Abstract
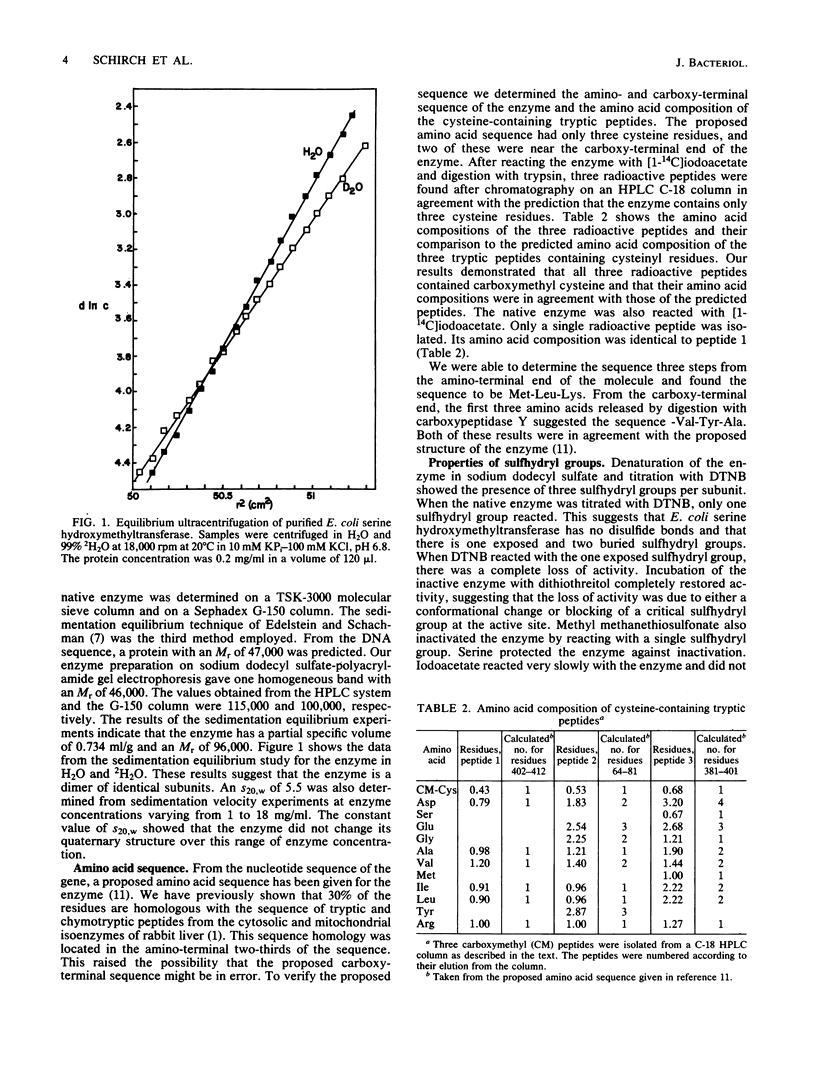
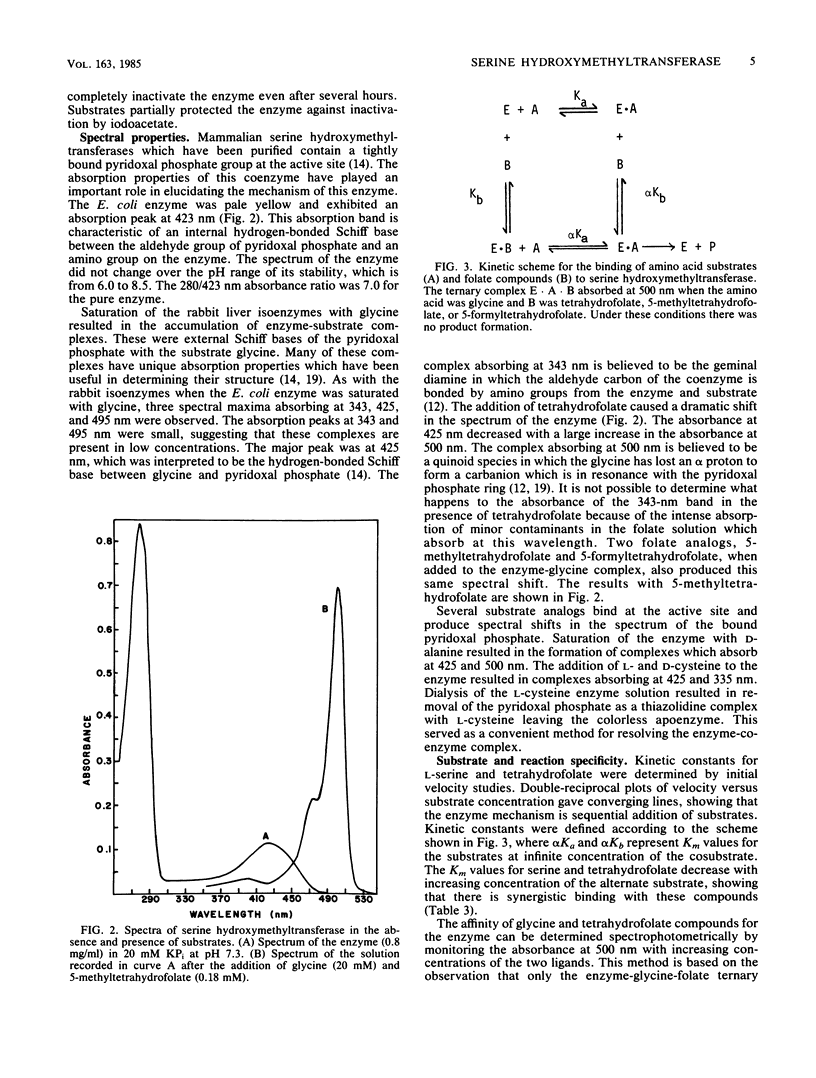
Serine hydroxymethyltransferase from Escherichia coli was purified to homogeneity. The enzyme was a homodimer of identical subunits with a molecular weight of 95,000. The amino acid sequence of the amino and carboxy-terminal ends and the amino acid composition of cysteine-containing tryptic peptides were in agreement with the primary structure proposed for this enzyme from the structure of the glyA gene (M. Plamann, L. Stauffer, M. Urbanowski, and G. Stauffer, Nucleic Acids Res. 11:2065-2074, 1983). The enzyme contained no disulfide bonds but had one sulfhydryl group on the surface of the protein. Several sulfhydryl reagents reacted with this exposed group and inactivated the enzyme. Spectra of the enzyme in the presence of substrates and substrate analogs showed that the enzyme formed the same complexes and in similar relative concentrations as previously observed with the cytosolic and mitochondrial rabbit liver isoenzymes. Kinetic studies with substrates showed that the affinity and synergistic binding of the amino acid and folate substrates were similar to those obtained with the rabbit liver isoenzymes. The enzyme catalyzed the cleavage of threonine, allothreonine, and 3-phenylserine to glycine and the corresponding aldehyde in the absence of tetrahydrofolate. The enzyme was also inactivated by D-alanine caused by the transamination of the active site pyridoxal phosphate to pyridoxamine phosphate. This substrate specificity was also observed with the rabbit liver isoenzymes. We conclude that the reaction mechanism and the active site structure of E. coli serine hydroxymethyltransferase are very similar to the mechanism and structure of the rabbit liver isoenzymes.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BLAKLEY R. L. The interconversion of serine and glycine: participation of pyridoxal phosphate. Biochem J. 1955 Oct;61(2):315–323. doi: 10.1042/bj0610315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra D., Martini F., Angelaccio S., Bossa F., Gavilanes F., Peterson D., Bullis B., Schirch L. Sequence homology between prokaryotic and eukaryotic forms of serine hydroxymethyltransferase. Biochem Biophys Res Commun. 1983 Nov 15;116(3):1007–1012. doi: 10.1016/s0006-291x(83)80242-3. [DOI] [PubMed] [Google Scholar]
- Ching W. M., Kallen R. G. Mechanism of serine hydroxymethylase catalyzed cleavage of L-erythro-beta-phenylserine: pH dependence of elementary kinetic processes from spectroscopic, pre-steady kinetic, and competitive inhibition studies. Biochemistry. 1979 Mar 6;18(5):821–830. doi: 10.1021/bi00572a013. [DOI] [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J. Regulation of synthesis of serine hydroxymethyltransferase in chemostat cultures of Escherichia coli. J Biol Chem. 1984 Jul 10;259(13):8394–8401. [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J. Role of methionine in the regulation of the synthesis of serine hydroxymethyltransferase in Escherichia coli. J Biol Chem. 1984 Jul 10;259(13):8402–8406. [PubMed] [Google Scholar]
- Edelstein S. J., Schachman H. K. The simultaneous determination of partial specific volumes and molecular weights with microgram quantities. J Biol Chem. 1967 Jan 25;242(2):306–311. [PubMed] [Google Scholar]
- Gavilanes F., Peterson D., Schirch L. Methyl methanethiosulfonate as an active site probe of serine hydroxymethyltransferase. J Biol Chem. 1982 Oct 10;257(19):11431–11436. [PubMed] [Google Scholar]
- Mansouri A., Decter J. B., Silber R. Studies on the regulation of one-carbon metabolism. II. Repression-derepression of serine hydroxymethyltransferase by methionine in Escherichia coli 113-3. J Biol Chem. 1972 Jan 25;247(2):348–352. [PubMed] [Google Scholar]
- Plamann M. D., Stauffer L. T., Urbanowski M. L., Stauffer G. V. Complete nucleotide sequence of the E. coli glyA gene. Nucleic Acids Res. 1983 Apr 11;11(7):2065–2075. doi: 10.1093/nar/11.7.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHIRCH L., JENKINS W. T. SERINE TRANSHYDROXYMETHYLASE. TRANSAMINATION OF D-ALANINE. J Biol Chem. 1964 Nov;239:3797–3800. [PubMed] [Google Scholar]
- Schirch L. Formyl-methenyl-methylenetetrahydrofolate synthetase from rabbit liver (combined). Evidence for a single site in the conversion of 5,10-methylenetetrahydrofolate to 10-formyltetrahydrofolate. Arch Biochem Biophys. 1978 Aug;189(2):283–290. doi: 10.1016/0003-9861(78)90214-x. [DOI] [PubMed] [Google Scholar]
- Schirch L., Gross T. Serine transhydroxymethylase. Identification as the threonine and allothreonine aldolases. J Biol Chem. 1968 Nov 10;243(21):5651–5655. [PubMed] [Google Scholar]
- Schirch L., Peterson D. Purification and properties of mitochondrial serine hydroxymethyltransferase. J Biol Chem. 1980 Aug 25;255(16):7801–7806. [PubMed] [Google Scholar]
- Schirch L., Ropp M. Serine transhydroxymethylase. Affinity of tetrahydrofolate compounds for the enzyme and enzyme-glycine complex. Biochemistry. 1967 Jan;6(1):253–257. doi: 10.1021/bi00853a039. [DOI] [PubMed] [Google Scholar]
- Schirch L. Serine transhydroxymethylase. Relaxation and transient kinetic study of the formation and interconversion of the enzyme-glycine complexes. J Biol Chem. 1975 Mar 10;250(5):1939–1945. [PubMed] [Google Scholar]
- Stauffer G. V., Plamann M. D., Stauffer L. T. Construction and expression of hybrid plasmids containing the Escherichia coli glyA genes. Gene. 1981 Jun-Jul;14(1-2):63–72. doi: 10.1016/0378-1119(81)90148-7. [DOI] [PubMed] [Google Scholar]
- Tarr G. E. Improved manual sequencing methods. Methods Enzymol. 1977;47:335–357. doi: 10.1016/0076-6879(77)47036-8. [DOI] [PubMed] [Google Scholar]
- Uyeda K., Rabinowitz J. C. Enzymes of the clostridial purine fermentation. Serine hydroxymethyltransferase. Arch Biochem Biophys. 1968 Feb;123(2):271–278. doi: 10.1016/0003-9861(68)90134-3. [DOI] [PubMed] [Google Scholar]



