Abstract
The population of elderly individuals has increased significantly over the past century and is predicted to rise even more rapidly in the future. Ageing is a major risk factor for many diseases such as neurodegenerative disease, diabetes and cancer. This highlights the importance of understanding the mechanisms involved in the ageing process. One plausible mechanism for ageing is accumulation of mutations in the mitochondrial genome. In this review, we discuss some of the most convincing data surrounding age-related mtDNA mutations and the evidence that these mutations contribute to the ageing process.
INTRODUCTION
Throughout the lifetime of an organism, the production of energy is a fundamental requirement. The majority of this energy, in the form of ATP, is produced through the process of oxidative phosphorylation (OXPHOS); the complexes involved are situated in the inner mitochondrial membrane. Mitochondria are intracellular organelles that contain their own DNA (mtDNA), which is distinct from nuclear DNA and can be replicated independently of the cell cycle. This allows the production of more mtDNA molecules capable of producing proteins for OXPHOS under high-energy demands. There is a small, ∼1 kb non-coding control region on the mitochondrial genome, but apart from this, the rest of the 16.5 kb genome is entirely transcribed. The mitochondrial genome codes for 37 genes, which includes 13 essential polypeptides of the OXPHOS system, 22 tRNAs and 2 rRNAs which are required for intramitochondrial protein synthesis. The vast majority of the proteins involved in OXPHOS are encoded by nuclear DNA, translated in the cytoplasm and imported into the mitochondria.
MtDNA is present in multiple copies within each cell; the actual number varies between cell type and on the energy demands within each tissue. The multi-copy nature of mtDNA means that any mutations occurring on the mitochondrial genome can exist amongst wild-type copies in a situation referred to as heteroplasmy (1). These mutant copies do not exert a biochemical phenotype on a cell until the mutant copies reach a certain level. This threshold of mutant: wild type can vary depending on the specific mutation and on cell type. The types of mtDNA mutations which can occur vary from single point mutations to large-scale rearrangements such as deletions and duplications (1). Mutations on the mitochondrial genome are thought to arise due to the close proximity of mtDNA to the OXPHOS system located on the inner mitochondrial membrane, making the mitochondrial genomes vulnerable to damage through the leakage of reactive oxygen species (ROS) during the OXPHOS process. Mitochondria are able to counteract the production of ROS with antioxidant defence systems which can detoxify the amount of ROS produced, however some ROS do evade these processes and are able to damage mtDNA as well as proteins and lipids. MtDNA molecules are contained within nucleoids, which contain essential maintenance proteins including the mitochondrial transcription factor A gene (TFAM) which effectively coats the mtDNA molecule. However, it is uncertain whether this offers any protection against ROS, and this along with limited repair capacity means that there is a high mutation rate. This increased susceptibility of mtDNA to ROS leading to mutations has led to the proposal of the mitochondrial theory of ageing (2,3). This theory suggests that damaged mitochondrial genomes lead to inefficient OXPHOS causing the production of more ROS which will further damage the mtDNA, resulting in a so-called ‘vicious cycle’.
Evidence for accumulation of mtDNA mutations with age
In support of a role for mtDNA mutations in ageing, both mtDNA point mutations and deletions have been described to accumulate on the mitochondrial genome with age in a variety of tissues (4–8). High levels of a m.414T>G transversion was found in fibroblasts from half of subjects older than 65 years but was absent from all younger individuals (6). The m.414T>G mutation has most recently been shown to occur significantly more frequently in fibroblasts taken from skin from sun-exposed skin sites suggesting that increased oxidative stress through ultraviolet (UV) radiation exposure results in the production of this mutation (9). The absence or marginal presence of the m.414T>G mutation in skeletal muscle, brain, heart, lymph nodes and spleen has led to the suggestion that it may be tissue specific (10–12). Suggestions of tissue-specific ‘hot-spots’ for point mutations on the mitochondrial genome have also been described (13), with the clonal expansion of point mutations accumulating with age in cardiomyocytes and buccal epithelium, but the distribution of these point mutations were described to be significantly different. The region 407–411 of mtDNA which is close to the age-associated m.414T>G mutation has also been suggested as being a hot-spot for mutations in muscle mtDNA (12). Work in our own laboratory on colonic epithelium has shown an age-dependent increase of clonally expanded point mutations, with ∼50% of colonocytes taken from aged patients harbouring a point mutation (7). However we have not observed any evidence for specific mutation ‘hot-spots’, with the distribution of the point mutations spread throughout the genome (7).
The age-related accumulation of mtDNA deletions has also been described. The initial observations were focused on a well-characterized 4977 bp deletion, the so-called ‘common deletion’. This deletion had previously been identified in a number of patients with mitochondrial disease (Kearns Sayre syndrome and chronic progressive external ophthalmoplegia). Studies on tissues from normal subjects showed that the common deletion was detectable in heart muscle, skeletal muscle, brain and other tissues of older human subjects, but not from the same tissues from young individuals (14–16). Technical difficulties in the study of mtDNA deletions resulted in the majority of early studies focussing on identifying single deletions in ageing tissues, such as the common deletion and the levels detected even in tissues from very elderly individuals rarely exceeded ∼1%. Thus, it was thought that these very low levels of mtDNA deletions associated with ageing were unlikely to contribute to the ageing process. However it was shown that if mtDNA mutations accumulate to high levels focally in a small subset of cells, then this leads to cells which have respiratory chain deficiency, detected by an absence of staining for cytochrome c oxidase (COX) activity (17,18).
Respiratory deficient cells have since been shown to accumulate with age in a number of tissues (4,7,19) (Figure 1). Also, it was apparent that the common deletion was only one of several different possible deletions present in ageing human tissues and the development of single-cell-based PCR techniques allowed the measurement of all deletions in single cells. Recently, we and colleagues were the first to report the age-related accumulation of mtDNA deletions to very high levels (∼50%) in substantia nigra neurons from both normal controls and individuals with Parkinson's disease (4,5). Analysis of mtDNA deletions using long-range PCR in individual neurons showed that each neuron harboured a different mtDNA deletion usually consisting of just one type, which suggests that these mtDNA deletions arose through clonal expansion (see later) (Figure 2). Respiratory chain-deficient neurons had significantly higher level of mtDNA deletions than neurons with normal COX activity, supporting the hypothesis that these deletions were the cause of the respiratory chain deficiency. Similar observations have also been made by analysing ageing muscle fibres. These studies also showed that in age-related respiratory chain-deficient fibres there was clonal expansion of mtDNA deletions, with over 80% of mtDNA in the affected fibres harbouring a clonally expanded deletion (20).
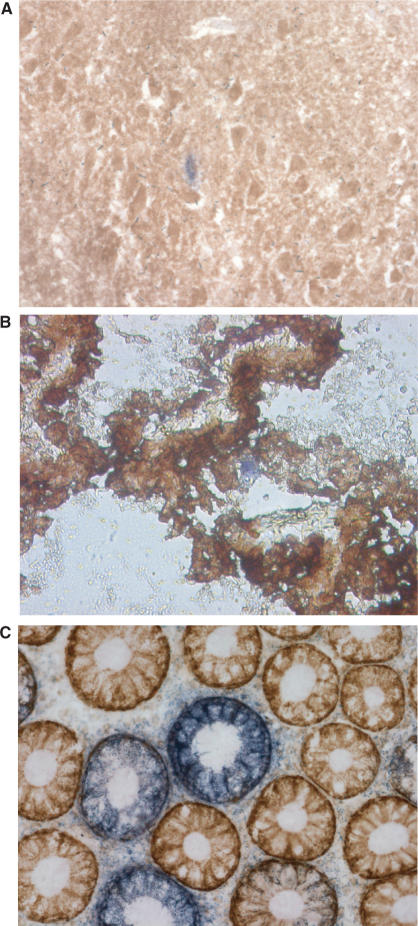
Figure 1.
Histochemical staining to observe the presence of respiratory chain-deficient cells (blue) amongst respiratory chain normal cells (brown) in aged: hippocampus (A), choroid plexus (B) and colonic epithelium (C).
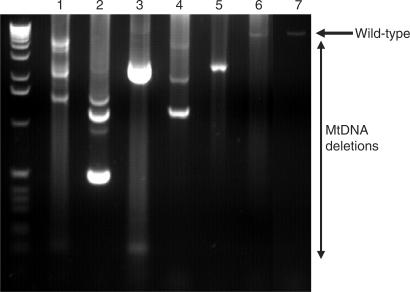
Figure 2.
Long-range PCR of substantia nigra neurons showing the presence of mtDNA deletions in 10 pooled neurons (lanes 1 and 2), five pooled neurons (lanes 3 and 4), a single neuron (lanes 5 and 6) and control DNA (lane 7). The ladder on the left of the gel is a 1 kb ladder.
Are mtDNA mutations important in human ageing?
In humans, the increase in mtDNA mutations with age does not prove that they have a role in the ageing process; they could just be biological markers of accumulating damage. It has been reported that mitochondria isolated from the fibroblasts of old animals that was subsequently microinjected into young cells can lead to cellular degeneration, similar results were also achieved from young, partially uncoupled mitochondria (21,22). Although the nature of the mtDNA abnormalities was not investigated in these studies, these results demonstrate that altered mitochondria may be playing a crucial role in the decline in cellular function. However, a recent study has shown that somatic mutation accumulation with age is dependent on nuclear genetic background in mice and not associated with increase in ROS or senescence (23). This suggests that the accumulation of the mtDNA mutations may not be as straightforward as first thought and that an individual's nuclear background may affect their susceptibility to the accumulation of mtDNA mutations and the resulting phenotype. This could have some effect on the interpretation of studies involving mtDNA mutations in cybrids, where there has been some evidence for increased ROS production with certain mtDNA mutations as these differences may vary in other nuclear backgrounds (24,25).
In the skin, changes associated with normal ageing such as wrinkles, changes in pigmentation and the increased incidence of skin cancer are accelerated by exposure to the potent mutagen UV radiation. MtDNA deletions occur more frequently in sun-exposed areas that have been subjected to photoageing (26–29). These observations support the theory that increased oxidative stress results in mtDNA damage and the production of mutations. It is unknown at present what effects these mtDNA deletions have on the normal functioning of the skin.
An important observation supporting a role of mtDNA mutations in age-related cellular degeneration has been shown by the age-associated loss of muscle mass, termed sarcopenia, which is a common feature in aged human subjects (30). Atrophy in skeletal muscle fibres from rhesus monkeys was shown to correlate in those sections with respiratory chain abnormalities and mtDNA deletions, compared to those sections without respiratory chain abnormalities (31), suggesting that mitochondrial dysfunction may be a contributing factor to sarcopenia. To strengthen this finding, in sarcopenic rats, mtDNA deletions have been shown to co-localize with respiratory chain abnormalities, atrophy and splitting (32,33). These observations provide a strong association that mtDNA deletions contribute to the age-related decline of muscle mass and function.
The age-associated accumulation of high levels of mtDNA deletions in the dopamine producing, pigmented neurons of the substantia nigra is additional support for a role of mtDNA mutations in the ageing process. This region of the brain loses neurons at a rate of ∼5% per decade (34) and there is an even greater neuronal loss in patients with Parkinson's disease. It is unknown at present whether the age-related loss of neurons in the substantia nigra is due to the accumulation of mtDNA deletions in these neurons. However, mtDNA dysfunction resulting from reduced mtDNA expression has been shown to cause a parkinsonian phenotype in mice (35). The cause of the reduced mtDNA expression in these mice is due to selective knockout of TFAM in dopamine producing neurons within the substantia nigra. The mice had respiratory chain deficiency accompanied by cell loss, selectively in this region and provides support that normal mtDNA expression is crucial for the functioning and viability of neurons within the substantia nigra. Recent studies have also shown that substantia nigra neurons have an unusual reliance on certain calcium channels to maintain their pacemaker currents and this seems likely to make them particularly sensitive to mitochondrial dysfunction (36).
Ageing mouse model
A major advance in the support for a direct role of mtDNA mutations in the ageing process was demonstrated by two groups who created mice with a premature ageing phenotype due to knock-in mutations in the exonuclease domain of polymerase gamma (POLG) (37,38). POLG is the only known polymerase to be targeted to mitochondria and as such is thought to be solely responsible for the maintenance of all aspects of mtDNA including replication and repair. In humans, POLG is a heterotrimeric enzyme consisting of a catalytic subunit (POLGA) and two identical accessory subunits (POLGB) (39). Therefore it was no surprise that a mutation in the exonuclease domain (proofreading) of POLGA resulted in the mice having increased mtDNA mutation rates as detected by cloning analysis (38). Some controversy exists over the validity of the mutation rates detected by the cloning methodology and a recent paper has re-analysed the mutation rates in the POLG mice using a random capture methodology (RMC) suggesting the mutation rates calculated by the cloning method are too high (40). However, even using the RMC method, the mutation rates of the POLG mice are higher than their wild-type counterparts.
A surprising observation in the POLG mice is that there is no evidence of increased ROS production (41), which is predicted by the mitochondrial theory of ageing. In addition, mRNA levels of antioxidant defence systems were unaffected, suggesting a lack of a ROS-induced stress response. The authors question the role of ROS in the premature ageing symptoms displayed by these mice and suggest that the respiratory chain deficiency may be the major factor involved in the age-associated decline in function, presumably caused through the generation of increased mtDNA mutations. An alternative explanation is that the generation of mtDNA mutations through inefficient POLG may be downstream from mechanisms that generate ROS (42). Therefore, it is possible that ROS can induce mtDNA mutations directly or through damaging POLG to make it error-prone, but the resulting mtDNA mutations do not increase the production of ROS. To further confuse the role of ROS in ageing is the observation that high oxidative damage levels are observed in the longest-living rodent, the naked mole rat (43). It has also been reported that heterozygous SOD2 knockouts have increased oxidative damage but normal lifespan (44). All these results seem to contradict the simplest assumption that increased ROS production promotes accelerated ageing. However, these results may be an indication that as discussed previously, other factors such as the nuclear genetic background play a crucial role in the susceptibility to the effects of increased oxidative damage.
Clonal expansion of mtDNA mutations
Whilst there remains some controversy over the best method to measure mutation rates, in terms of respiratory chain deficiency (and possibly ageing) the crucial factor in determining dysfunction is the clonal expansion of the mtDNA mutation within a cell. Clonal expansion of a specific mtDNA mutation has been described by many different authors in both disease and ageing tissues (4,5,7,13,20,45). In patients with primary mtDNA defects, the clonally expanded mutation is identical between different cells, whereas in ageing different mutations are seen in adjacent cells. In ageing it seems likely that the mutation was derived from a single event and this mutation subsequently expands to become the predominant species within an individual cell. The precise mechanism of clonal expansion is unknown, but several authors have proposed a random genetic drift model (46). An important research focus should be to understand the mechanism of clonal expansion in different tissues, especially post-mitotic tissues where the rate of replication of the mitochondrial genome is uncertain. Unfortunately, at present, methods to determine rates of mtDNA replication cannot be differentiated from those measuring repair. In post-mitotic tissues, DNA repair is an active process and may even be involved in the generation of mutations.
What do patients with mitochondrial diseases tell us about mitochondrial ageing?
An increasing number of patients are being described with primary or secondary defects of the mitochondrial genome. Secondary defects are those due to mutations in nuclear genes involved in mtDNA maintenance. These patients present with a variety of different clinical features, particularly involving tissues heavily dependent upon OXPHOS. These patients have been crucial in our understanding of human mitochondrial genetics, but also provide important clues as to the potential role of mtDNA in the ageing process. These include:
The concept of threshold for an individual mtDNA mutation leading to respiratory chain deficiency in an individual cell. This phenomenon is crucial to understanding the importance of clonal expansion in ageing tissues. Indeed an important part of the diagnosis of mtDNA mutations is that these mutations have to show a threshold to confirm the pathogenic nature of any specific point mutation. This confirms again the importance of clonal expansion of somatic mutations before a biochemical defect is seen.
MtDNA mutations lead to human pathology. In patients with mtDNA mutations, there is both clinical disease and evidence of cell loss and dysfunction. These changes are much more marked than those seen with ageing but are still relevant since we believe the respiratory chain-deficient cells are essentially the same in both mtDNA disease and ageing.
The clinical features of patients with mtDNA disease are very different from ageing although this is not surprising since these patients are born with high levels of a single mtDNA mutation. The phenotype will depend upon both the nature of the mtDNA mutation and the level in a number of different tissues. More relevant to ageing are those patients with primary nuclear mutations involved in mtDNA maintenance. These patients acquire mtDNA mutations throughout life, often mtDNA deletions. This is clearly similar to the observations in post-mitotic tissues in ageing individuals. The level of mtDNA mutations is much greater in tissues than seen in ageing since there is often severe disease leading to disability or death. Genetic defects of POLG are the most important of the defects leading to secondary mtDNA defects (47). Interestingly one of the phenotypes seen in these patients is Parkinsonism associated with loss of cells in the substantia nigra and high levels of deleted mtDNA (48–50). This is remarkably similar to recent observations in elderly tissues and confirms that defects of mtDNA can lead to cell loss in this region of the brain.
Whilst patients with either primary or secondary defects of mtDNA do not present good phenotypic models of human ageing, they do show that these genetic defects of the mitochondrial genome can lead to pathology and thus are supportive of a role of mtDNA mutations in human ageing.
Cell death
The ultimate insult that mtDNA mutations could have on a cell would be to impair the function so much as to lead to cell death. It is well known that mitochondria are central players in apoptosis, or programmed cell death. Therefore, if high levels of mtDNA mutations lead to mitochondrial dysfunction, it seems possible that this will compromise the cell and result in apoptosis. Respiratory chain deficiency, caused by reduced mtDNA expression, has been shown to be associated with increased apoptosis in TFAM knockout animal models (51). Mitochondrial dysfunction, induced by a variety of oxidative stresses has been shown to induce apoptosis in cultured neurons and cardiomyocytes (52–54), but evidence of underlying mtDNA mutations as the cause was not investigated in these studies. However, cleaved caspase-3 an indicator of apoptosis, was increased in a variety of tissues in the POLG mice (37), suggesting that the increased mtDNA mutation load in these animals could be responsible for the decline in tissue function with age. It has also been recently reported that genes involved in apoptosis are upregulated in the cochlear of the POLG-mutated mice and therefore the resulting effect could be the age-related hearing loss in these mice (55). In addition increased TUNEL staining was observed in the hippocampus in mitochondrial late-onset neurodegeneration (MILON) mice (56), which are mice with mtDNA depletion due to a knockout of TFAM. The TUNEL staining in these MILON mice was often observed before any obvious cell loss.
CONCLUSIONS
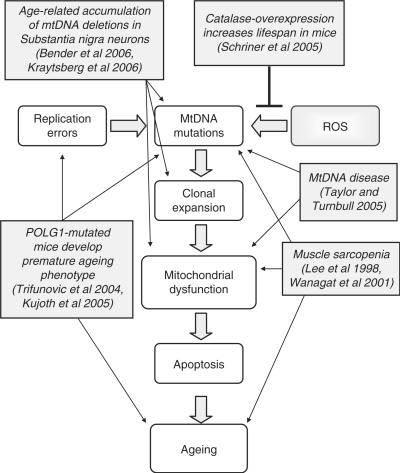
In this review, we have shown that there is increasing evidence to suggest that the accumulation of mtDNA mutations with age could play a role in the decline in cellular function within many tissues. We have summarized some of this evidence in Figure 3, which shows that mtDNA mutations could be caused by either replication errors or increased oxidative stress, if a mutated mtDNA molecule is then allowed to replicate and clonally expand within a cell, this cell may become respiratory chain deficient. This compromised energy production could lead to the demise of the cell, and if substantial cell loss is observed this may result in tissue dysfunction and the onset of ageing. If this is correct then by preventing the mtDNA mutations from arising should increase lifespan. Support for this theory has already been shown with the observation of an increase in lifespan of mice over-expressing human catalase localized to mitochondria (57). These mice had reduced cardiac pathology and cataract development along with reduced oxidative damage mirrored by a reduction in mtDNA deletions compared to wild-type mice. Catalase converts H2O2 into water and oxygen preventing it from becoming a highly reactive hydroxyl radical. Therefore, the resulting reduced oxidative damage followed by extension of lifespan in these mice supports the production of mitochondrial ROS as a factor in the ageing process. There are many challenges ahead. For example, we lack knowledge of several fundamental aspects of mitochondrial genetics such as the mechanisms causing mtDNA mutations and their subsequent clonal expansion. In addition, there are likely to be tissue-specific differences and influence on these processes of nuclear genetic factors.
Figure 3.
Schematic diagram displaying how mtDNA mutations could lead to the ageing process and the major evidence in the literature to support this role.
ACKNOWLEDGEMENTS
We are grateful for support from the Alzheimer's Research Trust, Food Standards Agency (UK), The Wellcome Trust and the Medical Research Council. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 4.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 5.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 6.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 7.Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 9.Birket MJ, Birch-Machin MA. Ultraviolet radiation exposure accelerates the accumulation of the aging-dependent T414G mitochondrial DNA mutation in human skin. Aging Cell. 2007;6:557–564. doi: 10.1111/j.1474-9726.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 10.Chinnery PF, Taylor GA, Howell N, Brown DT, Parsons TJ, Turnbull DM. Point mutations of the mtDNA control region in normal and neurodegenerative human brains. Am. J. Hum. Genet. 2001;68:529–532. doi: 10.1086/318204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, Yarasheski KE, Miller CA, Askanas V, Engel WK, et al. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc. Natl Acad. Sci. USA. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc. Natl Acad. Sci. USA. 2002;99:5521–5526. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat. Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 15.Cortopassi GA, Shibata D, Soong NW, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl Acad. Sci. USA. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat. Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 17.Brierley EJ, Johnson MA, Lightowlers RN, James OF, Turnbull DM. Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann.Neurol. 1998;43:217–223. doi: 10.1002/ana.410430212. [DOI] [PubMed] [Google Scholar]
- 18.Muller-Hocker J, Schneiderbanger K, Stefani FH, Kadenbach B. Progressive loss of cytochrome c oxidase in the human extraocular muscles in aging: a cytochemical-immunohistochemical study. Mutat. Res. 1992;275:115–124. doi: 10.1016/0921-8734(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 19.Cottrell DA, Blakely EL, Johnson MA, Ince PG, Borthwick GM, Turnbull DM. Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Aging. 2001;22:265–272. doi: 10.1016/s0197-4580(00)00234-7. [DOI] [PubMed] [Google Scholar]
- 20.Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbisier P, Remacle J. Influence of the energetic pattern of mitochondria in cell ageing. Mech. Ageing Dev. 1993;71:47–58. doi: 10.1016/0047-6374(93)90034-o. [DOI] [PubMed] [Google Scholar]
- 22.Corbisier P, Remacle J. Involvement of mitochondria in cell degeneration. Eur. J. Cell Biol. 1990;51:173–182. [PubMed] [Google Scholar]
- 23.Yao YG, Ellison FM, McCoy JP, Chen J, Young NS. Age-dependent accumulation of mtDNA mutations in murine hematopoietic stem cells is modulated by the nuclear genetic background. Hum. Mol. Genet. 2007;16:286–294. doi: 10.1093/hmg/ddl457. [DOI] [PubMed] [Google Scholar]
- 24.Jou MJ, Peng TI, Wu HY, Wei YH. Enhanced generation of mitochondrial reactive oxygen species in cybrids containing 4977-bp mitochondrial DNA deletion. Ann. NY Acad. Sci. 2005;1042:221–228. doi: 10.1196/annals.1338.024. [DOI] [PubMed] [Google Scholar]
- 25.Vives-Bauza C, Gonzalo R, Manfredi G, Garcia-Arumi E, Andreu AL. Enhanced ROS production and antioxidant defenses in cybrids harbouring mutations in mtDNA. Neurosci. Lett. 2006;391:136–141. doi: 10.1016/j.neulet.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 26.Berneburg M, Plettenberg H, Medve-Konig K, Pfahlberg A, Gers-Barlag H, Gefeller O, Krutmann J. Induction of the photoaging-associated mitochondrial common deletion in vivo in normal human skin. J. Invest. Dermatol. 2004;122:1277–1283. doi: 10.1111/j.0022-202X.2004.22502.x. [DOI] [PubMed] [Google Scholar]
- 27.Birch-Machin MA, Tindall M, Turner R, Haldane F, Rees JL. Mitochondrial DNA deletions in human skin reflect photo- rather than chronologic aging. J. Invest. Dermatol. 1998;110:149–152. doi: 10.1046/j.1523-1747.1998.00099.x. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan KJ, Birch-Machin MA. The incidence of both tandem duplications and the common deletion in mtDNA from three distinct categories of sun-exposed human skin and in prolonged culture of fibroblasts. J. Invest. Dermatol. 2006;126:408–415. doi: 10.1038/sj.jid.5700099. [DOI] [PubMed] [Google Scholar]
- 29.Yang JH, Lee HC, Wei YH. Photoageing-associated mitochondrial DNA length mutations in human skin. Arch. Dermatol. Res. 1995;287:641–648. doi: 10.1007/BF00371736. [DOI] [PubMed] [Google Scholar]
- 30.Short KR, Nair KS. Mechanisms of sarcopenia of aging. J. Endocrinol. Invest. 1999;22:95–105. [PubMed] [Google Scholar]
- 31.Lee CM, Lopez ME, Weindruch R, Aiken JM. Association of age-related mitochondrial abnormalities with skeletal muscle fiber atrophy. Free Radical Biol. Med. 1998;25:964–972. doi: 10.1016/s0891-5849(98)00185-3. [DOI] [PubMed] [Google Scholar]
- 32.Wanagat J, Cao Z, Pathare P, Aiken JM. Mitochondrial DNA deletion mutations colocalize with segmental electron transport system abnormalities, muscle fiber atrophy, fiber splitting, and oxidative damage in sarcopenia. FASEB J. 2001;15:322–332. doi: 10.1096/fj.00-0320com. [DOI] [PubMed] [Google Scholar]
- 33.Bua EA, McKiernan SH, Wanagat J, McKenzie D, Aiken JM. Mitochondrial abnormalities are more frequent in muscles undergoing sarcopenia. J. Appl. Physiol. 2002;92:2617–2624. doi: 10.1152/japplphysiol.01102.2001. [DOI] [PubMed] [Google Scholar]
- 34.Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 35.Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl Acad. Sci. USA. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson's disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- 37.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 38.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 39.Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 40.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 41.Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, et al. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl Acad. Sci. USA. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loeb LA, Wallace DC, Martin GM. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc. Natl Acad. Sci. USA. 2005;102:18769–18770. doi: 10.1073/pnas.0509776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andziak B, O’Connor TP, Qi W, DeWaal EM, Pierce A, Chaudhuri AR, Van Remmen H, Buffenstein R. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell. 2006;5:463–471. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 44.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 45.McDonald SA, Preston SL, Greaves LC, Leedham SJ, Lovell MA, Jankowski JA, Turnbull DM, Wright NA. Clonal expansion in the human gut: mitochondrial DNA mutations show us the way. Cell Cycle. 2006;5:808–811. doi: 10.4161/cc.5.8.2641. [DOI] [PubMed] [Google Scholar]
- 46.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum. Mol. Genet. 2006;15(Spec No 2):R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 48.Davidzon G, Greene P, Mancuso M, Klos KJ, Ahlskog JE, Hirano M, DiMauro S. Early-onset familial parkinsonism due to POLG mutations. Ann. Neurol. 2006;59:859–862. doi: 10.1002/ana.20831. [DOI] [PubMed] [Google Scholar]
- 49.Hudson G, Schaefer AM, Taylor RW, Tiangyou W, Gibson A, Venables G, Griffiths P, Burn DJ, Turnbull DM, et al. Mutation of the linker region of the polymerase gamma-1 (POLG1) gene associated with progressive external ophthalmoplegia and Parkinsonism. Arch. Neurol. 2007;64:553–557. doi: 10.1001/archneur.64.4.553. [DOI] [PubMed] [Google Scholar]
- 50.Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors A, Rautakorpi I, Peltonen L, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Silva JP, Gustafsson CM, Rustin P, Larsson NG. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc. Natl Acad. Sci. USA. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoyt KR, Gallagher AJ, Hastings TG, Reynolds IJ. Characterization of hydrogen peroxide toxicity in cultured rat forebrain neurons. Neurochem. Res. 1997;22:333–340. doi: 10.1023/a:1022403224901. [DOI] [PubMed] [Google Scholar]
- 53.Jang YM, Kendaiah S, Drew B, Phillips T, Selman C, Julian D, Leeuwenburgh C. Doxorubicin treatment in vivo activates caspase-12 mediated cardiac apoptosis in both male and female rats. FEBS Lett. 2004;577:483–490. doi: 10.1016/j.febslet.2004.10.053. [DOI] [PubMed] [Google Scholar]
- 54.Keller JN, Kindy MS, Holtsberg FW, St Clair DK, Yen HC, Germeyer A, Steiner SM, Bruce-Keller AJ, Hutchins JB, et al. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J. Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Someya S, Yamasoba T, Kujoth GC, Pugh TD, Weindruch R, Tanokura M, Prolla TA. The role of mtDNA mutations in the pathogenesis of age-related hearing loss in mice carrying a mutator DNA polymerase gamma. Neurobiol. Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.01.014. doi:10.1016/j.neurobiolaging.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson NG. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J. Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]