Abstract
The p53 tumor suppressor protein, often termed guardian of the genome, integrates diverse physiological signals in mammalian cells. In response to stress signals, perhaps the best studied of which is the response to DNA damage, p53 becomes functionally active and triggers either a transient cell cycle arrest, cell death (apoptosis) or permanent cell cycle arrest (cellular senescence). Both apoptosis and cellular senescence are potent tumor suppressor mechanisms that irreversibly prevent damaged cells from undergoing neoplastic transformation. However, both processes can also deplete renewable tissues of proliferation-competent progenitor or stem cells. Such depletion, in turn, can compromise the structure and function of tissues, which is a hallmark of aging. Moreover, whereas apoptotic cells are by definition eliminated from tissues, senescent cells can persist, acquire altered functions, and thus alter tissue microenvironments in ways that can promote both cancer and aging phenotypes. Recent evidence suggests that increased p53 activity can, at least under some circumstances, promote organismal aging. Here, we discuss the role of p53 as a key regulator of the DNA damage responses, and discuss how p53 integrates the outcome of the DNA damage response to optimally balance tumor suppression and longevity.
INTRODUCTION
Longevity assurance and tumor suppression
Cancer is a potentially lethal disease that poses a major challenge to longevity in organisms with renewable tissues. Cancers, or malignant tumors, contain cells that have acquired several aberrant properties, often by somatic mutation or epigenetic changes in gene expression. These properties include uncontrolled proliferation, resistance to cell death signals, inappropriate migratory and invasive capability, and the capacity to alter the tissue microenvironment to promote angiogenesis and evasion of immune surveillance (1). A number of mammalian genes function to prevent the development of cancer, some of which are directly implicated in longevity assurance. One such longevity assurance gene is TP53, which encodes the tumor suppressor protein p53. p53 promotes longevity by reducing somatic mutations and/or the survival or proliferation of mutant cells, thereby reducing the occurrence of cancer (2,3). Interestingly, however, recent findings suggest that too much of a good thing, even increased tumor suppression by p53, can have deleterious effects and promote selected aspects of the aging process (4–6). These findings suggest there is a tight balance between tumor suppression and long-term cell proliferative potential, which is essential for the longevity of organisms, such as mammals, with renewable tissues.
DNA damage, stem cells and differentiated somatic tissues
Cancer is a major age-related disease in mice, humans and many other mammals (7). In the absence of cancer, aging is mostly characterized by tissue atrophy and degeneration (8).
Most mammalian tissues can be described as being comprised of two major cellular components: stem or progenitor cells, which are responsible for regenerative capacity or repair after injury, and differentiated somatic cells, responsible for adult stem cell support and specialized tissue/organ functions. Based on this classification, two major mechanisms can account for tissue degeneration associated with age: loss of stem cell pool division potential (loss of regenerative capacity) and loss of differentiated somatic cell function, which directly leads to loss of organ function. Loss of differentiated somatic cell function can additionally indirectly affect adult stem and progenitor cells by altering the tissue microenvironment that is essential for stem cell support (the stem cell niche). In general, loss of stem cell pool division potential can occur through multiple mechanisms including stem cell senescence, death or dysfunction of the niche. One specific mechanism that can account for the loss of both stem cell and differentiated somatic cell function is the gradual accumulation of persistent DNA damage. Persistent DNA damage and its erroneous resolution include telomeric dysfunction (9–11) and somatic mutations (12), both of which increase with age; both also have been proposed to contribute to the loss of stem and differentiated somatic cell function with age (13,14). DNA damage accumulation in stem cells has been detected in mice and clearly contributes to the attrition of stem cell division potential during aging (15). Thus, it is likely that DNA damage contributes to aging by limiting stem cell division potential and by also interfering with somatic tissue functions, including stem cell niches.
How does a loss of division potential result from DNA damage? The answer lies in the signaling networks that link lesions in the DNA to the activation of DNA damage checkpoints, which in turn activate effectors of critical cell fate decisions. Interestingly, most regulators of the DNA damage response channel the damage signals through the well-known tumor suppressor protein, p53. Here, we discuss how DNA damage activates p53 and how this single protein integrates the outcome of the DNA damage response to optimally balance tumor suppression and longevity.
P53 AND DNA DAMAGE
p53: the quintessential tumor suppressor
p53 (Tumor protein p53, TP53, Li-Fraumeni syndrome 1) has been studied for nearly three decades, and is best known for its potent ability to suppress malignant tumorigenesis. Only recently has it been appreciated that p53 belongs to the ever-growing class of longevity assurance genes. The activities of genes in this family, mostly studied in lower organisms such as fruit flies and nematodes, ensure organismal longevity (3). p53 certainly promotes long life by reducing the risk of cancer, a major age-related disease. In addition, recent evidence suggests p53 can stimulate the expression of genes that reduce oxidative stress (16–18). Because oxidative damage is generally believed to be an important contributor to both the hyperproliferative age-related cancer and degenerative changes of aging (19,20), p53 may therefore also promote longevity by reducing oxidative stress. However, as discussed below, p53 also plays a crucial role in determining cell fate, and some cell fate decisions can lead to aging phenotypes in affected tissues. Thus, p53 might have both beneficial and deleterious effects, depending on the physiological context.
Mutations in p53 reduce the fidelity of genome maintenance. Genomic instability is a major cause of cancer and a hallmark of tumor progression; thus, the longevity of both humans and mice with germline deficiencies in p53 is curtailed by cancer (21,22). While it is clear that p53 suppresses cancer, there is no single mechanism by which this multifaceted protein suppresses malignant transformation. p53 lies at the hub of complex signaling networks that can integrate a multitude of potentially oncogenic stress signals including, but not limited to, oxidative stress (23), supraphysiological mitogenic signals or activated oncogenes (24–26), metabolic alterations (16,27) and DNA damage and repair, the focus of this review. The signaling pathways leading from DNA damage to p53 activation have been extensively investigated, and provide intriguing insights into how the activity of a tumor suppressor can promote lifespan by preventing cancer and promoting genome maintenance, while paradoxically reducing lifespan through promotion of cell fate decisions that can result in tissue degeneration.
The DNA damage response pathways leading to p53
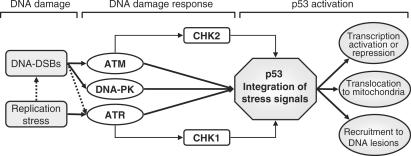
All cells must preserve genome integrity in order to guarantee proper genetic information propagation following division. Single cells respond to genotoxic stress, including DNA double-strand breaks (DSBs), by activating a signaling cascade known as the DNA damage response (DDR). The DDR is a complex interlaced network comprised of DNA damage repair factors and cell cycle regulators. Perhaps surprisingly for such an important mechanism, the DDR preferentially converges on a single protein, the tumor suppressor p53 (see Figure 1). While p53 is not the unique regulator and executor of the DDR, loss of p53 still compromises both proper repair of lesions and the timely execution of cell fate decisions.
Figure 1.
DNA damage response signaling pathways leading to p53 activation: DNA damage activates PIKK (ATM, ATR and DNA-PK), which leads to activation of checkpoint kinases (CHK1, CHK2) and p53. Activated p53 integrates the output of the DDR signaling network and triggers various cell fate decisions.
DNA damage can be caused by a multitude of factors, but ultimately takes the form of either chemical modification to a DNA base, the presence of single-stranded DNA (ssDNA) or ssDNA break or of a more severe DNA-DSB. The repair of lesions generated by chemical modifications to DNA bases, such as oxidative lesions or bulky adducts, are rapidly processed by the nucleotide excision repair (NER) and base excision repair (BER) pathways. For either BER or NER, it is believed that recruitment of the ATR/ATRIP (ATM and Rad3-related kinase/ATR-interacting protein) complex to a ssDNA intermediate in the repair process promotes initiation of the DDR. The DDR subsequently entails activation of the checkpoint kinase CHK1 and ultimately p53 activation (28–30). When the BER or NER pathways are deficient, the lesion likely converts into a DNA-DSB and initiates the DDR by activating the ATM (ataxia telangiectasia mutated) kinase (31,32) (Figure 1). Replication stress during S-phase can create ssDNA, which either promotes ATR activation as described above or progresses to DNA-DSB (29,30). Examples of replication stress include lesions that retard or interrupt replication fork progression, and the inappropriate firing of replicons, such as that caused by the hyperproliferation induced by certain oncogenes (25,26).
DNA-DSBs are caused by many stimuli including ssDNA breaks on opposite DNA strands, replication fork collapses, high energy radiation or dysfunctional telomeres, and are dangerous and potentially lethal lesions (33). DNA-DSBs require prompt repair, which can be achieved through two independent but not mutually exclusive mechanisms: error prone non-homologous end-joining (NHEJ) or relatively error-free homologous recombination (HR) (33). In response to DNA-DSBs both ATM and DNA-PK (DNA-dependent protein kinase) are activated. Activation of DNA-PK can lead to p53 phosphorylation on serine 15 and 37 (34), but mostly promotes repair of the DNA-DSB through NHEJ (28). On the other hand, activation of ATM leads to direct phosphorylation of p53 on serine 15 and 37 (34), as well as phosphorylation and activation of another checkpoint kinase (CHK2), which also phosphorylates p53 on serine 20 (28,34). It should be noted that while the pathways described above converge on p53, all three of the phosphoinositide 3-kinase-related kinases (PIKK)—ATM, ATR and DNA-PK—as well as the CHK1 and CHK2 kinases—can phosphorylate a multitude of downstream substrates with important functions in DNA repair and cell cycle arrest (28). One example of this feedback on repair from the DDR includes the phosphorylation of the histone variant H2AX at sites of DNA-DSBs by all three PIKKs. Phosphorylated H2AX (gamma-H2AX) is essential for stabilizing DNA repair complexes and promoting efficient repair (35). Nevertheless, p53 remains a key target of the DDR pathway. DDR-mediated multi-site p53 phosphorylations reduces binding of the H/MDM2 (human/mouse double minute 2) ubiquitin transferase to the p53 molecule, which in turn allows replacement of ubiquitin moieties by acetylation, resulting in p53 stabilization and full activation (36). Upon activation, depending on the severity of the stress signals, the pattern of post-transcriptional modifications and the cellular context, p53 acts as a potent transcriptional activator or repressor, can translocate to the mitochondria to induce apoptosis and finally, can localize directly to sites of DNA damage and promote proper repair (37–41) (Figure 1).
p53 activity and ensuing cellular outcomes: to be or not to be
Activation of p53 through the DDR usually leads to either proper repair of the lesion or elimination of the damaged cell from the proliferative cell pool. How stringent must this mechanism be to prevent pathological loss of cells and how does it impact organ homeostasis? The steps outlined below are general scenarios, based in some cases on indirect evidence and in all cases highly dependent on the cell, tissue and physiological context.
Once a cell has been damaged and the DDR and p53 are activated (Figure 1), a complex signaling network is engaged to result in a long-term cell fate decision. In normal mammalian cells, there are four such options. In all cases, initial processing of the DNA lesions begins immediately, while activation of cell cycle checkpoints by CHK1, CHK2 and p53 lead to transient cell growth arrest (28). While initial processing of the lesions continues, p53 physically localizes to sites of DNA damage to promote repair (39) and simultaneously stimulates the transcription of direct effectors of the cell growth arrest (e.g. the cyclin-dependent kinase inhibitor p21) as well as effectors required for efficient DNA repair of complex lesions that require longer processing (e.g. GADD45) (42). Once these early steps are in place, there are several potential cellular outcomes, most of which are heavily influenced by the cell type as well as the severity of the DNA lesions (see also Figure 2).
Figure 2.
The impact of p53-mediated cell fate decisions on tissue homeostasis and organismal longevity: In the presence of severe DNA damage, effectors triggered by p53 cause transient cell growth arrest, apoptosis or senescence, which in turn promote tissue atrophy and organismal aging. In contrast, loss of p53 function prevents critical cell fate decisions and dramatically favors cancer.
(i) Transient arrest and proper repair: when DNA damage is not severe; most of the time the result is proper repair of the lesions and recovery of the damaged cell with few, if any, consequences. Clearly this cellular outcome in which p53 promotes transient arrest and repair, is most favorable for the cell and organism.
(ii) Defective repair: alternatively, the repair attempt may fail. If the cell survives and does not arrest growth permanently (cell fate decisions 3 and 4 below), the result can be mutations, including chromosomal aberrations. This scenario, promotes the development of cancer.
(iii) Apoptosis: when the damage is very severe, p53 transcriptionally upregulates effectors of apoptosis such as proteins of the BH3-only family (PUMA, NOXA and BAX), and downregulates repressors of apoptosis such as BCL-2 and SURVIVIN (41). Newly expressed BH3-only proteins translocate to the mitochondria and promote an apoptotic cascade starting at the mitochondrial membrane (43). Moreover, p53 itself can translocate to the mitochondria and promote the mitochondrial apoptotic cascade by interfering directly with anti-apoptotic BCL-2 family members (38). Apoptosis is an important mechanism for eliminating supernumerary cells during embryonic development, and a potent tumor suppressor mechanism in neonatal and adult organisms. Because apoptosis irreversibly removes cells from tissues, this cell fate decision can also deplete tissues of stem or progenitor cells and contribute to organ degeneration (43).
(iv) Cellular senescence: cellular outcomes following DNA damage are highly dependent on cell type and physiological context (42). While lymphoid lineages usually undergo apoptosis following severe DNA damage (44), stromal and some epithelial lineages undergo senescence (45). Senescence is a complex genetic program and a final cell fate decision that establishes permanent cell growth arrest. The senescence response also causes dramatic alterations in cellular metabolism including the secretion of proteins that can modify the tissue microenvironment (46). In most cell types, activation of p53 is crucial for initiating the senescence response following DNA damage. In some cells, p53 is also important for maintaining the senescence growth arrest. In others, p53 is required only to establish the senescence growth arrest, which subsequently becomes irreversible and p53-independent (47). Senescent cells have been shown to accumulate with age in vivo (10,11,48–50). Because they have lost the ability to proliferate, senescent cells can no longer participate in tissue renewal and repair; thus, senescence can deplete both stem (51–53) and stromal (10,11) cell pools. Moreover, because senescent cells persist, they have the ability to alter the tissue microenvironment, and can therefore also promote the degeneration of organs and stem cell niches (14,46). Finally, senescent cells secrete factors such as matrix metalloproteinase-3 (MMP-3), which favors extra-cellular matrix remodeling, promotes defects in epithelial cell differentiation and stimulates cancer cell growth (46,54,55).
These four cellular outcomes and their potential impact on tissue homeostasis are summarized in Figure 2 and are all orchestrated by the p53 signaling network. There is now little doubt that activation of tumor suppression mechanisms such as apoptosis and senescence suppress the development of cancer and therefore promote longevity. In contrast, loss of p53 leads to a dramatic reduction in DNA damage-induced cell fate decisions (44,56–60) and this deficit shortens life span in mice and humans due to early cancer development (21,22). In light of the cellular outcome model above, the reasons for this are easily understood. Defects in p53-dependent cell fate decisions such as apoptosis and senescence can promote the survival of cells with potentially severe genotoxic damage, leading to excessive mutation accumulation and neoplastic transformation (see Figure 2).
While it is clear that reduced p53 activity shortens life span by promoting cancer, the model above paradoxically predict that loss of p53 should also prevent permanent cell fate decisions such as apoptosis and senescence that promote tissue and organ degeneration (Figure 2). Moreover, if p53 does promote tissue degeneration associated with aging, increased p53 activity should promote aging phenotypes further and do so despite a reduced incidence of cancer. Some of these possibilities can and have been tested in mice.
P53 AND AGING IN MOUSE MODELS
Recent studies indicate that proteins involved in DNA damage repair play an essential role in life span determination (61–64). Several of these studies implicate a role of p53 in establishing senescence and implicate it as a potential regulator of organismal aging (4,6). Although a physiological role for p53 in aging is controversial, studies with different mouse models indicate a delicate balance between the tumor suppressive and age-promoting functions of p53. Here, we discuss several of these mouse models (summarized in Table 1). Note that when referring to altered aging phenotypes in mouse models, in general, premature aging refers to phenotypes that occur very early (weeks) after birth, whereas accelarated aging refers to phenotypes that take longer to develop (months/years), but that would normally take much longer to develop in control animals.
Table 1.
Mouse models that alter p53 activity
| Mouse Model | Mutation type | Aging phenotype | References |
|---|---|---|---|
| Ku80−/− | Ku80 null | Enhanced | (61,66) |
| mTR−/− | Telomerase null | Enhanced | (71–73) |
| Zmpste24−/− | Zmpste24 null | Enhanced | (79) |
| Brca1Δ11/Δ11 | Hypomorphic mutation | Enhanced | (84,85) |
| p53+/m | p53 exon 1-6 plus 23 upstream genes deleted | Enhanced | (4,5) |
| P+/+mice | Expression of p53 isoform p44 | Enhanced | (6) |
| ‘Super–p53’ mouse | Extra copy of p53 gene | Normal | (89–91) |
| Mdm2puro/Δ7–12 | One null allele and another hypomorphic allele | Normal | (95) |
Mouse models affected by p53
Ku80 null mice
Ku80 is a DNA repair protein that associates with Ku70 to comprise the DNA-binding component of DNA-PK (65). Ku80 null mice exhibit accelerated aging phenotypes, including skin atrophy, osteopenia, hepatocellular degeneration and shortened life span (61). The authors speculated that a lack of NHEJ and therefore increased DNA damage load could account for the accelerated aging phenotype. In support of this hypothesis, mouse embryo fibroblasts (MEFs) derived from Ku80−/− mice clearly underwent p53-dependent premature replicative senescence. A requirement for p53 was illustrated by the limited proliferative capacity of Ku80−/− cells compared to Ku80−/− p53+/− fibroblasts (66). Thus, p53 deficiency rescues the premature replicative senescence of Ku80−/− MEFs. If, as suggested, increased DNA damage and increased p53 activity is responsible for the accelerated aging phenotype of Ku80−/− mice, crossing these animals with p53-deficient mice might rescue the accelerated aging. Interestingly, mice deficient in both Ku80 and p53 were much more cancer-prone than mice that were deficient only in p53 (66). The increase in cancer incidence was large enough to obscure any beneficial effect that loss of p53 might have conferred on the accelerated aging observed in Ku80-deficient mice. Clearly, loss of p53 cannot rescue accelerated aging phenotypes if cancer incidence is dramatically increased.
Telomerase-deficient mice
Telomerase is a key enzyme that maintains telomere length in stem cells and other proliferating cells. Defects in telomerase enzyme components lead to telomere dysfunction, resulting in premature aging phenotypes in mice and dyskeratosis congenita, which has aspects of premature aging, in humans (67). Dysfunctional telomeres are recognized by the DNA repair machinery as DNA-DSBs and thus activate the DDR and p53 (68–70). Mice deficient in the telomerase catalytic subunit (Terc) (mTR−/− mice) are prone to telomere erosion in all tissues, and late generation animals have a decreased life span and show decreased body weight, increased skin lesions and delayed wound healing (71,72). Because mice have long telomeres, early generation mTR−/− mice are phenotypically normal and have a normal life span. Importantly, some of the premature aging phenotypes observed in later generation mTR−/− mice could be rescued by p53 deficiency. This result indicates that, at least in a telomerase-deficient background, short telomeres activate p53, which contributes to premature aging (73).
Zmpste24 protease null mice
Zmpste24 is a metalloproteinase that participates in the maturation of lamin A. Lamin A is an essential component of the nuclear envelope, defects in which can cause premature aging phenotypes in humans and mice (74–76). Some humans with progeroid syndromes have been shown to carry mutations in the genes encoding Lamin A as well as ZMPSTE24 (77,78). Not surprisingly, mice deficient in Zmpste24 and lamin A exhibited many features of premature aging (74–76).
Microarray analysis of transcriptional alterations in tissues from Zmpste24−/− mice showed an upregulation of several p53 target genes (79). Although the levels of p53 itself were unchanged, it was proposed that a stress response triggered by nuclear envelope abnormalities in Zmpste24−/− mice activated p53. When compared to wild-type cells, fibroblasts from Zmpste24−/− mice showed a p53-dependent decrease in proliferative capacity and premature cellular senescence. Crossing Zmpste24−/− mice to p53−/− mice further tested the possibility that the progeroid symptoms observed in Zmpste24−/− mice were due to hyperactive p53. Indeed, loss of p53 in the Zmpste24−/− background partially rescued the aging phenotype characteristic of Zmpste24 deficiency, markedly increasing body weight and life span. Thus the premature aging syndrome caused by alterations in nuclear lamina structure triggers activation of p53, possibly through increased DNA damage and DDR signaling (80,81), which contributes to the premature aging of Zmpste24−/− mice.
BRCA1 hypomorphic mutant mice
BRCA1 is a tumor-suppressor protein that acts both as a checkpoint protein and DNA damage repair protein (82). About 98% of embryos carrying a hypomorphic mutation in Brca1 (BRCA1Δ11/Δ11) die between days 12 and 18 of gestation. Importantly, haploid loss of p53 completely suppressed this embryonic lethality, allowing Brca1 Δ11/Δ11 mice to survive to adulthood (83). In those adult animals, Brca1Δ11/Δ11 p53+/− females developed tumors whereas males exhibited accelerated aging phenotypes. These findings suggest that genomic instability and DNA damage due to BRCA1 deficiency lead to a rapid cancer phenotype in females. They also reveal a scenario in which increased DNA damage in cancer spared males may result in a remaining hyperactive p53 allele, which could promote accelerated aging (84). Recent findings show that indeed, the ATM-Chk2-p53 DDR pathway is activated upon BRCA1 deficiency in mice (85). Consistent with an important role for p53 in the DDR, complete or haploid loss of ATM or Chk2 rescued the Brca1 deficiency-associated embryonic lethality and accelerated aging in adult mutant mice. These findings are consistent with a model in which BRCA1 deficiency impairs proper DNA damage repair, which in turn promote p53 hyperactivity through the DDR components ATM and Chk2 (see Figure 1). In cancer spared animals, absence of ATM or Chk2 prevents the hyperactivity of p53 caused by BRCA1 deficiency, hence these mice are rescued from accelerated aging.
Mouse models that affect p53 directly
Short isoforms of p53 in mice: the p53+/m mice
The first evidence for a direct role for p53 in promoting aging phenotypes came from Tyner et al., who generated mice in which a spontaneous recombination event deleted a stretch of DNA upstream of the mouse p53 gene that included exons 1–6 of the p53 coding sequence (4). The mutant p53 allele (m allele) contained exons 7–11 and was presumed to be under the transcriptional control of a promoter from an upstream gene. Experiments with MEFS isolated from p53+/m mice indicated that the m allele product could enhance the stability and the transacivation activity of p53. Consistent with an increase in p53 activity, p53+/m mice were strikingly cancer resistant at 18 months of age compared to similarly aged p53+/+ mice. Surprisingly, however, the cancer resistant p53+/m mice had a 20–30% shorter life span. Moreover, the mutant mice exhibited several signs of accelerated aging including tissue atrophy (in the skin, skeletal muscle, liver and lymphoid organs). Thus, enhanced p53 activity could drive aging phenotypes at the cost of tumor suppression, at least in these mutant mice. The authors suggested that the accelerated aging was caused by an impaired ability of stem cells to produce adequate numbers of progenitor and mature differentiated cells. A recent analysis of hematopoietic stem cell dynamics in p53+/m mice confirmed that, compared to wild-type mice, these mice exhibit a reduced number of proliferating hematopoietic stem cells with age (5). Haploid insufficiency for p53 (p53+/−) partially rescued the reduced hematopoietic stem cell proliferative potential, presumably due to diminished p53 activity.
While the phenotypes of p53+/m mice suggest that increased p53 activity promotes tumor suppression at the cost of aging, there are two major caveats to consider. First, due to the low expression level of the m allele in p53+/m tissues, no m-derived protein could be detected. Second, and more important, the unknown stretch of DNA upstream of the p53 gene that was deleted in the m allele has recently been characterized and shown to include 24 upstream genes in addition to the p53 truncation (86). Absence of one or more of these 24 genes can in principle, give rise to the accelerated aging and cancer resistance of the p53+/m mice.
Short isoforms of p53 in mice: the P+/+ mice
Subsequent studies not only confirmed the idea that p53 status can modulate organismal aging but removed some of the ambiguities present in the above studies (6). Maier et al. created transgenic mice that overexpressed p44 (a short naturally occurring p53 isoform that lacks the main transactivation domain) (P+/+ mice). By 4 months of age, P+/+ mice showed selected signs of premature aging. By 1 year of age, most of the P+/+ mice had died, whereas the non-transgenic mice were still alive and healthy. P+/+ mice had a very low incidence of cancer, suggesting that p44 overexpression increased wild-type p53 activity. Indeed, the overexpressed p44 short isoform modulated p53 functions, resulting in enhanced expression or repression of certain p53 target genes. Moreover, the authors attributed the early premature aging phenotypes of p44-tg mice to p53 hyperactivity, which upregulated the activity of the IGF-signaling pathway. This pathway has been shown to drive aging in species as diverse as nematodes, fruit flies and mice (87,88).
The super-p53 mice
In line with phenotypes of p53+/m and P+/+ mice, Garcia-Cao et al. showed that transgenic mice carrying one or two extra copies of wild-type p53 genes (super-p53 or p53 tg mice) were remarkably cancer resistant. However, and in contrast to p53+/m and P+/+ mice, super-p53 mice showed no symptoms of premature or accelerated aging (89). Notably, cells from super-p53 mice expressed normal basal levels of p53, and displayed normal levels of p53 activity, in the absence of stress. In response to DNA damage, however, cells from super-53 mice displayed enhanced p53 activity. Upon whole body irradiation, super-53 mice showed elevated levels of p21 mRNA in all tissues and increased apoptosis in the thymus, compared to irradiated control mice. Super-p53 mice were also resistant to carcinogen-induced tumors. Despite enhanced p53-dependent apoptosis, super-p53 mice had a normal life span and showed no signs of premature or accelerated aging, as determined by normal hair growth and skin thickness and an absence of lordokyphosis and osteoporosis.
Why do these mice with increased p53 activity not show signs of premature/accelerated aging? The authors speculate that the constitutively high level of p53 activity in p53+/m or P+/+ mice, or a chronic stress signal is required to chronically activate p53 and induce premature/accelerated aging. This hypothesis raises the possibility that enhanced p53-dependent tumor suppression is possible without accelerating aging or increasing non-cancer mortality. Conversely, these results also suggest that super-p53 mice might age prematurely if chronically stressed. However, the accelerated aging shown by telomerase-null mice was not further accelerated when these mice were crossed with super-53 mice (90). The authors observed that cells harboring DNA damage due to defective telomeres were more efficiently eliminated in super-p53 mice as compared to wild-type mice, although p53 did not have any effect on telomere-driven aging. The authors suggested that the decrease in damaged cells was not of a sufficient magnitude to delay the pathologies associated with aging in mice. More recently, it was shown that the double super-p53/p19ARF mice can effectively eliminate cells harboring age-associated persistent DNA damage markers in the absence of any premature or accelerated aging phenotypes. In fact, the super-p53/p19ARF animals were cancer-free, showed increased resistance to oxidative stress and displayed general signs of delayed aging (91). The authors suggested that increased, but properly regulated p53 activity, provide both stress resistance and tumor suppression activity. It remains to be seen if super-p53 or super-p53/p19ARF mice display accelerated aging phenotypes when under other types of chronic stress.
The hypomorphic Mdm2 mice
H/Mdm2 is a powerful inhibitor of p53. Mdm2 binds the transcriptional activation domain of p53 and prevents its interaction with the transcriptional machinery. Mdm2 also facilitates the nuclear export of p53, and ubiquitinates p53 and promotes its degradation by the proteasome (36). Mdm2-deficient mice are embryonic lethal; deletion of p53 rescues this lethality, indicating that very high levels of p53 are incompatible with embryonic development (92,93). Similarly, acute activation of p53 in adult mice lacking Mdm2 but carrying an inducible p53 protein was incompatible with survival (94). Mendrysa et al. generated mice that express low levels of Mdm2 (mdm2 puro/Δ7−12mice) owing to the presence of one hypomorphic (puro) and one null allele (Δ7–12). These mice expressed 30% of the normal level of Mdm2 (95,96). As expected, these mice also showed increased basal levels of p53 and, upon activation, increased expression of p53 target genes as well as increased apoptosis in the small intestine. mdm2 puro/Δ7–12 mice were also highly resistant to cancer, presumably as a consequence of increased p53 activity. Despite having increased p53 activity, these mice had a normal life span, and showed no signs of accelerated aging.
Why do mdm2 puro/Δ7–12 mice escape accelerated aging despite constitutively high p53 activity? One possibility is that the p53 activity was not increased to the same extent in mdm2 puro/Δ7–12 mice as in p53+/m and P+/+ mice. Another possibility is that, in p53+/m and P+/+ mice, overexpression of an N-terminally truncated p53 causes p53 activity that is imbalanced or qualitatively altered, not simply quantitatively altered. Consistent with this idea, N-terminally truncated forms of p53 can alter the affinity of full-length p53 for different promoters (97). Thus, a balanced increase in the level of p53 activity and preservation of p53 specificity might be required to enhance tumor suppression without accelerating aging. As for super-p53 mice, it remains to be seen if mdm2 puro/Δ7–12 mice display accelerated aging phenotypes when under stress.
Aging and human p53 polymorphism at amino acid 72
Taken together, the mouse models support the idea that p53 is a potent tumor suppressor that can, under some circumstances, also promote aging. Insights into the role of p53 in aging are also derived from studies of human populations. Human p53 has a Pro/Arg polymorphism at amino acid residue 72 (98). Humans carrying one or two copies of the Pro or Arg p53 form have been studied for cancer susceptibility, mortality and survival. The results support a role for p53 in regulating life span in humans.
A recent systematic literature survey done by van Heemst et al. showed that humans with the Pro/Pro genotype had a higer risk of developing cancer compared to the Arg/Arg genotype (99). This finding agrees with a previous finding that p53Arg is a more potent inducer of apoptosis than the p53Pro form (100,101). A prospective study done by the same group in 1226 subjects aged 85 or over showed that the Pro/Pro carriers had a 41% increased survival, despite a 2.54-fold increase in cancer mortality, compared to the Arg/Arg carriers (99). The authors suggested that in older survivors, p53Arg protected against cancer more efficiently than p53Pro but at the cost of a diminished life span.
In another instance, Orsted et al. studied a large Danish population of 9219 participants aged 20–95 years for longevity, survival after cancer diagnosis, and risk of cancer (102). The authors found that overall 12-year survival was higher for Pro/Pro carriers compared to Arg/Arg carriers. Moreover, although Pro/Pro carriers had the same cancer risk as Arg/Arg carriers, the survival of Pro/Pro individuals after cancer or a life threatening disease was higher compared to Arg/Arg individuals. The authors suggested that the increased longevity of Pro/Pro individuals after a cancer diagnosis or other life threatening disease might also be due to an increase in survival. Again, carriers of a form of p53 that is a less potent inducer of apoptosis had increased survival after stress, suggesting that high p53 activity could be detrimental in humans. While limited in scope, correlations about this naturally occurring polymorphism and its impact on aging corroborates to some extent the data obtained from mouse models described above.
CONCLUSION: FINDING THE RIGHT BALANCE
The tumor suppressor p53 protects the genome by promoting the repair of potentially carcinogenic lesions in the DNA, thereby preventing mutations. In addition, p53 eliminates or arrests the proliferation of damaged or mutant cells by the processes of apoptosis and cellular senescence. However, there is mounting evidence that apoptosis and cellular senescence can lead to aging phenotypes, possibly by depletion of important stem cell and other stromal cell pools. Thus, integration of DNA damage signaling by p53 has been optimized to balance the beneficial effects of tumor suppression against the detrimental effects of tissue degeneration. Furthur studies will help us understand as to how exactly this balance is achieved and how it also varies among species having different life spans. It remains to be seen whether it will be possible in humans, as it appears to be possible in mice, to optimize tumor suppression without accelerating aging.
ACKNOWLEDGEMENTS
We thank members of our laboratories at the Lawrence Berkeley National Laboratory and Buck Institute for Age Research for ideas and stimulating discussions. Our special thanks to Chris Patil for critical reading of the manuscript and Kevin Peet for technical assistance. This work was supported by the National Institutes of Health, the California and Department of Defense Breast Cancer Research Programs, the Ellison Medical Foundation, the Department of Energy and the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. Funding to pay the Open Access publication charges for this article was provided by a NIH grant- contract number LB05-000446.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J. Cancer and ageing: rival demons? Nat. Rev. 2003;3:339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- 3.Vijg J., Suh Y. Genetics of longevity and aging. Annu. Rev. Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- 4.Tyner S.D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 5.Dumble M., Moore L., Chambers S.M., Geiger H., Van Zant G., Goodell M.A., Donehower L.A. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109:1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier B., Gluba W., Bernier B., Turner T., Mohammad K., Guise T., Sutherland A., Thorner M., Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balducci L., Ershler W.B. Cancer and ageing: a nexus at several levels. Nat. Rev. 2005;5:655–662. doi: 10.1038/nrc1675. [DOI] [PubMed] [Google Scholar]
- 8.Gessert C.E., Elliott B.A., Haller I.V. Dying of old age: an examination of death certificates of Minnesota centenarians. J. Am. Geriatr. Soc. 2002;50:1561–1565. doi: 10.1046/j.1532-5415.2002.50413.x. [DOI] [PubMed] [Google Scholar]
- 9.Sedelnikova O.A., Horikawa I., Zimonjic D.B., Popescu N.C., Bonner W.M., Barrett J.C. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat. Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 10.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J.M. Cellular senescence in aging primates. Science (New York, N.Y.) 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 11.Jeyapalan J.C., Ferreira M., Sedivy J.M., Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijg J., Dolle M.E. Large genome rearrangements as a primary cause of aging. Mech. Ageing Dev. 2002;123:907–915. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- 13.Bahar R., Hartmann C.H., Rodriguez K.A., Denny A.D., Busuttil R.A., Dolle M.E., Calder R.B., Chisholm G.B., Pollock B.H., et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 14.Carlson M.E., Conboy I.M. Loss of stem cell regenerative capacity within aged niches. Aging Cell. 2007;6:371–382. doi: 10.1111/j.1474-9726.2007.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I.L. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 16.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science (New York, N.Y.) 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 17.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 18.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balaban R.S., Nemoto S., Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Bokov A., Chaudhuri A., Richardson A. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Malkin D., Li F.P., Strong L.C., Fraumeni J.F., Jr, Nelson C.E., Kim D.H., Kassel J., Gryka M.A., Bischoff F.Z., et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science (New York, N.Y.) 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 22.Donehower L.A., Harvey M., Slagle B.L., McArthur M.J., Montgomery C.A., Jr, Butel J.S., Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 23.Wu C., Miloslavskaya I., Demontis S., Maestro R., Galaktionov K. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature. 2004;432:640–645. doi: 10.1038/nature03173. [DOI] [PubMed] [Google Scholar]
- 24.Mallette F.A., Gaumont-Leclerc M.F., Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre M., Nuciforo P.G., et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 26.Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 27.Green D.R., Chipuk J.E. p53 and metabolism: inside the TIGAR. Cell. 2006;126:30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- 28.Bakkenist C.J., Kastan M.B. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 29.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science (New York, N.Y.) 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 30.Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marini F., Nardo T., Giannattasio M., Minuzzo M., Stefanini M., Plevani P., Falconi M.M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl Acad. Sci. USA. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang G., Sancar A. Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell. Biol. 2006;26:39–49. doi: 10.1128/MCB.26.1.39-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassing C.H., Alt F.W. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Pluquet O., Hainaut P. Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett. 2001;174:1–15. doi: 10.1016/s0304-3835(01)00698-x. [DOI] [PubMed] [Google Scholar]
- 35.Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., et al. Genomic instability in mice lacking histone H2AX. Science (New York, N.Y.) 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavin M.F., Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 37.Marchenko N.D., Zaika A., Moll U.M. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 38.Murphy M.E., Leu J.I., George D.L. p53 moves to mitochondria: a turn on the path to apoptosis. Cell cycle (Georgetown, Tex.) 2004;3:836–839. [PubMed] [Google Scholar]
- 39.Al Rashid S.T., Dellaire G., Cuddihy A., Jalali F., Vaid M., Coackley C., Folkard M., Xu Y., Chen B.P., et al. Evidence for the direct binding of phosphorylated p53 to sites of DNA breaks in vivo. Cancer Res. 2005;65:10810–10821. doi: 10.1158/0008-5472.CAN-05-0729. [DOI] [PubMed] [Google Scholar]
- 40.Ho J., Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 41.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 42.Guillouf C., Grana X., Selvakumaran M., De Luca A., Giordano A., Hoffman B., Liebermann D.A. Dissection of the genetic programs of p53-mediated G1 growth arrest and apoptosis: blocking p53-induced apoptosis unmasks G1 arrest. Blood. 1995;85:2691–2698. [PubMed] [Google Scholar]
- 43.Danial N.N., Korsmeyer S.J. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 44.Lowe S.W., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 45.Di Leonardo A., Linke S.P., Clarkin K., Wahl G.M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 46.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Beausejour C.M., Krtolica A., Galimi F., Narita M., Lowe S.W., Yaswen P., Campisi J. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kishi S. Functional aging and gradual senescence in zebrafish. Ann. NY Acad. Sci. 2004;1019:521–526. doi: 10.1196/annals.1297.097. [DOI] [PubMed] [Google Scholar]
- 49.Melk A., Kittikowit W., Sandhu I., Halloran K.M., Grimm P., Schmidt B.M., Halloran P.F. Cell senescence in rat kidneys in vivo increases with growth and age despite lack of telomere shortening. Kidney Int. 2003;63:2134–2143. doi: 10.1046/j.1523-1755.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 50.Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Schulte B.A., LaRue A.C., Ogawa M., Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janzen V., Forkert R., Fleming H.E., Saito Y., Waring M.T., Dombkowski D.M., Cheng T., DePinho R.A., Sharpless N.E., et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 53.Molofsky A.V., Slutsky S.G., Joseph N.M., He S., Pardal R., Krishnamurthy J., Sharpless N.E., Morrison S.J. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parrinello S., Coppe J.P., Krtolica A., Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J. Cell Sci. 2005;118:485–496. doi: 10.1242/jcs.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D., Hornsby P.J. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- 56.Lowe S.W., Ruley H.E., Jacks T., Housman D.E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 57.Lotem J., Sachs L. Hematopoietic cells from mice deficient in wild-type p53 are more resistant to induction of apoptosis by some agents. Blood. 1993;82:1092–1096. [PubMed] [Google Scholar]
- 58.Christophorou M.A., Martin-Zanca D., Soucek L., Lawlor E.R., Brown-Swigart L., Verschuren E.W., Evan G.I. Temporal dissection of p53 function in vitro and in vivo. Nat. Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- 59.Christophorou M.A., Ringshausen I., Finch A.J., Swigart L.B., Evan G.I. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- 60.Komarova E.A., Kondratov R.V., Wang K., Christov K., Golovkina T.V., Goldblum J.R., Gudkov A.V. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23:3265–3271. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- 61.Vogel H., Lim D.S., Karsenty G., Finegold M., Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DePinho R.A. The age of cancer. Nature. 2000;408:248–254. doi: 10.1038/35041694. [DOI] [PubMed] [Google Scholar]
- 63.Kuro-o M. Disease model: human aging. Trends Mol. Med. 2001;7:179–181. doi: 10.1016/s1471-4914(01)01921-9. [DOI] [PubMed] [Google Scholar]
- 64.de Boer J., Andressoo J.O., de Wit J., Huijmans J., Beems R.B., van Steeg H., Weeda G., van der Horst G.T., van Leeuwen W., et al. Premature aging in mice deficient in DNA repair and transcription. Science (New York, N.Y.) 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 65.Chechlacz M., Vemuri M.C., Naegele J.R. Role of DNA-dependent protein kinase in neuronal survival. J. Neurochem. 2001;78:141–154. doi: 10.1046/j.1471-4159.2001.00380.x. [DOI] [PubMed] [Google Scholar]
- 66.Lim D.S., Vogel H., Willerford D.M., Sands A.T., Platt K.A., Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marrone A., Walne A., Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr. Opin. Genet. Dev. 2005;15:249–257. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Rodier F., Kim S.H., Nijjar T., Yaswen P., Campisi J. Cancer and aging: the importance of telomeres in genome maintenance. Int J. Biochem. Cell Boil. 2005;37:977–990. doi: 10.1016/j.biocel.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 69.d'Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 70.von Zglinicki T., Saretzki G., Ladhoff J., d'Adda di Fagagna F., Jackson S.P. Human cell senescence as a DNA damage response. Mech. Ageing Dev. 2005;126:111–117. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 71.Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 72.Rudolph K.L., Chang S., Lee H.W., Blasco M., Gottlieb G.J., Greider C., DePinho R.A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 73.Chin L., Artandi S.E., Shen Q., Tam A., Lee S.L., Gottlieb G.J., Greider C.W., DePinho R.A. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 74.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell. Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pendas A.M., Zhou Z., Cadinanos J., Freije J.M., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodriguez F., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 76.Bergo M.O., Gavino B., Ross J., Schmidt W.K., Hong C., Kendall L.V., Mohr A., Meta M., Genant H., et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl Acad. Sci. USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen L., Lee L., Kudlow B.A., Dos Santos H.G., Sletvold O., Shafeghati Y., Botha E.G., Garg A., Hanson N.B., et al. LMNA mutations in atypical Werner's syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 78.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varela I., Cadinanos J., Pendas A.M., Gutierrez-Fernandez A., Folgueras A.R., Sanchez L.M., Zhou Z., Rodriguez F.J., Stewart C.L., et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 80.Scaffidi P., Misteli T. Lamin A-dependent nuclear defects in human aging. Science (New York, N.Y.) 2006;312:1059–1063. [Google Scholar]
- 81.Liu B., Wang J., Chan K.M., Tjia W.M., Deng W., Guan X., Huang J.D., Li K.M., Chau P.Y., et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J., Powell S.N. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol. Cancer Res. 2005;3:531–539. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 83.Xu X., Qiao W., Linke S.P., Cao L., Li W.M., Furth P.A., Harris C.C., Deng C.X. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 84.Cao L., Li W., Kim S., Brodie S.G., Deng C.X. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao L., Kim S., Xiao C., Wang R.H., Coumoul X., Wang X., Li W.M., Xu X.L., De Soto J.A., et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. EMBO J. 2006;25:2167–2177. doi: 10.1038/sj.emboj.7601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gentry A., Venkatachalam S. Complicating the role of p53 in aging. Aging Cell. 2005;4:157–160. doi: 10.1111/j.1474-9726.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- 87.Guarente L., Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 88.Bluher M., Kahn B.B., Kahn C.R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science (New York, N.Y.) 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 89.Garcia-Cao I., Garcia-Cao M., Martin-Caballero J., Criado L.M., Klatt P., Flores J.M., Weill J.C., Blasco M.A., Serrano M. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia-Cao I., Garcia-Cao M., Tomas-Loba A., Martin-Caballero J., Flores J.M., Klatt P., Blasco M.A., Serrano M. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 2006;7:546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matheu A., Maraver A., Klatt P., Flores I., Garcia-Cao I., Borras C., Flores J.M., Vina J., Blasco M.A., et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 92.Jones S.N., Roe A.E., Donehower L.A., Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 93.Montes de Oca Luna R., Wagner D.S., Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 94.Ringshausen I., O'Shea C.C., Finch A.J., Swigart L.B., Evan G.I. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 95.Mendrysa S.M., O'Leary K.A., McElwee M.K., Michalowski J., Eisenman R.N., Powell D.A., Perry M.E. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mendrysa S.M., Perry M.E. Tumor suppression by p53 without accelerated aging: just enough of a good thing? Cell cycle (Georgetown, Tex.) 2006;5:714–717. doi: 10.4161/cc.5.7.2632. [DOI] [PubMed] [Google Scholar]
- 97.Bourdon J.C., Fernandes K., Murray-Zmijewski F., Liu G., Diot A., Xirodimas D.P., Saville M.K., Lane D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matlashewski G.J., Tuck S., Pim D., Lamb P., Schneider J., Crawford L.V. Primary structure polymorphism at amino acid residue 72 of human p53. Mol. Cell. Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bonafe M., Salvioli S., Barbi C., Trapassi C., Tocco F., Storci G., Invidia L., Vannini I., Rossi M., et al. The different apoptotic potential of the p53 codon 72 alleles increases with age and modulates in vivo ischaemia-induced cell death. Cell Death Differ. 2004;11:962–973. doi: 10.1038/sj.cdd.4401415. [DOI] [PubMed] [Google Scholar]
- 100.Dumont P., Leu J.I., Della Pietra A.C., III, George D.L., Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 101.van Heemst D., Mooijaart S.P., Beekman M., Schreuder J., de Craen A.J., Brandt B.W., Slagboom P.E., Westendorp R.G. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp. Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 102.Orsted D.D., Bojesen S.E., Tybjaerg-Hansen A., Nordestgaard B.G. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J. Exp. Med. 2007;204:1295–1301. doi: 10.1084/jem.20062476. [DOI] [PMC free article] [PubMed] [Google Scholar]