Abstract
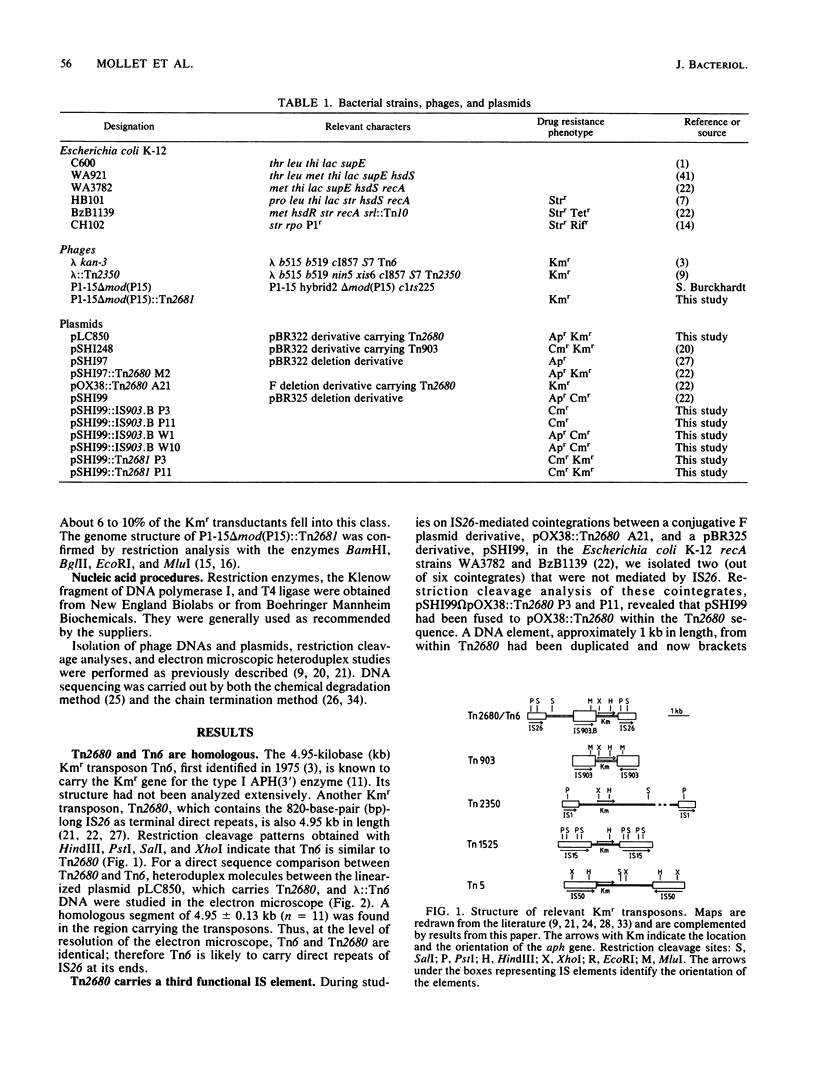
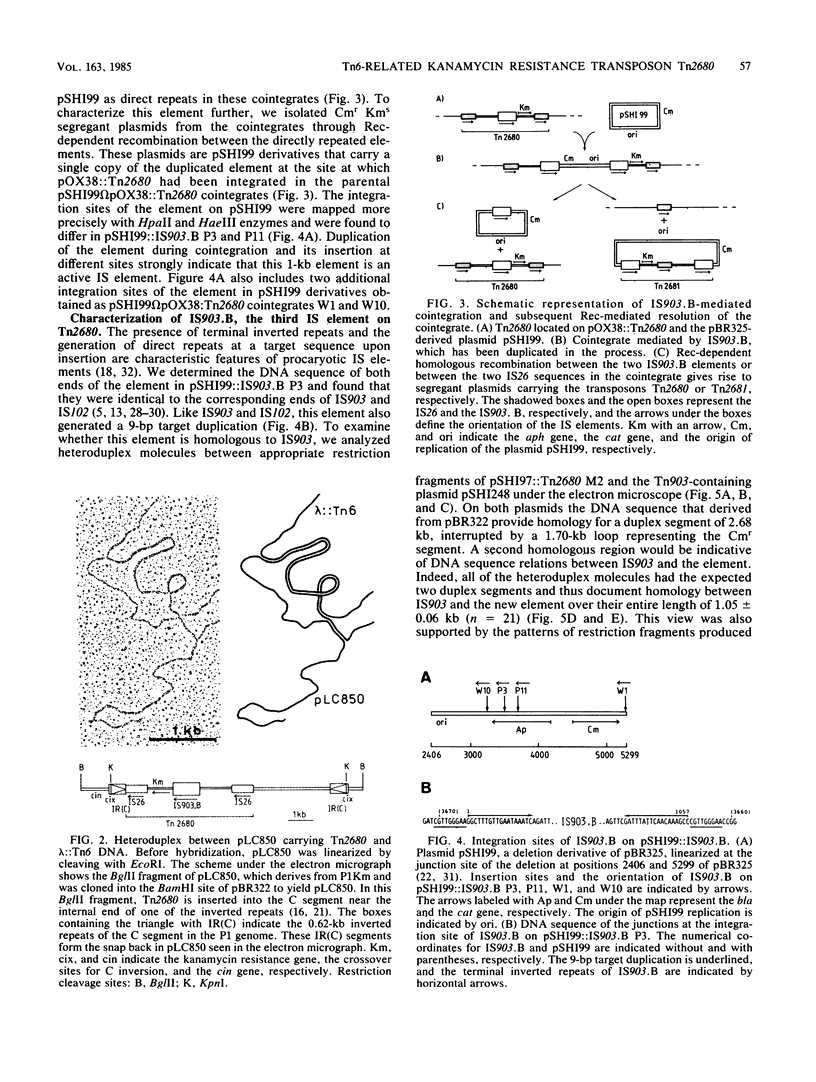
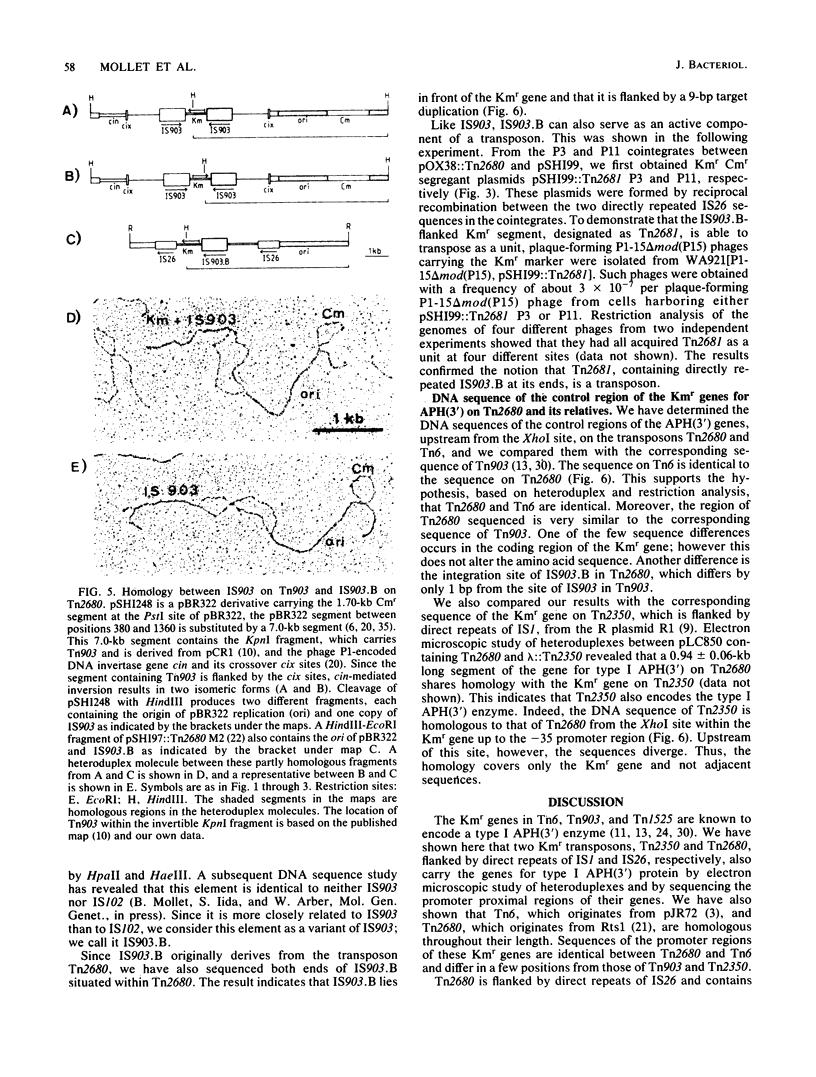
The kanamycin resistance transposon Tn2680, which originates from the R plasmid Rts1, is homologous to Tn6 and carries two directly repeated copies of IS26, one at each end. The kanamycin resistance gene codes for type I aminoglycoside-3'-phosphotransferase. Tn2680 also contains, in the middle of the transposon, an additional IS element homologous to IS903. This element, designated IS903.B, is flanked by a 9-base-pair direct target duplication. A novel kanamycin resistance transposon. Tn2681, can be generated from Tn2680 by IS903.B-mediated cointegration and subsequent reciprocal recombination between the directly repeated IS26 sequences. Tn2681 carries a single IS26 element in the middle of the transposon and is flanked by two directly repeated copies of IS903.B. Possible evolutionary relationships between Tn2680 and other kanamycin resistance transposons such as Tn903 and Tn2350 are discussed, based on the gene organization and DNA sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Egner C., Hirschel B. J., Howard J., Johnsrud L., Jorgensen R. A., Tlsty T. D. Insertion, excision, and inversion of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):115–123. doi: 10.1101/sqb.1981.045.01.020. [DOI] [PubMed] [Google Scholar]
- Bernardi A., Bernardi F. Complete sequence of an IS element present in pSC101. Nucleic Acids Res. 1981 Jun 25;9(12):2905–2911. doi: 10.1093/nar/9.12.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Clerget M. A 140 base-pair DNA segment from the kanamycin resistance region of plasmid R1 acts as an origin of replication and promotes site-specific recombination. J Mol Biol. 1984 Sep 5;178(1):35–46. doi: 10.1016/0022-2836(84)90229-8. [DOI] [PubMed] [Google Scholar]
- Clerget M., Chandler M., Caro L. Isolation of an IS1 flanked kanamycin resistance transposon from R1drd19. Mol Gen Genet. 1980;180(1):123–127. doi: 10.1007/BF00267360. [DOI] [PubMed] [Google Scholar]
- Coggins L. W., McCluskey E. Characterization of the ColE1-Km plasmids pCR1 and pCR11 by electron microscope and restriction endonuclease mapping. Plasmid. 1979 Jul;2(3):446–453. doi: 10.1016/0147-619x(79)90028-3. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Joyce C. M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänni C., Meyer J., Iida S., Arber W. Occurrence and properties of composite transposon Tn2672: evolution of multiple drug resistance transposons. J Bacteriol. 1982 Jun;150(3):1266–1273. doi: 10.1128/jb.150.3.1266-1273.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology. 1984 Apr 30;134(2):421–434. doi: 10.1016/0042-6822(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Arber W. Genesis and natural history of IS-mediated transposons. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):27–43. doi: 10.1101/sqb.1981.045.01.006. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Bächi B., Stålhammar-Carlemalm M., Schrickel S., Bickle T. A., Arber W. DNA restriction--modification genes of phage P1 and plasmid p15B. Structure and in vitro transcription. J Mol Biol. 1983 Mar 25;165(1):1–18. doi: 10.1016/s0022-2836(83)80239-3. [DOI] [PubMed] [Google Scholar]
- Iida S., Meyer J., Kennedy K. E., Arber W. A site-specific, conservative recombination system carried by bacteriophage P1. Mapping the recombinase gene cin and the cross-over sites cix for the inversion of the C segment. EMBO J. 1982;1(11):1445–1453. doi: 10.1002/j.1460-2075.1982.tb01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Meyer J., Linder P., Goto N., Nakaya R., Reif H. J., Arber W. The kanamycin resistance transposon Tn2680 derived from the R plasmid Rts1 and carried by phage P1Km has flanking 0.8-kb-long direct repeats. Plasmid. 1982 Sep;8(2):187–198. doi: 10.1016/0147-619x(82)90056-7. [DOI] [PubMed] [Google Scholar]
- Iida S., Mollet B., Meyer J., Arber W. Functional characterization of the prokaryotic mobile genetic element IS26. Mol Gen Genet. 1984;198(2):84–89. doi: 10.1007/BF00328705. [DOI] [PubMed] [Google Scholar]
- Iida S. On the origin of the chloramphenicol resistance transposon Tn9. J Gen Microbiol. 1983 Apr;129(4):1217–1225. doi: 10.1099/00221287-129-4-1217. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Replicon fusions promoted by the inverted repeats of Tn5. The right repeat is an insertion sequence. J Mol Biol. 1981 Jul 25;150(1):15–32. doi: 10.1016/0022-2836(81)90322-3. [DOI] [PubMed] [Google Scholar]
- Labigne-Roussel A., Briaux-Gerbaud S., Courvalin P. Tn1525, a kanamycin R determinant flanked by two direct copies of IS15. Mol Gen Genet. 1983;189(1):90–101. doi: 10.1007/BF00326060. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mollet B., Iida S., Shepherd J., Arber W. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 1983 Sep 24;11(18):6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Zenilman M., Ohtsubo E. Insertion element IS102 resides in plasmid pSC101. J Bacteriol. 1980 Oct;144(1):131–140. doi: 10.1128/jb.144.1.131-140.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Nomura N., Sugimoto, Sugisaki H., Takanami M. Nucleotide sequence at the insertion sites of a kanamycin transposon. Nature. 1978 Dec 21;276(5690):845–847. doi: 10.1038/276845a0. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Yin J. C., Yong-di Z., Johnson R. C., Reznikoff W. S. Genetic organization of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):99–105. doi: 10.1101/sqb.1981.045.01.018. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Terawaki Y., Kobayashi Y., Matsumoto H., Kamio Y. Molecular cloning and mapping of a deletion derivative of the plasmid Rts 1. Plasmid. 1981 Sep;6(2):222–234. doi: 10.1016/0147-619x(81)90068-8. [DOI] [PubMed] [Google Scholar]
- Terawaki Y., Takayasu H., Akiba T. Thermosensitive replication of a kanamycin resistance factor. J Bacteriol. 1967 Sep;94(3):687–690. doi: 10.1128/jb.94.3.687-690.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3'5"-aminoglycoside phosphotransferase type III. Gene. 1983 Sep;23(3):331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Labigne-Roussel A., Courvalin P. An IS15 insertion generates an eight-base-pair duplication of the target DNA. Gene. 1983 Sep;24(1):125–129. doi: 10.1016/0378-1119(83)90137-3. [DOI] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]