Abstract
Genome integrity is constantly threatened by DNA lesions arising from numerous exogenous and endogenous sources. Survival depends on immediate recognition of these lesions and rapid recruitment of repair factors. Using laser microirradiation and live cell microscopy we found that the DNA-damage dependent poly(ADP-ribose) polymerases (PARP) PARP-1 and PARP-2 are recruited to DNA damage sites, however, with different kinetics and roles. With specific PARP inhibitors and mutations, we could show that the initial recruitment of PARP-1 is mediated by the DNA-binding domain. PARP-1 activation and localized poly(ADP-ribose) synthesis then generates binding sites for a second wave of PARP-1 recruitment and for the rapid accumulation of the loading platform XRCC1 at repair sites. Further PARP-1 poly(ADP-ribosyl)ation eventually initiates the release of PARP-1. We conclude that feedback regulated recruitment of PARP-1 and concomitant local poly(ADP-ribosyl)ation at DNA lesions amplifies a signal for rapid recruitment of repair factors enabling efficient restoration of genome integrity.
INTRODUCTION
Genomic DNA is under constant surveillance and protection from mutagenic or clastogenic insults, which can result from environmental or endogenous threats such as ionizing radiation, genotoxic chemicals and free radicals. Specific proteins inspect the DNA for the presence of particular lesions such as base or nucleotide damage, single- or double-strand breaks and if necessary trigger appropriate repair mechanisms (1).
A growing number of proteins are known to be involved in these pathways enabling damage recognition, signaling of the damage, recruitment of other repair factors and finally restoration of the genetic and epigenetic information. A central surveillance factor, which is believed to play an important role in damage recognition and signaling is the poly(ADP-ribose) polymerase-1 (PARP-1). PARP-1 is the founding member of the PARP family encompassing 17 members involved in various biological processes such as DNA repair, transcription, mitotic segregation, telomere homeostasis and cell death (2). PARP-1 is a molecular sensor of single-strand DNA breaks (SSB) generated directly or resulting from the processing of damaged bases by the SSBR/BER pathway. The two C-X2-C-X28,30-H-X2-C zinc fingers of PARP-1 were shown to bind single-strand breaks in vitro and define a novel DNA interruptions binding module, present also in the SSBR/BER factor DNA ligase III (3,4). Upon binding to its DNA target, PARP-1 catalyzes the polymerization of ADP-ribose moieties from NAD+ on target proteins, a post-translational modification called poly(ADP-ribosyl)ation. Major targets of poly(ADP-ribose) (PAR) are PARP-1 itself and histones, mainly H1, leading to chromatin relaxation. In addition, PAR likely serves as a recruiting molecule, since several proteins were reported to interact with PAR or poly(ADP-ribosyl)ated PARP-1 (5). XRCC1, the non-enzymatic scaffold protein of SSBR/BER that interacts with and stimulates most of the SSBR/BER enzymes (6) was shown to interact preferentially with poly(ADP-ribosyl)ated PARP-1 (7). Recent studies demonstrated that XRCC1 is recruited to local damaged sites through a PAR- and PARP-1 dependent manner (8–10). However, the involvement of PARP-1 in DNA repair has been questioned by a study showing that BER is efficient in cells lacking PARP-1 (11).
One additional PARP, PARP-2 has been implicated in the cellular response to DNA damage (12,13). PARP-1 and PARP-2 deficient cellular and animal models indicated redundant but also complementary functions of the two enzymes in the surveillance and maintenance of genome integrity (14,15). PARP-1 and PARP-2 knock out mice are sensitive to ionizing radiation and alkylating agents (14,16–18), and embryonic fibroblasts derived from both genotypes showed a comparable delay in the repair of alkylated DNA (15,19). Yet, a recent report using siRNA suggested that PARP-2 depletion has only a minor impact on global SSBR rates (20).
Biochemical studies revealed that PARP-2, like PARP-1, interacts with the SSBR/BER repair factors XRCC1, DNA polymerase β and DNA ligase III (12,15). However, whether PARP-2 acts in a similar way as PARP-1 is still under debate. PARP-1 and PARP-2 can heterodimerize, but they recognize different targets within DNA (15). PARP-2 does not recognize SSBs, but gaps or flap structures which indicates that PARP-2 is probably involved in the later steps of the repair process (13).
As most data on the role and regulation of PARP-1 and PARP-2 are derived from biochemical experiments we systematically investigated the kinetics, role and interplay of PARP-1 and PARP-2 in living cells. With microirradiation and live cell microscopy we could show that both PARPs are recruited to DNA damage sites however with different kinetics and roles. Our data indicate that the initial step of the damage response is mediated by a feedback regulated accumulation of PARP-1 and concomitant local poly(ADP-ribosyl)ation leading to a rapid recruitment of repair factors.
MATERIALS AND METHODS
Cell culture and transfection
Hela cells stably expressing GFP-PARP-1 were generated by transfection of pEGFP-C3-hPARP-1 vector and selection of resistant clones with G418 (0.5 μg/ml). The activity of the recombinant fusion protein was verified by activity blot according to Dantzer et al. (21). Wild type, PARP-1 and PARP-2 deficient MEF cells were previously described (15,19). All cell lines were cultured in DMEM containing 50 μg/ml gentamicin supplemented with 10% FCS. Cells grown on μ-slides (Ibidi) or on gridded coverslips were cotransfected with jetPEI (PolyPlus Transfection) according to the manufacturer's instructions. For microirradiation experiments cells were either sensitized by incubation in medium containing BrdU (10 μg/ml) for 24–48 h, or incubated with Hoechst 33285 (10 μg/ml) for 10 min. NU1025 (Sigma) was added to the medium at least 1 h before microirradiation experiments in a final concentration of 200 μM.
Expression plasmids
Mammalian expression constructs encoding full length or truncated translational fusions of human PARP-2 were previously described (22). The GFP-PARP-1 expression vector was described in Maeda et al. (23). Mammalian expression constructs encoding truncated forms of human PARP-1 were generated by subcloning into the PstI site of pEGFP-C3 (Clontech). PstI/PstI fragments were isolated from the following pTG plasmids previously described: PARP-1C21G,C125G (4), PARP-1E988 (24), and PARP-11–373 (25). The GFP-XRCC1 expression construct was generated by subcloning the EcoRI/EcoRI fragment from pCD2E-XRCC1 into the EcoRI site of pEGFP-C2. A red variant of XRCC1 was generated by replacing GFP with RFP (26). In all cases expression was under the control of the CMV promoter. We tested all fusion proteins by expression in 293T cells followed by western blot analysis.
Immunofluorescence and detergent extraction
Cells were fixed in 3.7% formaldehyde for 10 min and permeabilized with ice-cold methanol for 5 min. The following primary antibodies (diluted in PBS containing 2% BSA) were used: anti-PAR (Trevigen) and anti-PARP-1 (C2-10) mouse monoclonal antibodies, and anti-PARP-2 rabbit polyclonal antibody (Yuc, Alexis). Primary antibodies were detected using secondary antibodies (diluted 1:400 in PBS containing 2% BSA) conjugated to Alexa Fluor 488, 555 or 647 (molecular probes). Cells were counterstained with DAPI and mounted in Vectashield (Vector Laboratories).
Live-cell microscopy, microirradiation and photobleaching experiments
Live cell imaging, microrirradiation and photobleaching experiments were carried out with a Leica TCS SP5/AOBS confocal laser scanning microscope equipped with a UV-transmitting HCX PL 63×/1.4 oil objective. Fluorophores were excited using a 488 nm Ar-laser line and a 561 nm DPSS laser line. The microscope was equipped with a heated environmental chamber set to 37°C. Confocal image series were typically recorded with a frame size of 256 × 256 pixels and a pixel size of 90 nm.
Microirradiation was carried out with a 405 nm diode laser set to 50% transmission. Preselected spots of ∼1 μm in diameter within the nucleus were microirradiated for 1 s. Before and after microirradiation confocal image series of one mid z-section were recorded at 2 s time interval (typically six preirradiation and 150 post-irradiation frames). For evaluation of the recruitment kinetics, fluorescence intensities of the irradiated region were corrected for background and for total nuclear loss of fluorescence over the time course and normalized to the preirradiation value. Data from microirradiation of individual cells obtained in at least two independent experiments performed on different days were averaged for evaluation and plotting of corresponding graphs.
For FRAP analysis, a region of interest was selected and photobleached for 300 ms with all laser lines of the Ar-laser and the 561 nm DPSS laser set to maximum power at 100% transmission. Before and after bleaching, confocal image series were recorded at 150 ms time intervals (typically 10 prebleach and 200 post-bleach frames). Mean fluorescence intensities of the bleached region were corrected for background and for total-nuclear loss of fluorescence over the time course and normalized to the mean of the last four prebleach values.
For the quantitative evaluation of microirradiation and photobleaching experiments, data of at least nine nuclei were averaged and the mean curve as well as the standard error of the mean calculated and displayed using Microsoft Excel software. The half-time of recovery was calculated from the average curves.
Images of fixed cells were taken with a Zeiss Axiophot 2 widefield epifluorescence microscope using a Zeiss Plan-Apochromat 63x/1.40 oil objective and a cooled CCD camera (Visitron Systems).
RESULTS
PARP-1 is recruited to DNA damage sites
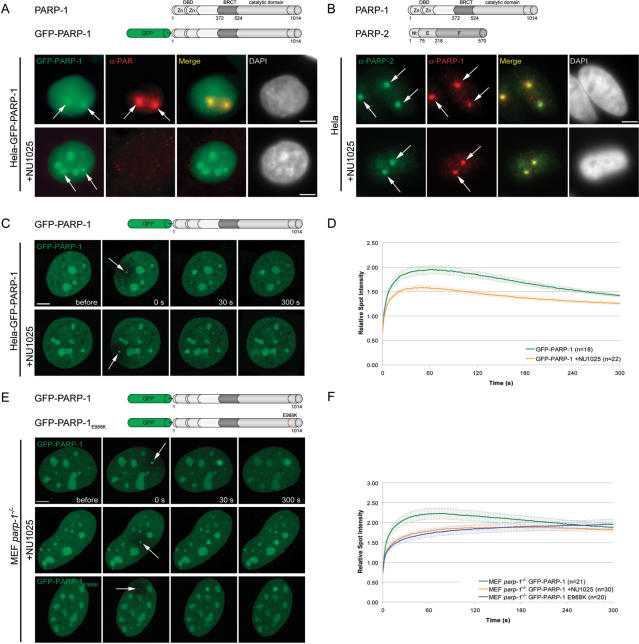
Various biochemical studies and knock out experiments have clearly shown the involvement of PARP-1 in DNA repair (2). However, whether and how PARP-1 is recruited to sites of DNA damage is still an open question. To investigate the dynamics of PARP-1 recruitment to DNA damage sites in living cells we generated DNA lesions at preselected subnuclear sites with a long wavelength UV diode laser in BrdU-sensitized cells, as described before (27,28). Immunofluorescence stainings with specific antibodies revealed that endogenous PARP-1 is recruited to microirradiated sites in Hela and MEF cells (Figure 1B and data not shown). When transiently or stably transfected in MEFs or Hela cells, GFP-PARP-1 was distributed throughout the nucleus and accumulated in nucleoli as previously described (22). For in vivo studies we determined the recruitment kinetics of PARP-1 in living cells by quantifying the amount of GFP-tagged PARP-1 accumulated at microirradiated sites. We observed a rapid accumulation of GFP-PARP-1 at DNA damage sites immediately after microirradiation (Figure 1C and D). Accumulation of PARP-1 at DNA damage sites was rather transient, as the fluorescence intensity gradually declined after reaching a maximum about 1 min after microirradiation (Figure 1C and D). Interestingly, we observed a similar fast recruitment of GFP-PARP-1 in cells undergoing mitosis (Supplementary Figure 1). To test whether PARP-1 recruitment is accompanied by poly(ADP-ribosyl)ation at microirradiated sites we performed immunostainings with specific antibodies against PAR. We found a strong PAR signal clearly colocalizing with GFP-PARP-1 at microirradiated sites (Figure 1A). Taken together, our results show a rapid but transient accumulation of PARP-1 at DNA damage sites colocalizing with sites of poly(ADP-ribosyl)ation.
Figure 1.
Recruitment of PARP-1 to DNA damage sites. (A) Immunostaining of PAR after microirradiation of Hela cells stably transfected with GFP-PARP-1. GFP-PARP-1 clearly colocalizes with PAR at microirradiated sites. Treatment of Hela GFP-PARP-1 cells with the PARP-1 inhibitor NU1025 results in loss of PAR signals at microirradiated sites, while GFP-PARP-1 accumulation is still present. (B) Immunostaining of PARP-1 and PARP-2 after microirradiation of Hela cells in the absence or presence of NU1025. (C) Live cell imaging of microirradiated Hela cells stably expressing GFP-PARP-1. Accumulation of GFP-PARP-1 can be observed immediately after microirradiation in untreated cells as well as in cells treated with the PARP inhibitor NU1025. (D) Quantitative evaluation of PARP-1 recruitment kinetics in the absence and presence of the PARP inhibitor NU1025. Inhibition of PARP activity does not prevent recruitment of PARP-1 but leads to a reduced accumulation at microirradiated sites. (E and F) Live cell imaging and quantitative evaluation of PARP-1 recruitment kinetics in the absence and presence of the PARP inhibitor NU1025 compared with the recruitment kinetics of the fluorescence tagged catalytic mutant PARP-1 after microirradiation of PARP-1 knock out cells. Error bars represent the SEM. Scale bar, 5 μm.
PARP activity enhances the recruitment of PARP-1 to DNA damage sites
It has previously been shown that PARP activity is required for the recruitment of the repair factor XRCC1 to DNA lesions (8–10). To address the question whether PARP activity has an effect on its own recruitment we tested the recruitment of GFP-PARP-1 in the presence of the PARP inhibitor NU1025. As expected, treatment with NU1025 efficiently inhibited poly(ADP-ribosyl)ation as no PAR signal could be detected after microirradiation of treated cells (Figure 1A). Interestingly, accumulation of endogenous and GFP-tagged PARP-1 at laser-induced DNA damage sites seemed not to be affected by this treatment (Figure 1A and B). Quantitative evaluation of live cell experiments, however, revealed that inhibition of PARP activity lead to a reduced recruitment efficiency in Hela cells (Figure 1C and D).
We then examined the recruitment of GFP-PARP-1 in MEFs lacking PARP-1. Whereas GFP-PARP-1 was efficiently but transiently recruited, similarly to what was observed in Hela cells, treatment of these parp-1−/− cells with NU1025 lead to a delayed and prolonged accumulation of GFP-PARP-1 (Figure 1E and F).
To further test the influence of the catalytic activity on the recruitment of PARP-1, we generated a catalytic mutant by replacing the central glutamic acid at aa position 988 by lysine (GFP-PARP-1E988K). This mutation, affecting the PAR chain elongation, converts PARP-1 into a mono-ADP-ribosyl-transferase (24). The inability of GFP-PARP-1E988K to synthesize PAR was verified by activity blot (data not shown). To circumvent side effects arising from endogenous PARP-1 dimerizing with the fusion protein, we performed the microirradiation experiments in parp-1−/− MEFs. The PARP-1E988K fusion protein showed a delayed accumulation and longer persistence at DNA damage sites in comparison to the wild-type protein (Figure 1E and F) which is in agreement with our data obtained from parp-1−/− MEFs treated with NU1025. Altogether, these results indicate that PARP activity is not essential for the initial recruitment of PARP-1 to DNA damage sites, but clearly enhances the recruitment efficiency.
Recruitment of PARP-1 to DNA damage sites is mediated by the DNA-binding domain and the BRCT domain
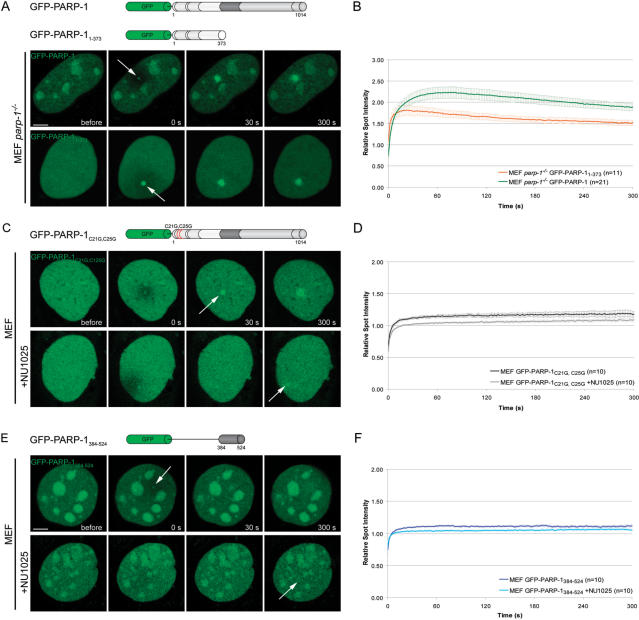
Having shown that PARP-1 accumulates at DNA damage sites, we determined which domain of PARP-1 mediates this recruitment in vivo. First we tested whether the two zinc finger containing DNA-binding domain of PARP-1 [DBD, residues 1–373, (4)] was sufficient for the recruitment to laser-induced DNA damage sites. We observed recruitment of GFP-PARP-11–373 in both parp-1−/− (Figure 2) and Hela cells (data not shown). A direct comparison of the recruitment kinetics of the DBD and the full-length PARP-1 revealed a fast but less efficient recruitment of the DNA binding domain (Figure 2A and B). Using half-nucleus FRAP experiments, we found that the initial, very fast, recruitment of the DBD is supported by an overall higher mobility of the isolated DBD (t1/2 = 3.75 s) in the nucleus compared to the full-length PARP-1 (t1/2 = 7.20 s) and PARP-1E988 (t1/2 = 7.25 s) harboring all interaction domains (Supplementary Figure 2).
Figure 2.
Mechanism of PARP-1 recruitment to DNA damage sites. (A) Live cell imaging of microirradiated PARP-1 knock out MEFs (MEF parp-1−/−) expressing either GFP-PARP-1 or the GFP-tagged DNA binding domain of PARP-1 (GFP-PARP-11–373). Accumulation of both, GFP-PARP-1 and GFP-PARP-11–373 can be observed immediately after microirradiation. (B) Quantitative evaluation of GFP-PARP-11–373 recruitment kinetics. For comparision, the recruitment kinetics of GFP-PARP-1 from Figure 1F are displayed. Time-matched controls are shown in Supplementary Figure 3. (C) Live cell imaging of microirradiated MEFs expressing a PARP-1 fusion protein containing two point mutations affecting the DNA binding capacities of PARP-1 (GFP-PARP-1C21G,C25G) in the absence or presence of the PARP inhibitor NU1025. (D) Quantitative evaluation of recruitment kinetics. (E) Live cell imaging of microirradiated MEFs expressing the GFP-tagged BRCT domain of PARP-1 (GFP-PARP-1384–524) in the absence or presence of the PARP inhibitor NU1025. (F) Quantitative evaluation of recruitment kinetics. Error bars represent the SEM. Scale bar, 5 μm.
The reduced and transient accumulation of the DBD suggests that another part of the protein could enhance the recruitment of PARP-1. To further test this hypothesis we mutated key residues within the DBD known to be essential for DNA binding, in the context of the full-length PARP-1. The C21G and C125G mutations target cysteine residues involved in zinc binding and abolish the binding to DNA (4). These mutations lead to a dramatically reduced, but still detectable recruitment of GFP-PARP-1C21G,C125G to DNA damage sites (Figure 2C and D). Interestingly, treatment with the PARP inhibitor NU1025 affected the recruitment of GFP-PARP-1C21G,C125G. (Figure 2C–F), indicating that PAR molecules synthesized at the damaged site by local PARP-1 are involved in this second wave of DBD-independent recruitment of PARP-1. Furthermore, we found that the BRCT domain alone (residues 384–524), which is involved in PARP-1 homodimerization (15) and PAR binding (data not shown), showed a weak accumulation at laser-induced DNA damage sites which was reduced in the presence of NU1025 (Figure 2E and F). Taken together, our results indicate that the DBD of PARP-1 is necessary and sufficient for recruitment of PARP-1 to DNA lesions. The catalytic activity of PARP-1 likely enhances the recruitment efficiency by locally generating PAR polymers, which are then recognized by the BRCT domain, recruiting more PARP-1 molecules.
The enzymatic activity is required for dissociation of PARP-1 from DNA damage sites
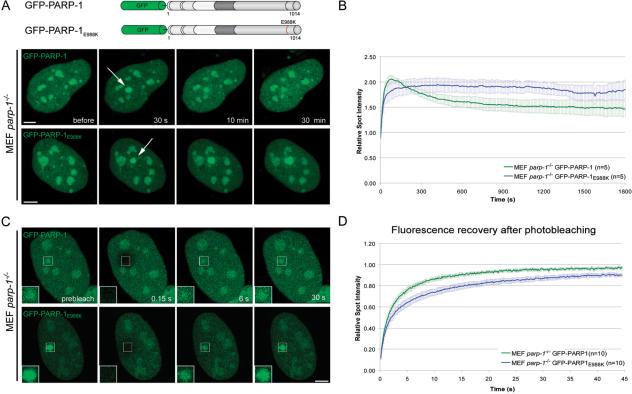
The longer persistence of the catalytic PARP-1 mutant at DNA damage sites (Figure 1E and F) was rather unexpected and led us to study this effect in more detail. We performed long-term live cell observations of microirradiated parp-1−/− MEFs transiently transfected with either GFP-PARP-1 or GFP-PARP-1E988K. In contrast to the very fast accumulation reaching a maximum about 1 min after microirradiation followed by the dissociation of PARP-1, GFP-PARP-1E988K showed a delayed accumulation and persisted at DNA repair sites during the observation period of 30 min (Figure 3A and B).
Figure 3.
The catalytic activity of PARP-1 is needed for dissociation from DNA damage sites. (A) Long-term observations of microirradiated PARP-1 knock out MEFs (MEF parp-1−/−) expressing either GFP-PARP-1 or a GFP-tagged catalytic mutant (GFP-PARP-1E988K). The catalytic mutant shows a prolonged association at DNA damage sites. (B) Quantitative evaluation of recruitment kinetics. (C) Mobility of GFP-PARP-1 and GFP-PARP-1E988K at DNA damage sites. The mobility of accumulated fluorescent fusion proteins was determined by bleaching the microirradiated site 5 min after microirradiation and subsequent recovery measurements. Inset shows the bleached microirradiated site. (D) FRAP data from 10 individual experiments are shown as mean curves. Error bars represent the SEM. Scale bar, 5 μm.
To analyze the mechanisms underlying these kinetic differences, we performed FRAP analysis. The irradiated region was bleached with a high-energy laser pulse 5 min after microirradiation and the fluorescence recovery was determined for GFP-PARP-1 and GFP-PARP-1E988K. We found a slower fluorescence recovery of GFP-PARP-1E988K (t1/2 = 2.25 s) in comparison to GFP-PARP-1 (t1/2 = 1.80 s), indicating a stronger binding of the catalytic mutant at DNA damage sites (Figure 3C and D). These results show that the catalytic activity of PARP-1 is not only needed for efficient targeting to but also for dissociation from DNA damage sites.
PARP-2 is recruited to DNA damage sites later than PARP-1
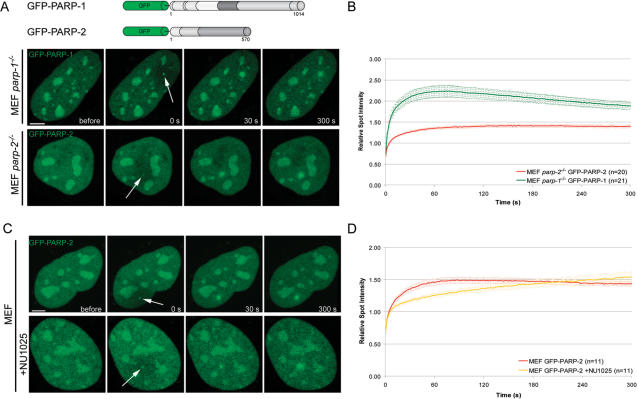
Besides PARP-1, PARP-2 is the only DNA-damage dependent PARP identified so far (12). PARP-2 is required for efficient single-strand break repair like PARP-1 (15), but its function(s) in the repair process are still largely unknown (2). When transiently expressed in MEFs or Hela cells, GFP-PARP-2 distributes throughout the nucleus and accumulates within the nucleoli, as previously described (22). Microirradiation of MEFs and Hela cells lead to the recruitment of GFP-PARP-2 to DNA damage sites. However, in comparison to PARP-1, PARP-2 was recruited slower but persisted longer at DNA repair sites (Figure 4A and B and Supplementary Figure 4). In addition, we could demonstrate recruitment of endogenous PARP-2 to laser-induced DNA damage sites (Figure 1B).
Figure 4.
Recruitment of PARP-2 to DNA damage sites in living cells. (A) Live cell imaging of microirradiated MEFs either expressing GFP-PARP-1 or GFP-PARP-2. Accumulation of GFP-PARP-1 and GFP-PARP-2 can be observed immediately after microirradiation. (B) Quantitative evaluation of GFP-PARP-2 recruitment kinetics. For comparision, the recruitment kinetics of GFP-PARP-1 from Figure 1F are displayed. Time-matched controls are shown in Supplementary Figure 3. (C and D) Live cell imaging of microirradiated MEFs reveals a slower accumulation of GFP-PARP-2 in the presence of NU1025. Error bars represent the SEM. Scale bar, 5 μm.
We next analyzed whether recruitment of PARP-2 depends on PARP activity or the presence of PARP-1. We found that recruitment of PARP-2 to DNA repair sites was less efficient in cells treated with NU1025 as well as in parp-1−/− cells, (Figure 4C and D and Supplementary Figure 4). Altogether, these results indicate that PARP-1 and PARP-2 show distinct recruitment and dissociation kinetics at DNA repair sites and that poly(ADP-ribosyl)ation enhances the recruitment efficiency of both.
The nucleolus is a storage of PARP-1 and PARP-2 for heavy DNA damage
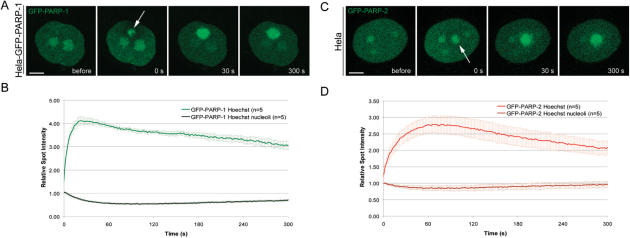
In the course of this study, we observed that microirradiation in the presence of the photosensitizer Hoechst leads to more DNA damage than sensitization with BrdU, which is likely due to more efficient absorption of the energy of the 405 nm laser. We therefore used Hoechst to determine the kinetics of GFP-PARP-1 and GFP-PARP-2 in response to heavy DNA damage. Microirradiation of Hoechst-sensitized cells resulted in massive recruitment of GFP-PARP-1 and GFP-PARP-2 from nucleoli to damage sites (Figure 5). This depletion of the nucleolar storage was transient and GFP-PARP-1 and GFP-PARP-2 reappeared in the nucleolus correlating with their dissociation from repair sites (Figure 5). These data suggest that the nucleolus serves as a storage supplying PARP-1 and PARP-2 in response to heavy DNA damage.
Figure 5.
The Nucleolus serves as a storage of PARP-1 and PARP-2 to cope with heavy DNA damage. (A and C) Live cell imaging of microirradiated Hela cells sensitized with Hoechst 33285. Microirradiation of Hoechst sensitized cells leads to massive recruitment and temporary depletion of PARP-1 and PARP-2 from the nucleolus. (B and D) Quantitative evaluation of recruitment and nucleolar depletion kinetics. Error bars represent the SEM. Scale bar, 5 μm.
Recruitment of XRCC1 to damage sites depends on PARP-1 but not on PARP-2
Recent studies have indicated that the recruitment of SSBR/BER factors, like XRCC1 depends on PARP activity (9,10). To analyze the effect of poly(ADP-ribosyl)ation on recruitment of XRCC1 in more detail, we microirradiated wild-type, parp-1−/− and parp-2−/− MEFs expressing GFP-XRCC1. We found a considerably reduced recruitment of GFP-XRCC1 in cells lacking PARP-1, whereas recruitment of GFP-XRCC1 in parp-2−/− MEFs was as in wild-type cells (Figure 6A and B). To elucidate the mechanisms underlying these different recruitment kinetics we performed FRAP analysis, 5 min after microiradiation. In wild-type cells as well as in cells lacking PARP-2 we found a slow turnover of GFP-XRCC1 at microirradiated sites (t1/2 = 3.3 s and t1/2 = 2.85 s, respectively) whereas in parp-1−/− cells GFP-XRCC1 fluorescence recovered much faster (t1/2 = 1.2 s), indicating a high mobility of XRCC1 at DNA damage sites (Figure 6C and D).
Figure 6.
Efficient recruitment of XRCC1 to DNA repair sites depends on the presence of PARP-1. (A) Live cell imaging of microirradiated wild-type, PARP-1 and PARP-2 knock out MEFs (MEF parp-1−/−, MEF parp-2−/−) expressing GFP-XRCC1. Accumulation of GFP-XRCC1 at DNA damage sites is dramatically reduced in the absence of PARP-1. (B) Quantitative evaluation of recruitment kinetics. (C and D) Mobility of GFP-XRCC1 at DNA damage sites. The mobility of accumulated fluorescent fusion proteins was determined by bleaching the microirradiated site 5 min after microirradiation and subsequent recovery measurements. Inset shows the bleached microirradiated site. FRAP data from 10 individual experiments are shown as mean curves. Error bars represent the SEM. Scale bar, 5 μm.
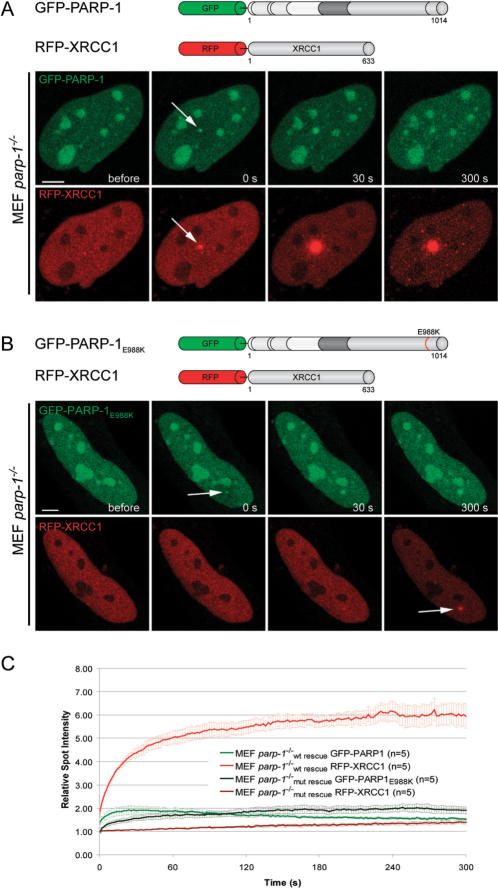
To test, whether the enzymatic activity of PARP-1 is needed for XRCC1 recruitment we cotransfected parp-1−/− MEFs with RFP-XRCC1 and GFP-tagged wild-type (GFP-PARP-1) or catalytically inactive PARP-1 (GFP-PARP1E988K). We found that RFP-XRCC1 is efficiently recruited to laser-induced DNA damage sites in parp-1−/− MEFs rescued with GFP-PARP-1 (Figure 7A and C). In contrast, recruitment of RFP-XRCC1 was dramatically reduced in parp-1−/− MEFs transfected with GFP-PARP-1E988K (Figure 7B and C). These results show that PARP-1 activity enhances the recruitment of repair factors to DNA damage sites by generating high-affinity binding sites.
Figure 7.
The catalytic activity of PARP-1 is needed for efficient recruitment of XRCC1 to laser-induced DNA damage sites. (A) Live cell imaging of microirradiated PARP-1 knock out MEFs (MEF parp-1−/−) coexpressing GFP-PARP-1 and RFP-XRCC1. Expression of GFP-tagged wild-type PARP-1 results in efficient recruitment of RFP-XRCC1. (B) Live cell imaging of microirradiated PARP-1 knock out MEFs (MEF parp-1−/−) coexpressing GFP-PARP-1E988K and RFP-XRCC1. Accumulation of RFP-XRCC1 at DNA damage sites is dramatically reduced in PARP-1 knock out MEFs expressing catalytically inactive GFP-PARP-1E988K. (C) Quantitative evaluation of recruitment kinetics. Error bars represent the SEM. Scale bar, 5 μm.
DISCUSSION
Genetic studies of knockout mice and cells have demonstrated the requirement of the two DNA-damage dependent PARPs, PARP-1 and PARP-2, for DNA repair (14–19). Based on their interaction with common proteins involved in genome restoration and binding to different DNA lesions and substrates, it was suggested that PARP-1 and PARP-2 have both overlapping and non-redundant functions (14,13). However, there have been reports questioning the importance of PARP-1 or PARP-2 for DNA repair (11,20). In this study, we compared the spatio-temporal redistribution of PARP-1 and PARP-2 in response to DNA damage induced by laser microirradiation in living cells. We observed a clear accumulation of both DNA-damage dependent PARPs at DNA damage sites. Consistent with distinct roles in DNA repair we found different recruitment kinetics for PARP-1 and PARP-2. While PARP-1 accumulated fast and transiently, PARP-2 showed a delayed and persistent accumulation at repair sites. The clear accumulation of PARP-2 at DNA damage sites together with biochemical and genetic data argues for an involvement of PARP-2 in DNA repair. Our kinetic studies suggest a role for PARP-2 in the latter steps of DNA repair, however the precise function of PARP-2 has to be elucidated in future studies.
Recruitment of PARP-1 is mainly mediated by its N-terminal DNA binding domain, as mutations of two cysteine residues within the Zn Finger domain dramatically reduced accumulation at repair sites, whereas the isolated DBD was sufficient for recruitment. Interestingly, the highly homologous Zn Finger domain of DNA ligase III, was neither necessary nor sufficient for recruitment to DNA repair sites, which was instead mediated by its BRCT domain binding to XRCC1 (28). Using a potent PARP inhibitor we could demonstrate that PARP activity is not essential for, but enhances the efficiency of, PARP-1 and PARP-2 recruitment to repair sites. This fits well with our observation that the second wave of PARP-1 recruitment relies on PAR binding via the BRCT domain of PARP-1. Interestingly, we found that the catalytic activity of PARP-1 is not only needed for efficient recruitment, but also for dissociation from DNA repair sites. This observation could be explained with earlier findings showing that automodification of PARP-1 abolishes DNA binding in vitro (29). These data argue for three distinct roles of PARP-1 in response to DNA damage: the detection and labeling of the damaged site, the local relaxation of chromatin structure and the recruitment of repair factors.
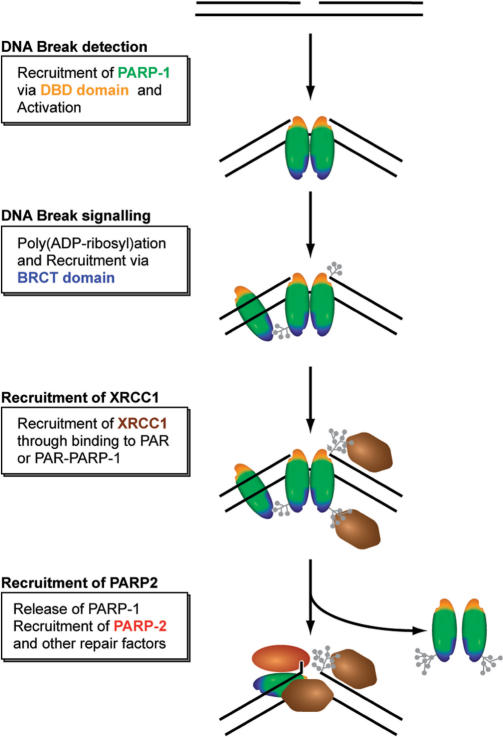
In summary, we propose the following model for the spatio-temporal accumulation of SSBR/BER factors at DNA strand breaks (Figure 8). Single-strand breaks are detected by the DNA binding domain of PARP-1. Poly(ADP-ribosyl)ation by PARP-1 leads to chromatin relaxation and attracts additional PARP-1 molecules via its BRCT domain. Further poly(ADP-ribosyl)ation at DNA lesions then leads to the release of PARP-1 through charge repulsion enabling a switch to the next step in DNA repair initiated by recruitment of the versatile loading platform XRCC1. Interestingly, PARP-2, which is required for DNA repair could not replace PARP-1 in the rapid recruitment of repair factors. However, we cannot exclude that PARP-2 could contribute to the slow recruitment of XRCC1 observed in parp1−/−MEFs.
Figure 8.
Simplified model for the recruitment of repair factors to SSB. See text for a detailed discussion of the role and regulation of PARPs.
This study of PARP-1 recruitment revealed a complex regulation of a repair factor in response to DNA damage. After detection of the DNA damage, PARP-1 activation and poly(ADP-ribosyl)ation leads to a positive feedback loop accumulating more PARP-1 and thus amplifying the signal for rapid recruitment of repair factors. Further accumulation is countered by a negative feedback resulting in the release of PARP-1 likely to protect against uncontrolled poly(ADP-ribosyl)ation which would disrupt cellular functions and lead to apoptosis. This feedback regulated recruitment of PARP-1 at DNA lesions thus allows a balance between signal amplification for rapid recruitment of repair factors and protection against extensive poly(ADP-ribosyl)ation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank G. de Murcia for helpful comments and suggestions. V.S. and J.C.A. are supported by funds from Centre National de la Recherche Scientifique, Association pour la Recherche contre le Cancer, Electricité de France, Ligue Nationale contre le Cancer (comité du Haut-Rhin) and Commissariat à l’Energie Atomique. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung to H.L. Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the last two authors should be regarded as joint Authors.
REFERENCES
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 3.Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 4.Gradwohl G, Menissier de Murcia JM, Molinete M, Simonin F, Koken M, Hoeijmakers JH, de Murcia G. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA. 1990;87:2990–2994. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 7.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber V, Ricoul M, Amé JC, Dantzer F, Meder VS, Spenlehauer C, Stiegler P, Niedergang C, Sabatier L, et al. PARP-2, structure-function relationship. In: Burkle A, editor. Poly(ADP-ribosyl)ation. Georgetown: Chapter 2. Landes Bioscience; 2004. pp. 13–31. [Google Scholar]
- 14.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo. J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 16.Masutani M, Nozaki T, Nishiyama E, Shimokawa T, Tachi Y, Suzuki H, Nakagama H, Wakabayashi K, Sugimura M. Function of poly(ADP-ribose) polymerase in response to DNA damage: gene-disruption study in mice. Mol. Cell Biochem. 1999;193:149–152. [PubMed] [Google Scholar]
- 17.Menissier de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Olivier FJ, Masson M, Dierich A, et al. Requirement of poly(ADP-ribose)polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher A, Hochegger H, Takeda S, Caldecott KW. Poly (ADP-ribose) polymerase-1 accelerates single-strand break repair in concert with poly (ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 22.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 23.Maeda Y, Hunter TC, Loudy DE, Dave V, Schreiber V, Whitsett JA. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J. Biol. Chem. 2006;281:9600–9606. doi: 10.1074/jbc.M510435200. [DOI] [PubMed] [Google Scholar]
- 24.Rolli V, O’Farrell M, Menissier-de Murcia J, de Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- 25.Molinete M, Vermeulen W, Burkle A, Menissier-de Murcia J, Kupper JH, Hoeijmakers JH, de Murcia G. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. Embo. J. 1993;12:2109–2117. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferro AM, Olivera BM. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. J. Biol. Chem. 1982;257:7808–7813. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.