Abstract
A small-plaque variant (SP) of West Nile virus (WNV) was isolated in Vero cell culture from kidney tissue of an American crow collected in New York in 2000. The in vitro growth of the SP and parental (WT) strains was characterized in mammalian (Vero), avian (DF-1 and PDE) and mosquito (C6/36) cells. The SP variant replicated less efficiently than did the WT in Vero cells. In avian cells, SP growth was severely restricted at high temperatures, suggesting that the variant is temperature sensitive. In mosquito cells, growth of SP and WT was similar, but in vivo in Culex pipiens (L.) there were substantial differences. Relative to WT, SP exhibited reduced replication following intrathoracic inoculation and lower infection, dissemination and transmission rates following oral infection. Analysis of the full length sequence of the SP variant identified sequence differences which led to only two amino acid substitutions relative to WT, prM P54S and NS2A V61A.
Keywords: West Nile virus, small plaque variant, wildtype, temperature sensitivity, Culex pipiens, vector competence
Introduction
West Nile virus (WNV) is an arthropod-borne virus belonging to the family Flaviviridae, genus Flavivirus that exists primarily in a mosquito-avian transmission cycle. WNV, which had been geographically limited to Africa, the Middle East, India, Australasia, and western and central Asia, was first introduced into the northeastern United States in 1999 (Lanciotti et al., 1999) and has since spread throughout the United States, into Canada, Mexico, the Caribbean, Latin America, and South America (Austin et al., 2004; Blitvich et al., 2003; Dupuis et al., 2005; Tachiiri et al., 2006). The single-stranded, positive-sense WNV genomic RNA is approximately 11 kb and encodes a single long polyprotein that is post-translationally processed by viral and host cellular proteases into three structural proteins and seven nonstructural proteins (Brinton, 2002). Comparisons of sequence data for full and partial genomes of WNV over the course of the virus spread throughout the U.S. demonstrate that there has been minimal genetic change since its introduction in 1999 (Davis et al., 2004). Although WNV has remained relatively genetically homogeneous in the U.S., a new WNV genotype emerged late in the 2001 transmission season and has gradually established itself as the dominant genotype of WNV in the U.S. (Davis et al., 2005; Ebel et al., 2004). Concurrent with the emergence of this new genotype was the rapid spread of WNV across the U.S. leading to the largest recorded epidemic of arboviral encephalitis in the western hemisphere (CDC 2002). Several studies have confirmed the presence of distinct WNV genetic variants existing at temporal and geographical foci (Beasley et al., 2003; Davis et al., 2004, 2005); for example, small plaque isolates of WNV that exhibited attenuated properties in vitro and in vivo were collected in Texas in 2003 (Davis et al., 2004).
Small-plaque variants of other flaviviruses have been isolated in the laboratory. An isolate of dengue virus type 2 exhibited a mixed plaque phenotype in LLC-MK cells, and a small plaque subline selected from the mixed population in primary green monkey kidney cells was temperature sensitive (ts) in LLC-MK cells and attenuated in suckling mice (Eckels et al., 1976). A small plaque antigenic variant of Japanese encephalitis virus (JEV) was selected in Vero cell culture in the presence of a neutralizing monoclonal antibody, E3.3; this variant exhibited resistance to MAb3.3, but showed neurovirulence and neuroinvasiveness similar to the parental strain (Wu et al., 1997). Chemically mutated JEV, tick-borne encephalitis virus (TBEV), and dengue virus type 4 (DENV-4) have all manifested small plaque phenotypes, ts, and attenuated pathogenicity in mice (Blaney et al., 2001, 2002, 2003; Eastman & Blair, 1985; Hanley et al., 2002; Rumyantsev et al., 2006). Neuroblastoma cell-adapted yellow fever 17D virus exhibited a small plaque morphology, as well as defective cell penetration, poor cell-to-cell spread and reduced growth efficiency, attributed to amino acid substitutions at positions 360 and 362 in the envelope protein (Chambers & Nickells, 2001; Nickells & Chambers, 2003; Vlaycheva & Chambers, 2002; Vlaycheva et al., 2004, 2005). An Ala to Pro single amino acid substitution in NS2A30 of WNVKUN strain resulted in abortive viral replication with low titer, absence of detectable plaques, and high level INFα/β production in A549 (human alveolar basal epithelial) cells; however, in BHK cells, the strain’s plaque size and replication efficiency were comparable to those of the wild type (Liu et al., 2006).
Here we report a small-plaque (SP) variant selected from the mutant spectrum of WNV isolated from an American crow collected in New York in 2000. This variant was characterized in vitro in mammalian, avian, and mosquito cell lines, and in vivo in Culex pipiens (L.) mosquitoes. We demonstrate that this SP variant is attenuated both in vitro and in vivo.
Results
Isolation and characterization of the small-plaque variant
The plaque morphology of the SP variant of WNV, isolated as described in Material and Methods, was compared to the parental wild type strain (WT). In Vero cells, at 72 hours post infection (hpi), WT exhibited a mixed-plaque phenotype, producing both large and small plaques while the SP virus had uniformly small plaques (Fig 1A). At this time point, SP virus plaques (mean: 0.44 mm in diameter, range: 0.20–0.83 mm) were significantly smaller than those of WT (mean: 1.62 mm, range: 0.34–2.87 mm; P< 0.001, T test; Fig 1B). In this analysis, 100 % of the SP population exhibited plaques less than 1.0 mm in diameter, whereas 91.5% of the WT plaques were larger than 1.0 mm.
Figure 1.
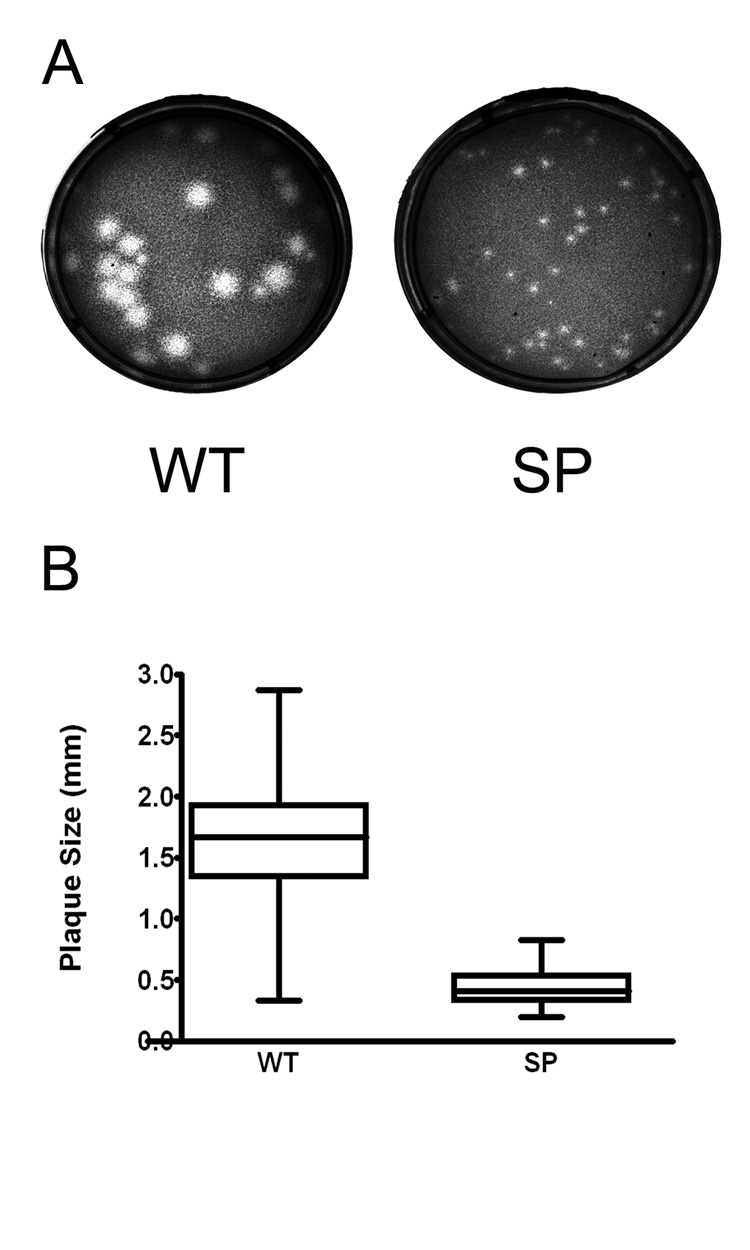
Plaque morphology of wild type (WT) and a small plaque (SP) variant WNV. Vero cells in 6-well plates were infected with Vero cell amplified WT or SP stock virus, and plaques were visualized 3 days post-infection after staining with neutral red. (A) Plaques were examined with a Zeiss Stemi 2000-C stereo microscope, and images were captured with a Zeiss Axiocam MRC digital camera. (B) Plaque sizes were measured using AxioVision 3.0 software (Zeiss, Germany). For each virus, 200 individual plaques were measured and analyzed by Graph Pad Prism (v.4.0). Bar within the box represents the median value, boxes extend from 25th to 75th percentile, and error bars represent highest and lowest values. Statistical analysis by t-test, P<0.0001.
In vitro growth kinetics
Growth of WT virus and the SP variant was compared in mammalian (Vero), avian (DF-1 and PDE) and mosquito (C6/36) cell lines. In Vero and DF-1 cells, at temperatures at or below 37°C, the SP variant grew to significantly lower titers than did the WT (P<0.01, t-test; Fig 2), but in PDE cells at 37°C and 39°C the titers of WT and SP were similar. At higher temperatures, the growth kinetics of SP were dependent on the cell type. In avian cells, SP growth was severely restricted at temperatures at or above 41°C, suggesting that the virus is ts in these cells. In Vero cells, although SP replicated more slowly than WT at 41°C and 42.5°C, the WT titers were equal to or lower than SP titers at later time points. This pattern could be due to high levels of WT viral replication resulting in increased cell mortality or deterioration of viable virus at high temperatures.
Figure 2.
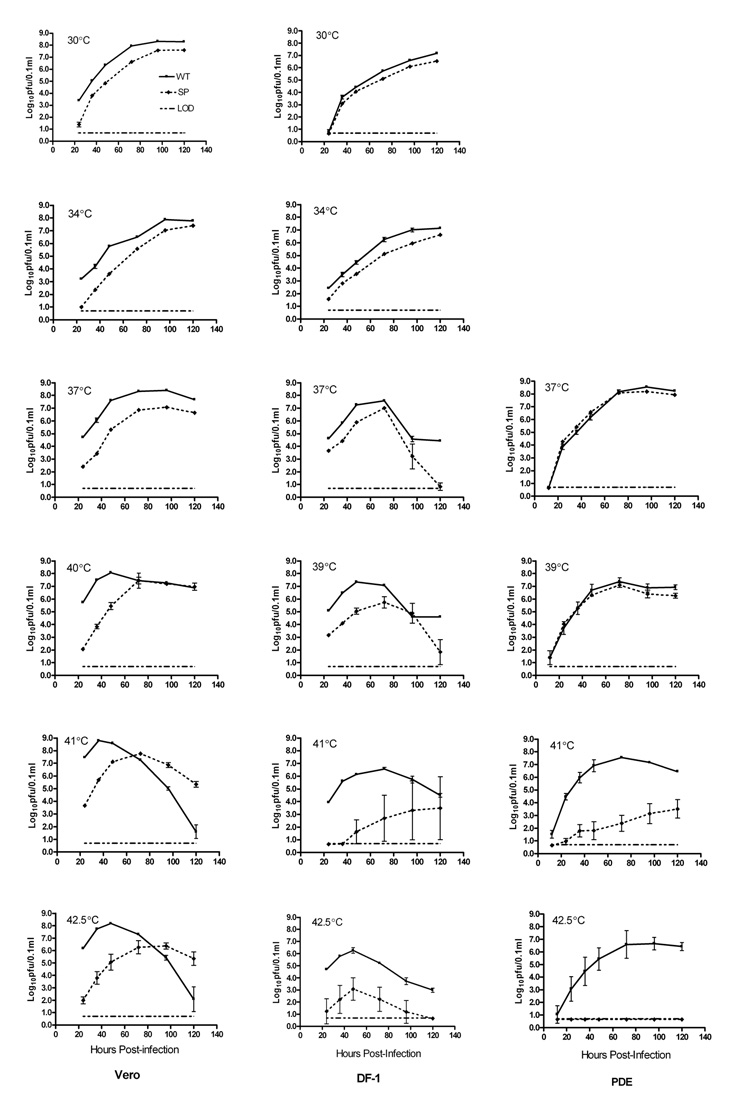
Comparison of growth of wild type (WT) and a small plaque (SP) variant WNV in three vertebrate cell lines. Vero, DF-1, and PDE cells in 6-well plates were infected with Vero cell amplified WT or SP stock virus in triplicate at an MOI of 0.01 pfu/cell, based on Vero cell titers. Both infection and incubation were carried out at the indicated temperatures. Virus was harvested from the medium at various time points after infection, and titers were determined by plaque assay on Vero cells. Symbols represent the mean titer and standard deviation for each time point. The limit of detection (LOD) was 0.7 log10pfu/0.1ml of supernatant of infected cell cultures; in cases where the sample had no detectable virus, a titer of 0.65 log10pfu/0.1ml was used for calculations.
The small-plaque phenotype was retained following growth of the SP variant in all cell types at 30°C, 34°C and 37°C. However, in Vero and DF-1 cells at or above 41°C, reversion to a mixed plaque phenotype was observed, with the proportion of plaques larger than 1.0 mm in diameter increasing with increasing temperature (data not shown). In PDE cells, growth of SP at temperatures up to 41°C did not result in a reversion to mixed plaque phenotype, and at 42.5°C no virus was recovered, indicating that SP failed to grow at this latter temperature.
In contrast to the results with mammalian and avian cells, SP exhibited replication efficiency similar to that of WT in C6/36 cells, with nearly equal virus titers at all incubation temperatures (Fig 3A). While peak virus titers of both viruses were reached by 96h at 30°C and 34°C, neither virus reached its peak titer in this cell line by 120h at 26°C, indicating that the replication of both virus strains was enhanced by increased incubation temperature. There was no reversion to a mixed plaque phenotype following growth of the SP variant in mosquito cells.
Figure 3.
Comparison of growth of wild type (WT) and a small plaque (SP) variant WNV in vitro in a mosquito cell line and in vivo in mosquitoes. (A) C6/36 cells in 6-well plates were infected with Vero cell amplified WT or SP stock virus in triplicate at an MOI of 0.01 pfu/cell. Virus was harvested from the medium at the indicated time points and titers determined by plaque assay on Vero cells. Symbols represent the mean titer and standard deviation for each time point. The limit of detection (LOD) was 0.7 log10pfu/0.1ml of supernatant of infected cell cultures; in cases where the sample had no detectable virus, a titer of 0.65 log10pfu/0.1ml was used for calculations. (B) Cx. pipiens mosquitoes were inoculated intrathoracically at 10× the ID50 of WT (5 pfu per mosquito) or SP (21 pfu per mosquito) with Vero cell amplified WT or SP stock virus, and were maintained at the indicated temperatures. Five mosquitoes per virus were harvested immediately following inoculation, and 10 mosquitoes were harvested at each time point thereafter. Viral titers in the mosquito bodies were determine by plaque titration on Vero cells. Symbols represent the mean viral titer and standard deviation for each time point. The limit of detection (LOD) was 0.7 log10pfu/mosquito, and in cases where the sample had no detectable virus, a titer of 0.65 log10pfu/mosquito was used for calculations.
In vivo growth kinetics in Cx. pipiens
The ID50 values of WT and SP WNV following intrathoracic (IT) inoculation of Cx. pipiens were not significantly different: 0.5 pfu /mosquito and 2.1 pfu/mosquito, respectively. Growth analyses were performed by inoculation of mosquitoes with ten times the ID50 of each virus to ensure that all mosquitoes became infected. SP grew more slowly than WT in mosquitoes at 15°C, 27°C and 32°C (Fig 3B), in contrast to the in vitro results in the mosquito cell line. The mean virus titer of SP at 27°C was significantly lower than that of WT at all time points from 2 to 28 days post-inoculation (dpi) (P<0.01, t-test). Peak titers, 6.0 and 7.4 log10 pfu/mosquito for SP and WT, respectively, were reached after 14 dpi. At 32°C, rates of replication of both SP and WT were accelerated compared to those at 27°C, and peak titers, which were similar to those found at 27°C, were reached earlier, by 6 dpi. At 15°C, WT and SP viruses replicated more slowly than at the higher temperatures (Fig 3B). WT replication, undetectable until 7 dpi, reached a mean titer of 6.3 log10pfu/mosquito on day 84 pi; this titer was still lower than the peak titers at 27°C and 32°C (7.4 log10 pfu/mosquito). Virus was not detected in the majority of SP-infected mosquitoes until 21 dpi; the maximum titer of 2.8 log10pfu/mosquito at 84 dpi was significantly lower than both the WT titer at 15°C and the SP titers at 27°C and at 32°C.
SP began to exhibit a mixed plaque phenotype in some individual mosquitoes after 2, 5, and 28 dpi at 32°C, 27°C, and 15°C, respectively, but the proportion of mosquitoes in which the phenotype reverted to WT, did not vary significantly among the temperatures examined (Table 1). At each time point, mosquitoes infected with SP that exhibited mixed plaque phenotype usually had higher virus titers than mosquitoes whose SP infection retained the small-plaque phenotype (data not shown).
Table 1.
Phenotypic reversion of SP variant of WNV recovered from infected Culex pipiensa
| Incubation temperature | No. virus-positive mosquitoesb | No. mosquitoes with virus that displayed a mixed-plaque phenotypec (% of virus-positive mosquitoes) |
|---|---|---|
| 15°C | 62 | 9 (14.5%) |
| 27°C | 105 | 14 (13.3%) |
| 32°C | 92 | 18 (19.5%) |
Mosquitoes were inoculated intrathoracically with Vero cell amplified SP virus at 21 pfu per mosquito, and maintained at the indicated temperatures
Number of SP-infected mosquitoes with detectable virus, evaluated by plaque assay on Vero cells (includes mosquitoes harvested at various times post inoculation)
SP virus recovered from mosquitoes had a mixed-plaque phenotype when 10 to 70% of its plaques were greater than 1.0 mm in diameter, measured at 72 hpi on Vero cells
Vector competence of Cx. pipiens
The vector competence of SP in Cx. pipiens was compared to that of WT. At both day 7 and day 14 following peroral infection, the percentage of mosquitoes infected by feeding on SP was significantly lower than for WT (P<0.01), and there was no significant increase in the rate of infection from day 7 to day 14 post-feeding (PF)(Table 2). Dissemination and transmission rates of SP-infected mosquitoes were similar to the rates for WT at day 7, but they were significantly lower than the rates for WT at day 14, resulting in a greater proportion of WT-infected mosquitoes with disseminated virus (71% vs. 12%; p<0.01) and transmitting virus (51% vs. 9%; p<0.01) than those infected with SP. The higher infection, dissemination, and transmission rates for WT were reflected in the titers of virus recovered from bodies of infected mosquitoes (Fig 4). Mean body titers were significantly greater for WT than for SP at both 7 and 14 days PF (P<0.01), and mean titers for both WT- and SP-infected mosquitoes increased significantly from day 7 to day 14. Mosquitoes with disseminated infections generally had higher body titers, although there were exceptions among both SP- and WT-infected individuals.
Table 2.
Vector Competence of SP and WT strains of WNV in Culex pipiens
| Virus | Day PFa | No. fed b | No. infected (%)c | No. disseminated (%)d | No. transmitting (%)e |
|---|---|---|---|---|---|
| SP | 7 | 50 | 21 (42)f | 3 (14)f | 1 (5)f |
| 14 | 65 | 34 (52)f | 7 (21)f | 3 (9)f | |
| WT | 7 | 50 | 36 (72)g | 6 (17)f | 1 (3)f |
| 14 | 41 | 35 (85)g | 25 (71)g | 18 (51)g |
Day post-feeding with the C6/36 cell amplified WT or SP virus, oral dose of 108.4 pfu/ml in blood meal.
Number of fully engorged mosquitoes
Number of mosquitoes with detectable virus in their bodies (expressed as percentage of those fed)
Number of mosquitoes with detectable virus in their legs (expressed as percentage of those infected)
Number of mosquitoes secreting virus in saliva (expressed as percentage of those infected)
Within each column, any two values followed by the same letter are not significantly different from each other (P<0.01).
Figure 4.
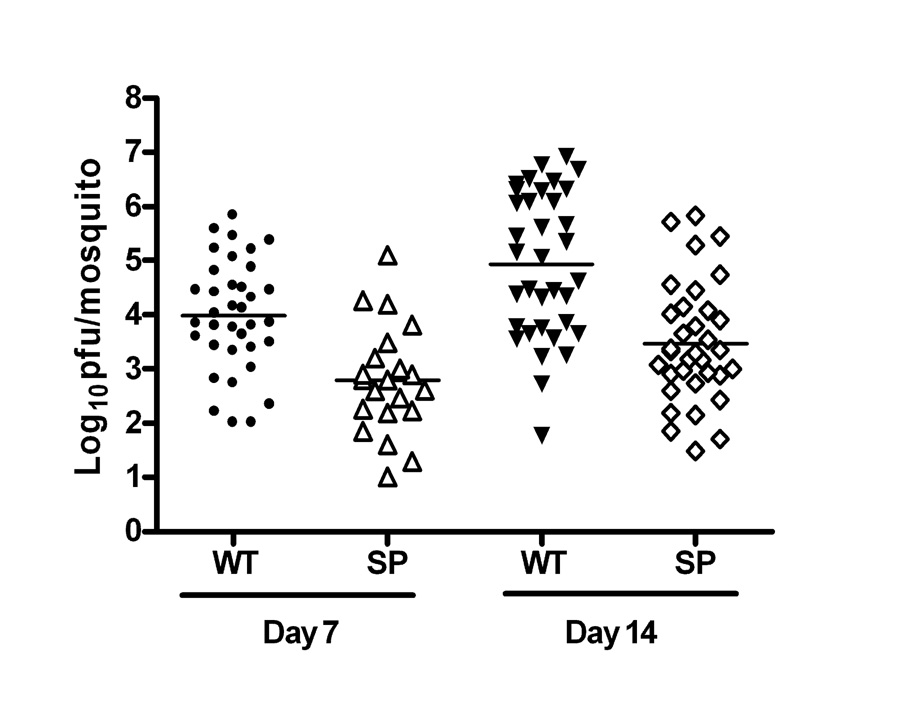
Virus titers of wild type (WT) and small plaque (SP) variant WNV in Cx. pipiens bodies at 7 and 14 days after oral infection with the C6/36 cell amplified virus. Mosquitoes were fed on defibrinated goose blood containing 108.4 pfu/ml of either WT or SP virus, incubated at 27°C, and harvested at 7 or 14 days post-feeding (PF). Virus recovered from each mosquito body was titered on Vero cells; each symbol represents the titer from an individual mosquito. The mean titer of SP was significantly less than WT at both 7 and 14 days PF by t-test (P7 = 0.001; P14=0.001). For both SP and WT the mean titer at 7 day PF was significantly lower than at 14 day PF by t-test (PSP=0.012; PWT=0.001).
Comparison of WT and SP sequences
The complete genome of the SP variant was sequenced and compared to the consensus sequence of the parental WT in order to identify mutations that could be responsible for the observed phenotypic differences. Four nucleotide differences between SP and WT were detected. Two (C1599U and C9660U) of these nucleotide changes were translationally silent, while two (C625U and U3707C) resulted in amino acid changes: a proline to serine change at position 54 (P54S) in the pr region of prM and a valine to alanine change at position 61 (V61A) in the NS2A protein (Table 3).
Table 3.
Nucleotide and amino acid sequence changes for WNV SP variant, relative to the parental WT sequence
| Nucleotide | Nucleotide | |||
|---|---|---|---|---|
| Gene/Region | |
AA Change | ||
| Position | WTa | SPb | ||
| 625 | Pr-54 | C | U | Proline→Serine |
| 1599 | E211 | C | U | |
| 3707 | NS2A-61 | U | C | Valine→Alanine |
| 9660 | NS5 | C | U | |
WT: WNV 3356K isolated from the kidney of an American crow collected in New York in 2000, and amplified twice in Vero cells
SP: A small plaque variant (SP), isolated from the first Vero amplified WT stock, followed by 2 sequential rounds of plaque purification and two rounds of amplification in Vero cells.
We also analyzed the prM and NS2A sequences of 70 individual plaques of mosquito- and cell culture-derived SP viruses that exhibited phenotypic reversion. From the mosquito-derived viruses, we picked 30 large plaques and 30 small plaques; however, from the cell culture-derived viruses only large plaques were selected as there were no small plaques available for analysis after incubation at 42.5°C). All large plaques from mosquitoes held at 27°C and 32°C, as well as the large plaques from the cell culture-derived viruses, had reverted from the SP sequence to the WT sequence at prM 625 (Table 4); however, half of the small plaques from these mosquitoes also contained the WT sequence at prM 625. Both large and small plaques from one mosquito held at 15°C had reverted to the WT sequence at prM 625. Three large and two small plaques from a second mosquito held at 15°C had reverted to the WT sequence at NS2A 3707 (Table 4), while one large plaque had reverted at both positions. Four plaques from this mosquito, including one large plaque, contained the SP sequence at both prM 625 and NS2A 3707.
Table 4.
Sequence analysis of revertant viruses derived from SP-infected mosquitoes or cells
| Source of virus | Incubation temperature | Plaques picked | prM 625 (U to C) reversion | NS2A 3707 (C to U) reversion |
|---|---|---|---|---|
| 2 Mosquitoes | 15°C | 10 lpa | 6c | 4c |
| 10 spb | 5 | 2 | ||
| 2 Mosquitoes | 27°C | 10 lp | 10 | 0 |
| 10 sp | 4 | 0 | ||
| 2 Mosquitoes | 32°C | 10 lp | 10 | 0 |
| 10 sp | 5 | 0 | ||
| Vero | 42.5°C | 5 lp | 5 | 0 |
| DF-1 | 42.5°C | 5 lp | 5 | 0 |
Ten-fold serial dilutions of virus recovered from SP infected mosquitoes and Vero or DF-1 cells were plated on Vero cell monolayers in 6-well plates. Well-isolated plaques were picked from the wells containing only 1 or 2 plaques at 72 hpi.
lp: Plaque diameter equal to or larger than 1mm
sp: Plaque diameter smaller than 1mm
Includes one large plaque with double reversion
Discussion
Flaviviruses frequently demonstrate mixed-plaque morphology in cell culture (Butrapet et al., 2000; Davis et al., 2004). Experimentally mutagenized flaviviruses have been found to yield small plaque variants that are ts in cell culture (Eckels et al., 1976) and attenuated in mice (Blaney et al., 2001, 2002, 2003; Hanley et al., 2002; Liu et al., 2006). In this study, SP, a small-plaque viral variant, was selected from the mixed-plaque population of WNV isolated from an American crow collected in NY in 2000. Its replication efficiency in Vero cells was significantly lower than that of WT, and this difference was retained across a wide temperature range. In avian cell lines, SP growth was equal to or slower than WT growth at low temperatures, but at temperatures above 39°C, SP exhibited temperature sensitivity. Both viruses grew equally well in the mosquito cell line, C6/36. Thus, it was surprising to observe that both IT and peroral infection of Cx. pipiens mosquitoes resulted in significantly lower virus titers and infection rates for SP than for WT. Interestingly, although other small-plaque WNV strains have demonstrated a ts phenotype in Vero cell culture (Davis et al., 2004; Wicker et al., 2006), the SP variant characterized here did not.
WNV is maintained in a transmission cycle primarily between ornithophilic mosquitoes and passeriform birds. Worldwide, Culex spp. are considered to be the primary vectors for WNV transmission; members of the Cx. pipiens complex are the principal vectors of WNV in northeastern and north central US. The SP variant described here was attenuated in Cx. pipiens following both parenteral and peroral infection, and it exhibited lower dissemination and transmission rates at 14 days following feeding as compared to WT. These results suggest that SP has a defect that blocks cell-to-cell spread. Future studies are planned using the SP variant as a tool to help elucidate the determinants of WNV spread in vivo.
The observation that WNV exists as a genetically diverse population in its natural mosquito and avian hosts (Jerzak et al., 2005) is most likely a consequence of the high mutation rate resulting from the error-prone RNA-dependent RNA polymerase (Domingo et al., 2005). In studies of other flaviviruses where mutations were chemically induced, the small-plaque phenotypes did not appear to be stable with in vitro passaging, but rather demonstrated high rates of reversion. More than 2/3 of ts and/or small-plaque mutants of DENV-4 lost these phenotypes after two rounds of biological cloning (Blaney et al., 2002); similarly, >80% of JEV clones lost the ts phenotype during three rounds of plaque purification (Eastman & Blair, 1985). The WNV SP variant described here was stable through three rounds of plaque purification and two amplification rounds that were carried out before the current experiments were performed. During subsequent growth assays, the small-plaque phenotype was retained in all cell lines except Vero and DF-1 grown at temperatures at or above 41°C. The correlation of the change to a mixed plaque size phenotype with in vitro growth under the stressful condition of high temperature suggests a selective advantage for the larger-plaque phenotype. It was interesting that larger plaques were not generated when SP was grown in PDE cells at 41°C; the lack of change in plaque phenotyope may explain why no virus growth was detected in this cell culture when the temperature was raised to 42.5°C. It is not clear why SP reverted to a mixed-plaque phenotype in DF-1 but not in PDE, since both cell lines are of avian origin. In contrast to the in vitro results, phenotypic reversion to WT plaque sizes was observed at all incubation temperatures, from 15°C to 32°C, after infection of live mosquitoes with SP. SP virus that exhibited a mixed plaque phenotype after recovery from mosquitoes often had titers similar to those of WT, although phenotypic reversion of SP in vitro did not appear to restore replication efficiency to WT levels.
Small plaque variants commonly are found in natural populations. We have isolated seven additional small plaque variants from other WNV isolates collected in NY from 2000–2004, each having mutations different from those detected in the particular SP variant characterized in this study (data not shown). Using site-directed mutagenesis, Davis et al. demonstrated that a mutation in the NS4B protein (E249G) in combination with either a mutation in the NS5 protein (A804V) or three mutations in the 3′ UTR (A10596G, C10774U, A10799G) were sufficient to produce the small plaque, ts, and attenuated phenotypes exhibited by six viruses isolated in Texas in 2003 (Davis et al., 2004; Davis et al., 2007). Other work has shown that WNV containing only the E249G mutation in NS4B exhibited smaller plaques, slower growth kinetics, and decreased RNA synthesis as compared to the wild-type virus (Puig-Basagoiti et al, 2007). These data illustrate that different mutations in the WNV genome can produce virus variants with small plaque phenotypes.
The SP sequence contained four nucleotide changes from the sequence of WT. Two of these changes resulted in amino acid substitutions, a P54S change in the prM protein and a V61A change in the NS2A protein. The prM protein plays a crucial role in virus assembly and transportation and exhibits a chaperone-like activity critical for proper folding and maturation of the viral E protein (Brinton, 2002). A deletion at the cleavage site of prM of TBEV did not influence virus replication and release from infected BHK cells, but the progeny virus lost the ability to infect new BHK cells; infectivity was restored by the addition of trypsin (Elshuber et al., 2003). A different mutation in prM of TBEV reduced particle secretion, but did not affect envelope protein folding or the chaperone-like activity of prM (Yoshii et al., 2004). NS2A is a small hydrophobic protein that plays an important role in virus replication (Brinton, 2002) and is a major inhibitor of interferon promoter-driven transcription (Liu et al., 2005). A single amino acid substitution in the NS2A (A30P) region of a Kunjin isolate (WNVKUN) resulted in attenuation of both neurovirulence and neuroinvasiveness (Liu et al., 2006).
Sequencing of the SP mixed plaque population revealed that 35 out of 40 large plaques from mosquitoes and cells had reverted to the WT sequence at prM 625, three reverted to the WT sequence at NS2A 3707, and one reverted at both positions (Table 4). These data suggest that the prM mutation in SP may contribute to its small plaque phenotype, although the NS2A mutation may also be involved. However, 14 out of 30 small plaques also reverted at prM 625 and two reverted at NS2A 3707, indicating that genotypic reversion at either of these positions is not sufficient to restore the WT plaque size phenotype. In addition, as the entire genomes of the plaque-purified viruses were not sequenced, we cannot evaluate the potential contributions of any other sequence changes that may be present. Studies are currently underway to determine the relative contributions of the prM and NS2A amino acid changes to the SP phenotypes observed here; this information should enable the potential mechanism of attenuation of the SP variant to be clarified.
Materials and Methods
Viruses and Cells
African green monkey kidney cells (Vero, ATCC #CCL-81) were grown in minimal essential medium (MEM; Gibco, Invitrogen Corp., Carlsbad CA) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT), 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 100 U/ml of penicillin and 100µg/ml of streptomycin. Aedes albopictus cells (C6/36; ATCC #CRL-1660) were grown in MEM supplemented with 10% FBS, 2mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids, 100 U/ml of penicillin and 100 µg/ml of streptomycin. Chicken embryo fibroblasts (DF-1; ATCC #CRL-12203) were grown in Dulbecco’s Modified Eagle's medium (DMEM; ATCC, Manassas, VA) with 10% FBS, 4mM L-glutamine, 4.5g/L glucose, 1.5 g/L sodium bicarbonate, 100 U/ml of penicillin and 100µg/ml of streptomycin. Peking duck embyro fibroblasts (PDE; ATCC #CCL-141) were grown in MEM supplemented with 10% FBS, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acids, 1.0 mM sodium pyruvate, 100 U/ml of penicillin and 100µg/ml of streptomycin.
The WT WNV strain 3356K was isolated in Vero cell culture from the kidney of an American crow (Corvus brachyrhynchos) collected in New York in 2000 and passed twice in Vero cells to amplify the virus. The SP variant was selected from the first Vero cell passage, and then subjected to two sequential rounds of plaque purification (total of three plaque-to-plaque transfers), followed by two rounds of amplification in Vero cells. The cell culture supernatants from the second round Vero cell amplification of WT and SP were clarified by centrifugation at 3220×g for 10 min, supplemented with 20% FBS, and aliquots were stored at −80°C as virus stocks. All in vitro experiments and in vivo mosquito inoculation studies were performed using the second round Vero-amplified virus stocks. For the mosquito vector competence studies, second round Vero-amplified virus stocks of WT and SP were amplified once in C6/36 cells at a multiplicity of infection (MOI) of 0.01 plaque forming units (pfu)/cell, based on Vero cell titer, and were harvested at 96 hrs post-infection, as described above.
Mosquitoes
Culex pipiens were originally collected as egg rafts in Pennsylvania in 2004 by M Hutchinson (Pennsylvania Department of Environmental Protection) and were colonized at the Arbovirus Laboratories, Wadsworth Center. Mosquitoes were maintained as previously described (Ebel et al., 2005) in 12”×12”×12” cages at 27°C with 16:8 hours light:dark photoperiod and provided 10% sucrose ad lib.
Measurement of Plaque Size
WT and SP viral stocks were inoculated onto Vero cells in 6-well culture plates as described previously (Payne et al., 2006). After staining with neutral red, plaques were examined at 72 hours post-infection using a Zeiss Stemi 2000-C stereo microscope, and images were captured with a Zeiss Axiocam MRC digital camera. Plaque sizes were measured using AxioVision 3.0 software (Zeiss, Germany). For analysis, 200 individual plaques were measured for each virus.
In vitro growth kinetics
Vero, C6/36, DF-1, and PDE cells were seeded in Costar 6-well culture cluster plates (Corning Incorporated, Corning, NY) at 37°C, 28°C, 39°C, and 37°C respectively, in 5% CO2. Confluent cell monolayers were inoculated with virus at an MOI of 0.01, based on Vero cell titer. After 1 hr of viral adsorption, the inoculum was removed and the monolayers were washed and overlayed with 3 ml of the appropriate maintenance medium containing 2% FBS. Vertebrate cells were incubated at each of the following temperatures: 30°C, 34°C, 37°C, 40°C (39°C for DF-1 and PDE), 41°C, and 42.5°C (Vero, DF-1, PDE). Mosquito cells were incubated at 26°C, 28°C, 30°C, and 34°C. The temperatures for virus adsorption were the same as the incubation temperature used subsequently in all growth curve experiments. Samples were harvested at 12 (for PDE), 24, 36, 48, 72, 96, and 120 hrs post-infection into media containing 20% FBS and were stored at −80°C. All growth curves were performed using triplicate wells. Plaque assays were conducted as described above. Viral titers were calculated and expressed as log10pfu/0.1ml.
Determination of mosquito ID50
Four groups (n=20 each) of six-day old Cx. pipiens mosquitoes were inoculated IT essentially as previously described (Rosen & Gubler, 1974) with approximately 10, 1, 0.1 or 0.01 pfu of either SP or WT. Inoculated mosquitoes were maintained at 27°C, with a 16:8 light:dark photoperiod, and were provided with a 10% sucrose solution ad lib on cotton wicks. On day 7 post-inoculation, mosquitoes were individually frozen in 1.0 ml of mosquito diluent (MD: PBS with 20% FBS, 100units/ml penicillin, 100µg/ml streptomycin, 0.25µg/ml fungizone, and 10µg/mL gentamicin) for subsequent trituration and screening on Vero cell culture to determine infection rates at each dose.
In vivo growth kinetics in Cx. pipiens
Six-day old Cx. pipiens mosquitoes were inoculated IT with WT or SP at ten times the ID50 and maintained at 15°C, 27°C or 32°C, as described above. Immediately following inoculation, 5 mosquitoes were placed individually in 1.0 ml mosquito diluent and stored at −80°C for subsequent trituration and titration on Vero cells. At each subsequent timepoint, 10 mosquitoes per virus strain were harvested, as described above.
Vector competence of Cx. pipiens
Four- to six-day old female Cx. pipiens mosquitoes were deprived of sucrose solution for approximately 44 hrs and then allowed to feed for one hr on gauze pledgets soaked with defibrinated goose blood containing 108.4 pfu/ml of either WT or SP virus supplemented with 2.5% sucrose. Fully engorged mosquitoes were removed to pint cartons, incubated at 27°C under a 16:8 light:dark photoperiod, and provided a 10% sucrose solution ad lib on cotton wicks.
After 7 and 14 days post feeding (PF), mosquito salivary secretions were collected in capillary tubes charged with 50% sucrose plus fetal bovine serum (1:1), as previously described (Ebel et al., 2005) to estimate transmission rates. Bodies and legs were placed in separate microfuge tubes with 1.0 ml mosquito diluent, homogenized using a rapid high speed mechanical homogenizer (Mixer Mill MM300; Qiagen, Valencia, CA) for 30 s at 24 cycles/s, and clarified by centrifugation (4 min at 5796 × g). Bodies, legs, and salivary secretions were screened or titrated in Vero cell culture to determine infection, dissemination, and transmission rates, respectively.
Genome sequencing and analysis
Nucleotide sequencing of the full genome of SP and WT was conducted as described (Davis et al., 2005). Briefly, viral RNA was extracted directly from 100 µl of the second round Vero-amplified virus stocks using the QiaAMP viral RNA extraction kit (Qiagen,Valencia, CA). The genomic RNA was reverse transcribed and amplified in 16 overlapping fragments using one-step RT-PCR (Qiagen, Valencia, CA). RT-PCR products were electrophoretically separated on a 1.5% agarose gel, and bands of the appropriate size were excised and gel purified using the QiaQuick Gel Extraction Kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. Full-length sequencing was carried out using multiple pairs of overlapping primers and was performed using an ABI 373 or 377 automated sequencer according to the manufacturer protocols. A comprehensive description of the amplification and sequencing primers is available from the corresponding author.
Sequences were compiled and edited using the SeqMan module of the DNAStar software package (DNAStar, Inc., Madison, WI) with a minimum of two-fold redundancy throughout for sequence data to be considered complete. Sequences were aligned using MegAlign within DNAStar, and changes between the SP variant and WT sequence were documented.
Sequence analysis of revertant SP virus from mosquitoes and cells
Virus derived from SP infected mosquitoes by IT inoculation (two mosquitoes from each incubation temperature) or harvested from Vero or DF-1 cells incubated at 42.5°C with reverted plaque phenotype, were plated on Vero cells. Ten plaques (five large and five small plaques) were picked from each mosquito-derived virus, and five large plaques were picked from cell culture-derived viruses. Viral RNA extraction, RT-PCR, sequencing, and sequence analysis were conducted as described above.
Acknowledgments
The authors would like to acknowledge Pamela Chin for overseeing mosquito rearing and the excellent technical assistance provided by Amy Lovelace, Alexander Ciota, Linda Styer, and Ann Payne, as well as other members of Arbovirus Laboratories, in assisting with cell culture work. We thank the Wadsworth Center Media and Tissue Culture Facility for providing cells and media for this work and the Wadsworth Center Sequencing Core for providing sequencing data. This work was supported partially by federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health (contract number NO1-AI-25490).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin RJ, Whiting TL, Anderson RA, Drebot MA. An outbreak of West Nile virus-associated disease in domestic geese (Anser anser domesticus) upon initial introduction to a geographic region, with evidence of bird to bird transmission. Canadian Veterinary Journal. 2004;45:117–123. [PMC free article] [PubMed] [Google Scholar]
- Beasley DW, Davis CT, Guzman H, Vanlandingham DL, Travassos da Rosa APA, Parsons RE, Higgs S, Tesh RB, Barrett ADT. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology. 2003;309:190–195. doi: 10.1016/s0042-6822(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Blaney JE, Jr, Johnson DH, Manipon GG, Firestone CY, Hanson CT, Murphy BR, Whitehead SS. Genetic basis of attenuation of dengue virus type 4 small plaque mutants with restricted replication in suckling mice and in SCID mice transplanted with human liver cells. Virology. 2002;300:125–139. doi: 10.1006/viro.2002.1528. [DOI] [PubMed] [Google Scholar]
- Blaney JE, Jr, Manipon GG, Murphy BR, Whitehead SS. Temperature sensitive mutations in the genes encoding the NS1, NS2A, NS3, and NS5 nonstructural proteins of dengue virus type 4 restrict replication in the brains of mice 4. Archives of Virology. 2003;148:999–1006. doi: 10.1007/s00705-003-0007-y. [DOI] [PubMed] [Google Scholar]
- Blaney JE, Jr, Johnson DH, Firestone CY, Hanson CT, Murphy BR, Whitehead SS. Chemical Mutagenesis of Dengue Virus Type 4 Yields Mutant Viruses Which Are Temperature Sensitive in Vero Cells or Human Liver Cells and Attenuated in Mice. Journal of Virology. 2001;75:9731–9740. doi: 10.1128/JVI.75.20.9731-9740.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich BJ, Fernandez-Salas I, Contreras-Cordero JF, Marlenee NL, Gonzalez-Rojas JI, Komar N, Gubler DJ, Calisher CH, Beaty BJ. Serological evidence of West Nile virus infection in horses, Coahuila State, Mexico. Emerging Infectious Diseases. 2003;9:853–856. doi: 10.3201/eid0907.030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA. The molecular biology of West Nile virus: a new invader of the Western Hemisphere. Annual Review of Microbiology. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Butrapet S, Huang CY, Pierro DJ, Bhamarapravati N, Gubler DJ, Kinney RM. Attenuation markers of a candidate dengue type 2 vaccine virus, strain 16681 (PDK-53), are defined by mutations in the 5' noncoding region and nonstructural proteins 1 and 3. Journal of Virology. 2000;74:3011–3019. doi: 10.1128/jvi.74.7.3011-3019.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Nickells M. Neuroadapted yellow fever virus 17D: genetic and biological characterization of a highly mouse-neurovirulent virus and its infectious molecular clone. Journal of Virology. 2001;75:10912–10922. doi: 10.1128/JVI.75.22.10912-10922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CT, Beasley DW, Guzman H, Siirin M, Parsons RE, Tesh RB, Barrett AD. Emergence of attenuated West Nile virus variants in Texas, 2003. Virology. 2004;330:342–350. doi: 10.1016/j.virol.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett AD. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Davis CT, Galbraith SE, Zhang S, Whiteman MC, Li L, Kinney RM, Barrett AD. A combination of naturally occurring mutations in north american west nile virus nonstructural protein genes and in the 3' untranslated region alters virus phenotype. Journal of Virology. 2007;81:6111–6116. doi: 10.1128/JVI.02387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Escarmis C, Lazaro E, Manrubia SC. Quasispecies dynamics and RNA virus extinction. Virus Research. 2005;107:129–139. doi: 10.1016/j.virusres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Dupuis AP, Marra PP, Reitsma R, Jones MJ, Louie KL, Kramer LD. Serologic evidence for West Nile virus transmission in Puerto Rico and Cuba. American Journal of Tropical Medicine and Hygiene. 2005;73:474–476. [PubMed] [Google Scholar]
- Eastman PS, Blair CD. Temperature-sensitive mutants of Japanese encephalitis virus. Journal of Virology. 1985;55:611–616. doi: 10.1128/jvi.55.3.611-616.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000–2003. American Journal of Tropical Medicine and Hygiene. 2004;71:493–500. [PubMed] [Google Scholar]
- Ebel GD, Rochlin I, Longacker J, Kramer LD. Culex restuans (Diptera:culicidae) relative abundance and vector competence for West Nile virus. Journal of Medical Entomology. 2005;42:838–843. doi: 10.1603/0022-2585(2005)042[0838:CRDCRA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Eckels KH, Brandt WE, Harrison VR, McCown JM, Russell PK. Isolation of a temperature-sensitive dengue-2 virus under conditions suitable for vaccine development. Infect Immun. 1976;14:1221–1227. doi: 10.1128/iai.14.5.1221-1227.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- Hanley KA, Lee JJ, Blaney JE, Murphy BR, Whitehead SS. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. Journal of Virology. 2002;76:525–531. doi: 10.1128/JVI.76.2.525-531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. Journal of General Virology. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, Hall RA, MacKenzie JS, Cropp CB, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. Journal of Virology. 2006;80:2396–2404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of 21 West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. Journal of Virology. 2005;79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickells M, Chambers TJ. Neuroadapted yellow fever virus 17D: determinants in the envelope protein govern neuroinvasiveness for SCID mice. Journal of Virology. 2003;77:12232–12242. doi: 10.1128/JVI.77.22.12232-12242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay. Journal of Virological Methods. 2006;134:183–187. doi: 10.1016/j.jviromet.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Puig-Basagoiti F, Tilgner M, Bennett CJ, Zhou Y, Munoz-Jordan JL, Garcia-Sastre A, Bernard KA, Shi PY. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology. 2007;361:229–241. doi: 10.1016/j.virol.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L, Gubler D. The use of mosquitoes to detect and propagate dengue viruses. American Journal of Tropical Medicine and Hygiene. 1974;23:1153–1160. doi: 10.4269/ajtmh.1974.23.1153. [DOI] [PubMed] [Google Scholar]
- Rumyantsev AA, Murphy BR, Pletnev AG. A tick-borne Langat virus mutant that is temperature sensitive and host range restricted in neuroblastoma cells and lacks neuroinvasiveness for immunodeficient mice. Journal of Virology. 2006;80:1427–1439. doi: 10.1128/JVI.80.3.1427-1439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiiri K, Klinkenberg B, Mak S, Kazmi J. Predicting outbreaks: a spatial risk assessment of West Nile virus in British Columbia. International Journal of Health Geographics. 2006;5:21. doi: 10.1186/1476-072X-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaycheva L, Nickells M, Droll DA, Chambers TJ. Yellow fever 17D virus: pseudo-revertant suppression of defective virus penetration and spread by mutations in domains II and III of the E protein. Virology. 2004;327:41–49. doi: 10.1016/j.virol.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Vlaycheva L, Nickells M, Droll DA, Chambers TJ. Neuroblastoma cell-adapted yellow fever virus: mutagenesis of the E protein locus involved in persistent infection and its effects on virus penetration and spread. J Gen Virol. 2005;86:413–421. doi: 10.1099/vir.0.80314-0. [DOI] [PubMed] [Google Scholar]
- Vlaycheva LA, Chambers TJ. Neuroblastoma cell-Adapted yellow fever 17D virus: characterization of a viral variant associated with persistent infection and decreased virus spread. Journal of Virology. 2002;76:6172–6184. doi: 10.1128/JVI.76.12.6172-6184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker JA, Whiteman MC, Beasley DW, Davis CT, Zhang S, Schneider BS, Higgs S, Kinney RM, Barrett AD. A single amino acid substitution in the central portion of the West Nile virus NS4B protein confers a highly attenuated phenotype in mice. Virology. 2006;349:245–253. doi: 10.1016/j.virol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Wu SC, Lian WC, Hsu LC, Liau MY. Japanese encephalitis virus antigenic variants with characteristic differences in neutralization resistance and mouse virulence. Virus Research. 1997;51:173–181. doi: 10.1016/s0168-1702(97)00098-1. [DOI] [PubMed] [Google Scholar]
- Yoshii K, Konno A, Goto A, Nio J, Obara M, Ueki T, Hayasaka D, Mizutani T, Kariwa H, Takashima I. Single point mutation in tick-borne encephalitis virus prM protein induces a reduction of virus particle secretion. Journal of General Virology. 2004;85:3049–3058. doi: 10.1099/vir.0.80169-0. [DOI] [PubMed] [Google Scholar]


