age-related increase in oxidative DNA damage as seen through EndoIII- and Fpg-sensitive sites in mouse aortic cells. Likewise, a significant reduction in the age-related increase in oxidative DNA damage was seen by Wolf et al. (109) in the brain, skeletal muscle, heart, liver, tenuum mucosa and lymphocytes from rats. One major problem often ignored in measuring oxidative damage to DNA is the potential of generating oxidative damage during the isolation of DNA. Our group showed that 8-hydroxydeoxyguanosine generation during DNA isolation was eliminated using the sodium iodide isolation method (110). Using this method of DNA isolation, we measured the levels of 8-hydroxydeoxyguanosine in nuclear DNA from a large number tissues from both mice and rats (111). A significant increase in oxo8dG levels in nDNA was observed with age in all tissues studied and all strains of rodents. Thus, an age-related increase in DNA oxidation (i.e. 8-hydroxydeoxyguanosine levels) appears to be a universal phenomenon, at least in rodents. CR significantly reduced the age-related accumulation of 8-hydroxydeoxyguanosine levels in nDNA of all tissues of mice and all tissues of rats, except liver and kidney (111). In a subsequent study, our group showed that CR did indeed reduce the 8-hydroxydeoxyguanosine levels in livers and kidneys of rats (112). Thus, these data are consistent with the view that CR reduces oxidative damage in DNA in most, if not all tissues of rodents.
In addition to the protection conferred by long-term CR on nuclear DNA from various endogenous and exogenous factors and oxidative damage, CR has also been shown to protect mitochondrial DNA from oxidative damage. A 20–25% reduction in the level of 8-hydroxydeoxyguanosine in rat liver mitochondrial DNA has been shown by CR (103). Using the sodium iodide isolation method, we observed an age-related increase in 8-hydroxydeoxyguanosine levels in mtDNA isolated from the livers of rats and mice, and this increase was significantly reduced by CR (111). Moreover, Melov et al. (113) showed that CR significantly reduced the age-related accumulation of mitochondrial DNA mutations in heart tissue of mice. More recently, Kang et al. (114) witnessed a reduction in age-related mitochondrial DNA deletions in the rat liver by CR. In contrast, the brain of these animals exhibited no alteration in the level of mitochondrial DNA deletions by CR. Monica Lopez-Torres et al. (115) showed that CR for 12 months reduced oxidative damage to mitochondrial DNA by 46%. Short-term CR (6 months) reduced mitochondrial ROS formation by 23%, and reduced significantly 8-hydroxydeoxyguanosine levels in mitochondrial DNA. However, no difference was observed in the oxidative damage in nuclear DNA compared to the old ad libitum control animals. They suggested that the difference in oxidative damage to nuclear DNA and mitochondrial DNA could be due to the efficient repair of nuclear DNA and that the reduced 8-hydroxydeoxyguanosine by CR in mitochondrial DNA was due to the reduced ROS production.
Overview and future directions
The concept of the ‘anti-aging’ and ‘anti-cancer’ actions of CR has come a long way since its discovery some 70 years ago. It is now clear that the increase in the survival of rodents by CR is due to the restriction of calories (14). Studies over the last three decades have refuted some of the previously proposed mechanisms, e.g. alteration in growth, body weight and metabolism, as the sites of CR action. Currently, the major hypotheses advanced to explain the mechanistic basis for the protective effects of CR are: oxidative damage attenuation, altered glucose–insulin system, and hormesis (14). Perhaps the strongest evidence in support of the above proposed mechanisms is data from studies in which aging has been retarded by CR or studies of genetically accelerated aging. Based on the data collected over the last few decades, it is becoming clearer that perhaps the anti-aging action of CR is due to the collective actions of different mechanisms, impacting the capacity to adapt to environmental stimuli by enhancing repair/maintenance status such as antioxidant defenses and DNA repair capacity. In line with this view, the hormesis hypothesis has become an attractive proposal explaining CR's ability to ‘increase longevity, retard senescent deterioration, retard age-associated diseases, and enhance coping with environmental stimuli’ (14). In other words, the hormesis hypothesis embraces many of other proposed mechanisms, suggesting that while high intensity stress may trigger pathology and death, low intensities or concentrations of environmental challenges would lead to a beneficial change. Thus, under hormesis, DNA repair capacity would become a relevant mechanism where CR is capable of enhancing stress response genes such as DNA polymerase β, leading to resistance to stress and enhanced longevity (14,33,116).
As outlined in this review in support of hormesis hypothesis, many laboratories have shown the role of DNA repair capacity as a beneficial action of CR through increased DNA repair in cells from rodents (Table 1), reduced levels of oxidative damage in DNA, and reduced levels of mutations (Table 2). Therefore, under hormesis, CR attenuates the rate of accumulation of damage by enhancing DNA repair response through upregulation in the expression of stress response genes, thereby retarding senescent deterioration and perhaps extending life span. However, if CR's ability to extend life span is due to hormesis, will CR enhance the stress response even in young mice where no aging phenotypes are observed? Interestingly, in an extensive study, Cabelof et al. (33) examined the effect of CR on the BER pathway in young mice and rats. While CR was shown to reverse the age-related decline in BER, as well as DNA polymerase β activity, protein, and mRNA levels (33), CR also reversed mutation frequency in young animals exposed to DNA damaging agents. In support of Cabelof's finding and to address whether CR is beneficial earlier in maturation, Shima et al. (117) measured the effects of CR during murine development, and discovered that during development, CR is moderately protective against chemically induced mutagenesis. Thus, perhaps the low-intensity environmental challenge exerted by CR even in young mice resulted in beneficial effects via enhancement of the stress response genes in DNA repair pathways. It becomes attractive to suggest that CR may confer protection from endogenous DNA damage at a young age; i.e. at a very early stage of initiation, CR can prevent fixation of DNA damage, which might then delay and/or prevent initiation of cancer and delay aging. Consistent with this suggestion is the observation that a decrease in spontaneous mutation frequency is observed in 6-month-old CR mice (33). Intriguingly, the capacity of short-term CR to provoke phenotypic changes noted by long-term CR (118) suggests that genomic imprinting occurs early with the inception of CR and that the early alterations in expression of key DNA repair genes observed are a means to the life-extending capability of CR. With respect to DNA repair, and excision repair in particular, the possibility that CR has immediate and long-term effects on the protection of genomic integrity is compelling. It suggests that CR is not acting solely through reversal of age-accumulated changes, but that it has specific and direct effects on expression and activity of repair/maintenance pathways, e.g. the BER pathway, which would protect organism from endogenous and environmental damages as well as support life-long maintenance of genomic integrity. Thus, the use of animals with targeted disruptions in specific DNA repair pathways, e.g., BER genes, will allow further analysis of the role that DNA repair may play in the anti-tumorigenic and anti-aging actions of CR. Further, it will be possible using transgenic models to determine whether CR prolongs lifespan through alterations in the DNA repair machinery of the animals. The data from such experiments will provide investigators with first-hand knowledge of the mechanism by which CR reduces DNA damage/mutations and prolongs life span. In summary, the recent studies on the effects of CR on DNA repair/damage are quite interesting and most results are in favor of the view that changes in DNA repair/damage play an important role in aging. However, additional experiments that employ more accurate assays of DNA repair are required to establish definitively that CR alters DNA repair/damage.
ACKNOWLEDGMENTS
This research was supported by NIH grants R01 CA121298 (A.R. Heydari); R37 AG26577 and P01-AG19316 (A. Richardson) and The San Antonio Nathan Shock Aging Center grant, 1P30-AG13319. Funding to pay the Open Access publication charges for this article was provided by NIH-NIA: R37AG26577.
Conflict of interest statement. None declared.
REFERENCES
- 1.McCay C.M., Crowell M.F., Maynard L.A. The effect of retarded growth upon the length of life span and upon the ultimate body size. J. Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 2.Yu B.P., Masoro E.J., Murata I., Bertrand H.A., Lynd F.T. Life span study of SPF Fischer 344 male rats fed ad libitum or restricted diets: Longevity, growth, lean body mass and disease. J. Gerontol. 1982;37:130–141. doi: 10.1093/geronj/37.2.130. [DOI] [PubMed] [Google Scholar]
- 3.Weindruch R., Walford R.L., Fligiel S., Guthrie D. The retardation of aging in mice by dietary restriction: Longevity, cancer, immunity and lifetime energy intake. J. Nutr. 1986;116:654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 4.Masoro E.J. Food restriction in rodents: An evaluation of its role in the study of aging. J. Gerontol. 1988;43:B59–B64. doi: 10.1093/geronj/43.3.b59. [DOI] [PubMed] [Google Scholar]
- 5.Masoro E.J., Shimokawa I., Yu B.P. Retardation of the aging processes in rats by food restriction. Ann. N. Y. Acad. Sci. 1991;621:337–352. doi: 10.1111/j.1749-6632.1991.tb16990.x. [DOI] [PubMed] [Google Scholar]
- 6.Masoro EJ. Dietary restriction: An experimental approach to the study of the biology of aging. In: Masoro EJ, Austad SN, editors. Handbook of the Biology of Aging. 5th edn. San Diego, CA: Academic Press; 2001. pp. 396–420. [Google Scholar]
- 7.Masoro EJ. Assessment of nutritional components in prolongation of life and health by diet. Proc. Soc. Exp. Biol. Med. 1990;193:31–34. doi: 10.3181/00379727-193-42985. [DOI] [PubMed] [Google Scholar]
- 8.Kubo C, Johnson BC, Gajjar A, Good RA. Crucial Dietary Factors in Maximizing Life Span and Longevity in Autoimmune-Prone Mice. J. Nutr. 1987;117:1129–1135. doi: 10.1093/jn/117.6.1129. [DOI] [PubMed] [Google Scholar]
- 9.Masoro EJ. Nutrition and aging: A current assessment. J. Nutr. 1985;115:842–848. doi: 10.1093/jn/115.7.842. [DOI] [PubMed] [Google Scholar]
- 10.Yu BP, Masoro EJ, Maman CA. Nutritional influences on aging of Fischer 344 rats: I. Physical, metabolic, and longevity characteristics. J. Gerontol. 1985;40:657–670. doi: 10.1093/geronj/40.6.657. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman JA, Malloy V, Krajck R, Orentreich N. Nutritional control of aging. Exp. Gerontol. 2003;38:47–52. doi: 10.1016/s0531-5565(02)00149-3. [DOI] [PubMed] [Google Scholar]
- 12.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-1 and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masoro EJ, Iwasaki L, Gleiser CA, McMahan CA, Seo E, Yu. BP. Dietary modulation of the progression of nephropathy in aging rats: An evaluation of the importance of protein. Am. J. Clin. Nutr. 1989;49:217–227. doi: 10.1093/ajcn/49.6.1217. [DOI] [PubMed] [Google Scholar]
- 14.Masoro E.J. Overview of caloric restriction and ageing. Mech. Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Masoro E.J. Potential role of the modulation of fuel use in the antiaging action of dietary restriction. Ann. N. Y. Acad. Sci. 1992;663:403–411. doi: 10.1111/j.1749-6632.1992.tb38684.x. [DOI] [PubMed] [Google Scholar]
- 16.Masoro E.J. Retardation of aging processes by food restriction: An experimental tool. Am. J. Clin. Nutr. 1992;55 doi: 10.1093/ajcn/55.6.1250S. [DOI] [PubMed] [Google Scholar]
- 17.Heydari A.R., Richardson A. Does gene expression play any role in the mechanism of the antiaging effect of dietary restriction. Ann. N. Y. Acad. Sci. 1992;663:384–395. doi: 10.1111/j.1749-6632.1992.tb38682.x. [DOI] [PubMed] [Google Scholar]
- 18.Masoro E.J. Antiaging action of caloric restriction: endocrine and metabolic aspects. Obes. Res. 1995;3 doi: 10.1002/j.1550-8528.1995.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 19.Heydari A.R., You S., Takahashi R., Gutsmann A., Sarge K.D., Richardson A. Effect of caloric restriction on the expression of heat shock protein 70 and the activation of heat shock transcription factor 1. Dev. Genet. 1996;18:114–124. doi: 10.1002/(SICI)1520-6408(1996)18:2<114::AID-DVG4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 20.Berg B.N., Simms H.S. Nutrition and longevity in the rat: II. Longevity and onset of disease with different levels of food intake. J. Nutr. 1960;71:255–263. [PubMed] [Google Scholar]
- 21.Sacher G.A. Life table modification and life prolongation. In: Finch C.E., Hayflick L., editors. Handbook of the biology of aging. New York: Van Nostrand Reinhold; 1977. pp. 582–638. [Google Scholar]
- 22.McCarter R.J., Masoro E.J., Yu B.P. Does food restriction retard aging by reducing the metabolic rate? Am. J. Physiol. 1985;248:E488–E490. doi: 10.1152/ajpendo.1985.248.4.E488. [DOI] [PubMed] [Google Scholar]
- 23.McCarter R.J., McGee J.R. Transient reduction of metabolic rate by food restriction. Am. J. Physiol. 1989;257:E175–E179. doi: 10.1152/ajpendo.1989.257.2.E175. [DOI] [PubMed] [Google Scholar]
- 24.Yu B.P. Aging and oxidative stress: modulation by dietary restriction. Free Radic. Biol. Med. 1996;21:651–668. doi: 10.1016/0891-5849(96)00162-1. [DOI] [PubMed] [Google Scholar]
- 25.Zainal T.A., Oberley T.D., Allison D.B., Szweda L.I., Weindruch R. Caloric restriction of rhesus monkeys lowers oxidative damage in skeletal muscle. FASEB. 2000;14:1825–1836. doi: 10.1096/fj.99-0881com. [DOI] [PubMed] [Google Scholar]
- 26.Feuers R.J., Weindruch R., Hart R.W. Caloric restriction, aging, and antioxidant enzymes. Mutat. Res. 1993;295:191–200. doi: 10.1016/0921-8734(93)90020-4. [DOI] [PubMed] [Google Scholar]
- 27.Hyun D-H., Emerson S.S., Jo D-G., Mattson M.P., De Cabo R. Calorie restriction up-regulates the plasma membrane redox system in brain cells and suppresses oxidative stress during aging. Proc. Natl Acad. Sci. 2006;103:19908–19912. doi: 10.1073/pnas.0608008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Cabo R., Cabello R., Rios M., Lopez-Lluch G., Ingram D.K., Lane M.A., Navas P. Calorie restriction attenuates age-related alterations in the plasma membrane antioxidant system in rat liver. Exp. Gerontol. 2004;39:297–304. doi: 10.1016/j.exger.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Gou Z.M., Yang H., Hamilton M.L., VanRemmen H., Richardson A. Effects of age and food restriction on oxidative DNA damage and antioxidant enzyme activities in the mouse aorta. Mech. Ageing Dev. 2001;122:1771–1786. doi: 10.1016/s0047-6374(01)00298-6. [DOI] [PubMed] [Google Scholar]
- 30.Armeni T., Pieri C., Marra M., Sacucci F., Principato G. Studies on the life prolonging effects of food restriction: glutathione levels and glyoxylase enzymes in rat liver. Mech. Ageing Dev. 1998;101:101–110. doi: 10.1016/s0047-6374(97)00167-x. [DOI] [PubMed] [Google Scholar]
- 31.Xia E., Rao G., VanRemmen H., Heydari A.R., Richardson A. Activities of antioxidant enzymes in various tissues of male Fischer 344 ratas are altered by food restriction. J. Nutr. 1995;125:195–201. doi: 10.1093/jn/125.2.195. [DOI] [PubMed] [Google Scholar]
- 32.Guo Z.M., Heydari A.R., Richardson A. Nucleotide excision repair of actively transcribed versus nontranscribed DNA in rat hepatocytes. Exp. Cell. Res. 1998;245:228–238. doi: 10.1006/excr.1998.4269. [DOI] [PubMed] [Google Scholar]
- 33.Cabelof D.C., Yanamadala S., Raffoul J.J., Guo Z., Soofi A., Heydari A.R. Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair. 2003;2:295–307. doi: 10.1016/s1568-7864(02)00219-7. [DOI] [PubMed] [Google Scholar]
- 34.Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA. Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J. 2004;18:595–597. doi: 10.1096/fj.03-0890fje. [DOI] [PubMed] [Google Scholar]
- 35.Masoro E.J., McCarter R.J.M., Katz M.S., McMahan C.A. Dietary restriction alters the characterize of glucose fuel use. J. Gerontol. Biol. Sci. 1992;47:208. doi: 10.1093/geronj/47.6.b202. [DOI] [PubMed] [Google Scholar]
- 36.Kemnitz J.W., Roecker E.B., Weindruch R., Elson D.F., Baum S.T., Bergmann R.N. Dietary restriction increases insulin sensitivity and lowers blood glucose in Rhesus monkeys. Am. J. Physiol. 1994;266:E540–E547. doi: 10.1152/ajpendo.1994.266.4.E540. [DOI] [PubMed] [Google Scholar]
- 37.Cefalu W.T., Wagner J.D., Wang Z.Q., Bell-Farrow A.D., Collins J., Haskell D., Bechtold R., Morgan T. A study of caloric restriction and cardiovascular aging in cynomolgus monkeys (macaca facicularis): a potential model for aging research. J. Gerontol. Biol. Sci. 1997;52A:19. doi: 10.1093/gerona/52a.1.b10. [DOI] [PubMed] [Google Scholar]
- 38.Bluher M., Kahn B.B., Kahn R.C. Extended longetivity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 39.Ingram D.K., Zhu M., Mamczarz J., Zou S., Lane M.A., Roth G.S., DeCabo R. Calorie restriction memetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 40.D’costa A.P., Lenham J.E., Ingram R.L., Sonntag W.E. Moderate caloric restriction increases type 1 IGF receptors and protein synthesis in aging rats. Mech. Ageing. Dev. 1993;71:59–71. doi: 10.1016/0047-6374(93)90035-p. [DOI] [PubMed] [Google Scholar]
- 41.Coschigano K.T., Clemmons D., Bellush M.E., Kopchick J.J. Assessment of growth parameters and lifespan of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 42.Heydari A.R., Wu B., Takahashi R., Strong R., Richardson A. Expression of heat shock protein 70 is altered by age and diet at the level of transcription. Mol. Cell. Biol. 1993;13:410–418. doi: 10.1128/mcb.13.5.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., De Oliveira K.M., Leid M., McBurney M.W., Guarente L. Sirtl promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and Sir 2 for life-span extension by caloric restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 45.Anderson R.M., Bitterman K.J., Wood J.G., Medvedik O., Sinclair D.A. Nicotinamide and PNC1 govern lifespan extension by caloric restriction in Saccharomyces cervisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Um J.H., Kim S.J., Kim D.W., Ha M.Y., Jang J.H., Kim D.W., Chung B.S., Kang C.D., Kim S.H. Tissue-specific changes of DNA repair protein Ku and mtHSP70 in aging rats and their retardation by caloric restriction. Mech. Ageing Dev. 2003;124:967–975. doi: 10.1016/s0047-6374(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 47.Raffoul J.J., Guo Z., Soofi A., Heydari A.R. Caloric restriction and genomic stability. J. Nutr. Health Aging. 1993;3:102–110. [PubMed] [Google Scholar]
- 48.Haley-Zitlin V., Richardson A. Effect of dietary restriction on DNA repair and DNA damage. Mutat. Res. 1993;295:237–245. doi: 10.1016/0921-8734(93)90023-v. [DOI] [PubMed] [Google Scholar]
- 49.Tannenbaum A. The initiation and growth of tumors. Introduction. I. Effects of underfeeding. Am. J. Cancer. 1940;38:335–350. [Google Scholar]
- 50.Tannenbaum A. The initiation and growth of tumors. II. Effects of caloric restriction per se. Cancer Res. 1942;2:460–467. [Google Scholar]
- 51.Saxton J.A., Jr., Boon M.C., Furth J. Observations on the inhibition of development of spontaneous leukemia in mice by underfeeding. Cancer Res. 1944;4:401–409. [Google Scholar]
- 52.Visscher R.L., Ball Z.B., Barnes R.H., Sivertsen I. The influence of caloric restriction upon the incidence of spontaneous mammary carcinoma in mice. Surgery. 1942;11:48–55. [Google Scholar]
- 53.Weraarchakul N., Strong R., Wood W.G., Richardson A. The effect of aging and dietary restriction on DNA repair. Exp. Cell Res. 1989;181:204. doi: 10.1016/0014-4827(89)90193-6. [DOI] [PubMed] [Google Scholar]
- 54.Szilard L. On the nature of aging process. Proc. Natl. Acad. Sci. USA. 1959;45:30–45. doi: 10.1073/pnas.45.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander P. The role of DNA lesions in processes leading to aging in mice. Sym. Soc. Exp. Biol. 1967;21:29–51. [PubMed] [Google Scholar]
- 56.Mullaart E., Lohman P.H., Berends F., Vijg J. DNA damage metabolism and aging. Mutat. Res. 1990;37:189–210. doi: 10.1016/0921-8734(90)90001-8. [DOI] [PubMed] [Google Scholar]
- 57.Bohr V.A., Anson R.M. DNA damage, mutation, and fine structure DNA repair in aging. Mutat. Res. 1995;338:25–34. doi: 10.1016/0921-8734(95)00008-t. [DOI] [PubMed] [Google Scholar]
- 58.Koshland D.E., Jr. Molecule of the year: the DNA repair enzyme. Science. 1994;266:1925. doi: 10.1126/science.7801114. [DOI] [PubMed] [Google Scholar]
- 59.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 60.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 61.Hanawalt P.C. Evolution of concepts in DNA repair. Environ. Mol. Mutagen. 1994;23 (Suppl. 24):78–85. doi: 10.1002/em.2850230617. [DOI] [PubMed] [Google Scholar]
- 62.Hanawalt P.C., Donahue B.A., Sweder K.S. Repair and transcription. Collision or collusion? Current Biol. 1994;4:518–521. doi: 10.1016/s0960-9822(00)00112-3. [DOI] [PubMed] [Google Scholar]
- 63.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 64.Holmquist F.P. Endogenous lesions, S-phase-independent spontaneous mutations, and evolutionary strategies for base excision repair. Mutat. Res. 1998;400:59–68. doi: 10.1016/s0027-5107(98)00051-7. [DOI] [PubMed] [Google Scholar]
- 65.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision- repair. Mutat. Res. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 66.Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase β mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc. Natl. Acad. Sci. USA. 1990;87:5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto Y., Bogenhagen D.F. Repair of a synthetic abasic site involves concerted reactions of DNA synthesis followed by excision and ligation. Mol. Cell. Biol. 1991;11:4441–4447. doi: 10.1128/mcb.11.9.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dianov G., Price A., Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992;12:1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sancar A. DNA excision repair. Ann. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 70.Hart R.W., Setlow R.B. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc. Natl. Acad. Sci. USA. 1974;71:2169–2173. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hall K.Y., Hart R.W., Benirschke A.K., Walford R.L. Correlation between ultraviolet-induced DNA repair in primate lymphocytes and fibroblasts and species maximum achievable life span. Mech. Ageing Dev. 1984;24:163–173. doi: 10.1016/0047-6374(84)90068-x. [DOI] [PubMed] [Google Scholar]
- 72.Kato H., Hasoda M., Tsuchiya K., Mariwaki K. Absence of correlation between DNA repair in ultraviolet irradiated mammalian cells and life span of donor species. Jpn. J. Genet. 1980;55:99–108. [Google Scholar]
- 73.Walter C.A., Grabowski D.T., Street K.A., Conrad C.C., Richardson A. Analysis and modulation of DNA repair in aging. Mech. Ageing Dev. 1997;98:203–222. doi: 10.1016/s0047-6374(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 74.Sobol R.W., Horton J.K., Kuhn R, Hua G., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H. Requirement of mammalian DNA polymerase β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 75.Cabelof D.C., Ikeno Y., Nyska A., Busuttil R.A., Anyangwe N., Vijg J., Matherly L.H., Tucker J.D., Wilson S.H., et al. Haploinsufficiency in DNA polymerase β increases cancer risk with age and alters mortality rate. Cancer Res. 2006;66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- 76.Giese H., Dolle M.E.T., Hezel A., Van Steeg H., Vijg J. Accelerated accumulation of somatic mutations in mice deficient in the nucleotide excision repair gene XPA. Oncogene. 1999;18:1257–1260. doi: 10.1038/sj.onc.1202404. [DOI] [PubMed] [Google Scholar]
- 77.De Vries A., Van Oostrom C.T.M., Dortant P.M., Beems R.M., Van Kreijl C.F., Capel P.J.A., Van Steeg H. Spontaneous liver tumors and benzo[a]pyrene-induced lymphomas in XPA-deficient mice. Carcinogenesis. 1997;19:46–53. [PubMed] [Google Scholar]
- 78.Wijnhoven S.W.P., Kool H.J.M., Mullenders L.H.F., Van Zeeland A.A, Friedberg E.C., Van der Horst G.T.J, Van Steeg H., Vrieling H. Age-dependent spontaneous mutagenesis in Xpc mice defective in nucleotide excision repair. Oncogene. 2000;19:5037. doi: 10.1038/sj.onc.1203844. [DOI] [PubMed] [Google Scholar]
- 79.Friedberg E.C., Bond J.P., Burns D.K., Cheo D.L., Greenblatt M.S., Meira L.B., Nahari D., Reis A.M. Defective nucleotide excision repair in Xpc mutant mice and its association with cancer predisposition. Mutat. Res. 2000;459:99–108. doi: 10.1016/s0921-8777(99)00068-3. [DOI] [PubMed] [Google Scholar]
- 80.Hasty P., Campisi J., Hoeijmakers J., Van steeg H., Vijg J. Aging and Genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 81.Dolle M.E.T., Busuttil R.A., Garcia A.M., Wijnhoven S., Van Drunen E., Niedernhofer L.J., Van der Horst G., Hoeijmakers J.H.J., Van Steeg H., et al. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat. Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 82.Licastro F., Weindruch R., Davis L.J., Walford R.L. Effect of dietary restriction upon the age-associated decline of lymphocyte DNA repair activity in mice. Age. 1988;11:48–52. [Google Scholar]
- 83.Srivastava V.K., Busbee D.L. Decreased fidelity of DNA polymerases and decreased DNA excision repair in aging mice: effects of caloric restriction. Biochem. Biophys. Res. Commun. 1992;182:712–721. doi: 10.1016/0006-291x(92)91790-w. [DOI] [PubMed] [Google Scholar]
- 84.Tilley R., Miller S., Srivastava V., Busbee D. Enhanced unscheduled DNA synthesis by secondary cultures of lung cells established from calorically restricted aged rats. Mech. Ageing Dev. 1992;63:165–176. doi: 10.1016/0047-6374(92)90062-i. [DOI] [PubMed] [Google Scholar]
- 85.Lipman J.M., Turturro A., Hart R.W. The influence of dietary restriction on DNA repair in rodents: A preliminary study. Mech Ageing Dev. 1989;48:135–143. doi: 10.1016/0047-6374(89)90045-6. [DOI] [PubMed] [Google Scholar]
- 86.Asakura S., Sawada S., Daimon H., Fukuda T., Ogura K., Yamatsu K., Furihata C. Effects of dietary restriction on induction of unscheduled DNA synthesis (UDS) and replicative DNA synthesis (RDS) in rat liver. Mutation Res. 1994;322:257–264. doi: 10.1016/0165-1218(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 87.Shaddock J.G., Feuers R.J., Chou M.W., Casciano D.A. Evidence that DNA repair may not be modified by age or chronic caloric restriction. Mutation Res. 1993;301:261–266. doi: 10.1016/0165-7992(93)90067-6. [DOI] [PubMed] [Google Scholar]
- 88.Bohr V.A. Carcinogenesis. 1991;12:1983–1992. doi: 10.1093/carcin/12.11.1983. [DOI] [PubMed] [Google Scholar]
- 89.Bohr V.A., Smith C.A., Okumoto D.S., Hanawalt P.C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 90.Mellon I., Hanawalt P.C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 91.Sweder K.S., Hanawalt PC. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl. Acad. Sci. USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo Z., Heydari A.R., Wu W., Yang H., Sabia M.R., Richardson A. Characterization of gene-specific DNA repair by primary cultures of rat hepatocytes. J Cell Physiol. 1998;176:314–322. doi: 10.1002/(SICI)1097-4652(199808)176:2<314::AID-JCP9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 93.Mellon I., Spivak G., Hanawalt P.C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 94.Mellon I., Bohr V.A., Smith C.A., Hanawalt P.C. Preferential DNA repair of an active gene in human cells. Proc. Natl. Acad. Sci. USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venema J., Van Hoffen A., Karkagi V., Natarajan A.T., van Zeeland A.A., Mullender L.H.F. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srivastava V.K., Miller S., Schroeder M.D., Hart R.W., Busbee D. Age-related changes in expression and activity of DNA polymerase: Some effects of dietary restriction. Mutation Res. 1993;295:265–280. doi: 10.1016/0921-8734(93)90025-x. [DOI] [PubMed] [Google Scholar]
- 97.Prapurna D.R., Rao K.S. Long-term effects of caloric restriction initiated at different ages on DNA polymerases in rat brain. Mech. Ageing Dev. 2003;92:133–142. doi: 10.1016/s0047-6374(96)01815-5. [DOI] [PubMed] [Google Scholar]
- 98.Hasty P. The impact energy metabolism and genome maintenance have on longevity and senescence: lessons from yeast to mammals. Mech. Ageing Dev. 2001;122:1651–1662. doi: 10.1016/s0047-6374(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 99.Tsao J.L., Dudley S., Kwok B., Nickel A.E., Laird P.W., Siegmund K.D., Liskay R.M., Shibata D. Diet, cancer and aging in DNA mismatch repair deficient mice. Carcinogenesis. 2002;23:1807–1810. doi: 10.1093/carcin/23.11.1807. [DOI] [PubMed] [Google Scholar]
- 100.Dempsey J.L., Pfeiffer M., Morley A.A. Effect of dietary restriction on in vivo somatic mutation in mice. Mutat. Res. 1993;291:141–145. doi: 10.1016/0165-1161(93)90153-q. [DOI] [PubMed] [Google Scholar]
- 101.Casciano D.A., Chou M., Lyn-Cook L.E., Aidoo A. Calorie restriction modulates chemically induced in vivo somatic mutation frequency. Environ. Mol. Mutagen. 1996;27:162–164. doi: 10.1002/(SICI)1098-2280(1996)27:2<162::AID-EM10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 102.Chung M.H., Kasai H., Nishimura S., Yu B.P. Protection of DNA damage by dietary restriction. Free Radic. Biol. Med. 1992;12:523–525. doi: 10.1016/0891-5849(92)90105-p. [DOI] [PubMed] [Google Scholar]
- 103.Djuric Z., Lu M.H., Lewis S.M., Luongo D.A., Chen X.W., Heilbrun L.K., Reading B.A., Duffy P.H., Hart R.W. Oxidative DNA damage levels in rats fed low-fat, high-fat, or calorie-restricted diets. Toxicol. Appl. Pharmacol. 1992;115:156–160. doi: 10.1016/0041-008x(92)90318-m. [DOI] [PubMed] [Google Scholar]
- 104.Sohal R.S., Agarwal S., Candas M., Forster M.J., Lal H. Effect of age and caloric restriction on DNA oxidative damage in different tissues of C57BL/6 mice. Mech Ageing Dev. 1994;76:215–224. doi: 10.1016/0047-6374(94)91595-4. [DOI] [PubMed] [Google Scholar]
- 105.Kaneko T., Tahara S., Matsuo M. Retarding effect of dietary restriction on the accumulation of 8-hydroxy-2′-deoxyguanosine in organs of Fischer 344 rats during aging. Free. Radic. Biol. Med. 1997;23:76–81. doi: 10.1016/s0891-5849(96)00622-3. [DOI] [PubMed] [Google Scholar]
- 106.Simic M.G., Bergtold D.S. Dietary modulation of DNA damage in human. Mutat. Res. 1991;250:17–24. doi: 10.1016/0027-5107(91)90158-k. [DOI] [PubMed] [Google Scholar]
- 107.Simic M.G. DNA markers of oxidative processes in vivo: relevance to carcinogenesis and anticarcinogenesis. Cancer Res. 1994;54:1918–1923. [PubMed] [Google Scholar]
- 108.Fu C.S., Harris S.B., Wilhelmi P., Walford R.L. Lack of effect of age and dietary restriction on DNA single-stranded breaks in brain, liver, and kidney of (C3H x C57BL/10)F1 mice. J. Gerontol. 1991;46:B78–B80. doi: 10.1093/geronj/46.2.b78. [DOI] [PubMed] [Google Scholar]
- 109.Wolf F.I., Fasanella S., Tedesco B., Cavallin G., Donati A., Bergamini E., Cittadini A. Peripheral lymphocyte 8-OHdG levels correlate with age-associated increase of tissue oxidative DNA damage in Sprague-Dawley rats. Protective effects of caloric restriction. Exp. Gerontol. 2005;40:181–188. doi: 10.1016/j.exger.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 110.Hamilton M.L., Guo Z.M., Fuller C.D., Van Remmen H., Ward W.F., Austad S.N., Troyer D.A., Thompson I., Richardson A. A reliable assessment of 8-Oxo-3-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hamilton M.L., Van Remmen H., Drake J.A., Yang H., Guo Z.M., Kewitt K., Walter C.A., Richardson A. Does oxidative damage to DNA increase with age? Proc. Natl Acad. Sci., USA. 2001;98:10469–10474. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ward W.F., Qi W., Van Remmen H., Zackert W.E., Roberts L.J, II, Richardson A. Effects of age and caloric restriction on lipid peroxidation: Measurement of oxidative stress by F2-isoprostane levels. J. Gerontol. 2005;60:847–851. doi: 10.1093/gerona/60.7.847. [DOI] [PubMed] [Google Scholar]
- 113.Melov S., Hinerfeld D., Esposito L., Wallace D.C. Multi-organ characterization of mitochondrial genomic rearrangements in ad libitum and caloric restricted mice show striking somatic mitochondrial DNA rearrangements with age. Nucleic Acids Res. 1997;25:974–982. doi: 10.1093/nar/25.5.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kang C.M., Kristal B.S., Yu BP. Age-related mitochondrial DNA deletions: effect of dietary restriction. Free Radic. Biol. Med. 1998;24:148–154. doi: 10.1016/s0891-5849(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 115.Lopez-Torres M., Gredilla R., Sanz A., Barja G. Influence of aging and long-term caloric restriction on oxygen radical generation and oxidative DNA damage in rat liver mitochondria. Free Radic. Biol. Med. 2002;32:882–889. doi: 10.1016/s0891-5849(02)00773-6. [DOI] [PubMed] [Google Scholar]
- 116.Timiras P.S., Yaghmaie F., Saeed O., Thung E., Chinn G. The ageing phenome: caloric restriction and hormones promote neural cell survival, growth and de-differentiation. Mech. Ageing Dev. 2005;126:3–9. doi: 10.1016/j.mad.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 117.Shima N., Swiger R.R., Heddle J.A. Dietary restriction during murine development provides protection against MNU-induced mutations. Mutat. Res. 2000;470:189–200. doi: 10.1016/s1383-5718(00)00104-2. [DOI] [PubMed] [Google Scholar]
- 118.Cao S.X., Dhahbi J.M., Mote P.L., Spindler S.R. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc. Natl Acad. Sci. USA. 2001;98:10630–10635. doi: 10.1073/pnas.191313598. [DOI] [PMC free article] [PubMed] [Google Scholar]